As revealed by population genetic analyses, different human interferon genes evolved under distinct selective constraints and signatures of positive selection vary according to geographic region, suggesting that some sequence changes may have conferred an advantage by increasing resistance to viral infection.
Abstract
Interferons (IFNs) are cytokines that play a key role in innate and adaptive immune responses. Despite the large number of immunological studies of these molecules, the relative contributions of the numerous IFNs to human survival remain largely unknown. Here, we evaluated the extent to which natural selection has targeted the human IFNs and their receptors, to provide insight into the mechanisms that govern host defense in the natural setting. We found that some IFN-α subtypes, such as IFN-α6, IFN-α8, IFN-α13, and IFN-α14, as well as the type II IFN-γ, have evolved under strong purifying selection, attesting to their essential and nonredundant function in immunity to infection. Conversely, selective constraints have been relaxed for other type I IFNs, particularly for IFN-α10 and IFN-ε, which have accumulated missense or nonsense mutations at high frequencies within the population, suggesting redundancy in host defense. Finally, type III IFNs display geographically restricted signatures of positive selection in European and Asian populations, indicating that genetic variation at these genes has conferred a selective advantage to the host, most likely by increasing resistance to viral infection. Our population genetic analyses show that IFNs differ widely in their biological relevance, and highlight evolutionarily important determinants of host immune responsiveness.
IFNs are helicoidal cytokines released by host cells in response to the presence of pathogens or tumor cells. Human IFNs have been classified into three major types on the basis of the cognate receptors through which they signal, gene sequence similarity, and chromosomal location (Pestka et al., 2004). Type I IFNs include 17 subtypes (13 subtypes of IFN-α and IFNs β/ε/κ/ω), all of which bind to a receptor composed of two chains, IFNAR1 and IFNAR2 (Uzé et al., 2007). The genes encoding type I IFNs are intronless and are located in a region spanning ∼400 kb on chromosome 9, with the exception of IFNK, which is located ∼6 Mb away from the other type I IFN genes (Trent et al., 1982; Henco et al., 1985; Díaz et al., 1994). There is only one type II IFN, IFN-γ, which signals via a receptor composed of the IFN-γR1 and IFN-γR2 subunits (Wheelock and Sibley, 1965; Pestka et al., 2004). The more recently described type III IFNs constitute a group of three cytokines, IL-28A, IL-28B, and IL-29 (also known as IFN-λ2, IFN-λ3, and IFN-λ1, respectively), the genes for which are clustered in an ∼50-kb region of chromosome 19 (Kotenko et al., 2003; Sheppard et al., 2003). These IFNs activate a signaling pathway similar to that of type I IFNs, but act via a different receptor composed of the type III IFN-specific IL-28RA and the IL-10RB, the latter subunit being also used by the IL-10 and IL-22 receptor (Kotenko et al., 1997; Xie et al., 2000). There is increasing evidence to suggest that type I and III IFNs have a different role from the type II IFN: IFN-α/β and IFN-λ appear to have potent antiviral activities, whereas IFN-γ has antibacterial, antiparisitic, and antifungal properties (Pestka et al., 2004; Zhang et al., 2008).
In recent years, human genetics studies of both Mendelian and complex diseases have identified several variants affecting the production of, or the response to, IFNs, shedding light on the genuine functions of IFNs in the natural setting (Zhang et al., 2008). Disorders or specific mutations in genes involved in the IFN-γ circuit, such as in IFNGR1 and IFNGR2, confer a Mendelian predisposition to mycobacterial disease (Filipe-Santos et al., 2006), whereas the disorders or specific mutations in patients with impaired type I or type III responses are associated with a stronger predisposition to viral infections (Dupuis et al., 2003; Chapgier et al., 2006; Minegishi et al., 2006). Likewise, mutations affecting type I or type III IFN responses have been associated with various autoimmune pathologies (Crow et al., 2006a,b; Glocker et al., 2009; Rice et al., 2009). Several epidemiological genetics studies have recently shown that genetic variants in the region encompassing the type III IFN IL28B gene are associated with the spontaneous clearance of hepatitis C virus (HCV) and the response to HCV therapeutic treatment (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Thomas et al., 2009).
Our understanding of the mechanisms controlling IFN production, the downstream signaling pathways associated with these molecules, and their involvement in physiology and pathology is starting to be fully appreciated, but several biological questions remain unanswered. Given that multiple IFN molecules signal through the same receptor (e.g., IFN-α/β and IFN-λ), are all IFNs equally relevant to host survival? Are some IFN genes more essential for immunity to infection whereas others display immunological redundancy? Does IFN-γ, which is not a prototypic antiviral cytokine, have a distinctive evolutionary signature? Has genetic variation at specific IFN gene loci conferred a selective advantage to the host, associated with an increase in resistance to infectious disease? Here, we tackled these questions using an evolutionary genetics approach, which investigates the way in which infections have shaped the variability of host defense genes by natural selection (Sabeti et al., 2006; Nielsen et al., 2007; Barreiro and Quintana-Murci, 2010). This approach, which has been shown to be an indispensable complement to clinical and epidemiological genetics, and to immunological studies (Casanova and Abel, 2007; Quintana-Murci et al., 2007; Casanova et al., 2011), should help to determine the biological relevance of IFNs in the setting of a natural ecosystem governed by natural selection.
RESULTS
Full sequencing of genes encoding the human IFNs and their receptors
To obtain insight into the selective forces that have driven the evolution of the three families of IFNs in humans, we characterized the levels of sequence-based diversity in the 21 genes encoding the IFNs and the 6 genes encoding their receptor chains by full resequencing in a panel of 186 healthy individuals originating from sub-Saharan Africa, Europe, and Asia. We sequenced a total of 81.5 kb in each individual—24% of which corresponded to protein-coding regions, the rest comprising noncoding exons, introns, and promoter regions (Table S1)—and identified 1,066 polymorphisms, including 988 single-nucleotide polymorphisms (SNPs) and 78 insertions/deletions (Table S2). This resequencing dataset was used to estimate several population genetic parameters and summary statistics that were, when relevant, compared with available genome-wide datasets based on genotyping or resequencing. These analyses allowed us to explore the effects of natural selection on IFN evolution since the divergence of the human and chimpanzee lineages and within different human populations.
Naturally occurring genetic diversity varies between IFNs and populations
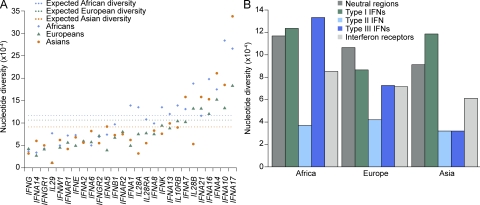
We observed remarkable differences in the levels of nucleotide diversity within the population between the genes encoding the various IFNs and their receptors and between IFN families (Fig. 1 and Table S3). The extremely low level of nucleotide diversity observed for the type II IFNG, which was uniform across populations, contrasted with several type I IFNA genes, such as IFNA4, IFNA7, IFNA10, IFNA16, IFNA17, and IFNA21, which displayed high levels of diversity (Fig. 1 A). In accordance with the “Out of Africa” model (Lewin, 1987) and genome-wide datasets (i.e., HapMap and 1,000 Genomes Project; Altshuler et al., 2010; Durbin et al., 2010), African populations generally displayed the highest levels of diversity. However, one third of the IFN genes (mostly type I IFNA genes: IFNA4, IFNA5, IFNA6, IFNA7, IFNA14, and IFNA17) were most diverse in the Asian population, which in turn presented the lowest diversity for the three members of the type III IFN family (Fig. 1 and Table S3).
Figure 1.
Patterns of nucleotide diversity for the three families of IFN genes and for the genes encoding their receptors in human populations. (A) Nucleotide diversity levels for the individual genes in populations representing major ethnic groups. The expected diversity (dotted lines) corresponds to the mean diversity levels observed for 20 autosomal noncoding regions in each geographic area (Laval et al., 2010). (B) Comparison of nucleotide diversity between IFN families and receptors.
Because the type I IFNA genes, as well as the three type III IFN genes, display high levels of sequence identity and are organized into two distinct clusters of paralogous genes (Pestka et al., 2004; Woelk et al., 2007), gene conversion is likely to have been an important mechanism for the evolution of these gene families. Indeed, in multigene families, gene conversion among paralogous loci has been shown to play an important role in the introduction of genetic variation to each gene (Innan and Kondrashov, 2010; Ohta, 2000, 2010). We thus evaluated the extent to which gene conversion has contributed to the levels of nucleotide diversity observed at these two groups of IFN genes. To do so, we screened highly homologous regions among paralogs for the presence of human-specific sites (polymorphic or fixed) that have been most likely introduced by gene conversion events rather than by point mutations (Materials and methods). This analysis allowed us to detect a substantial number of putative gene conversion events (Table S4), 30 of which corresponded to polymorphic amino acid–altering mutations (Table S5). As gene conversion is usually disregarded in population genetics tests, owing to the uncertainty associated with the underlying models of gene conversion, all variants identified as resulting from gene conversion were not considered in the statistical analyses to detect natural selection.
Functional diversity is not evenly distributed between human IFNs
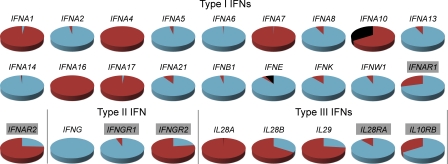
We identified 245 SNPs in coding regions, including 164 nonsynonymous and 8 nonsense mutations present in the general human population. The occurrence and frequency distribution of these variants differed markedly between the various IFNs and between populations (Fig. 2, Table 1, and Table S3). IFNs with very low levels of amino acid–altering variation are represented by IFNG, in which no nonsynonymous mutations were observed, and by a group of type I IFNs (IFNA2, IFNA5, IFNA6, IFNA8, IFNA13, IFNA14, IFNA21, IFNB1, IFNK, and IFNW1) and the two receptor subunits IFNGR1 and IL28RA, which presented nonsynonymous mutations at a low frequency within the population. In contrast, we found that 13 genes accumulated nonsynonymous variants at very high frequency in the human population (∼30–100%; Fig. 2 and Table 1). Most of these variants were predicted to be benign by the PolyPhen algorithm (Adzhubei et al., 2010), but some genes, such as IFNA10, IFNA16, IFNA17, IFNAR1, IL28A, and IL29, presented high frequencies of missense mutations predicted to alter protein function (i.e., damaging mutations; Table 1 and Table S3). The most extreme cases were those of IFNA10 and IFNE, for which nonsense mutations were present in the homozygous state, at high frequency, in several populations. For example, one of the nonsense mutations of IFNA10 (SNP 60T>A, C20STOP, rs10119910), which is located in the signal peptide, abolishes the translation of the entire protein. Surprisingly, this stop mutation has attained a worldwide frequency of 34%, ranging from 18% in Europeans to 54% in Asians. The IFNE nonsense mutation (SNP 211C>T, Q71STOP, rs2039381) decreases the length of the protein by two thirds and has attained a worldwide frequency of 7%, increasing to 15% in Asia. Such high frequencies of nonsynonymous or nonsense mutations in some IFN genes may reflect either a relaxation of selective constraints caused by the redundancy of the genes concerned, or a selective advantage accounted for by the higher frequency of functionally advantageous variants.
Figure 2.
Proportion of chromosomes carrying at least one nonsynonymous or nonsense variant in the general human population. The red portion of the pie charts corresponds to the proportion of chromosomes carrying at least one nonsynonymous polymorphism, the black portion to the proportion of chromosomes carrying at least one nonsense polymorphism, and the blue portion to the proportion of chromosomes carrying neither nonsynonymous nor nonsense polymorphisms. Genes shaded in gray correspond to those encoding the receptor subunits of each IFN family.
Table 1.
Coding variation at IFN genes and their receptors in human populations
| Number of polymorphismsa | Highest derived allele frequencyb | Proportion of chromosomesc | |||||||||
| Gene | Silent Sites | NS Sites | NS | PRD | STOP | NS | PRD | STOP | NS | PRD | STOP |
| IFNA1 | 1835 | 440 | 5 | 1 | 0 | 94.62 | 0.27 | 0 | 97.85 | 0.27 | 0 |
| IFNA2 | 2030 | 438 | 3 | 0 | 0 | 1.08 | 0 | 0 | 2.69 | 0 | 0 |
| IFNA4 | 1839 | 439 | 10 | 1 | 0 | 99.73 | 0.27 | 0 | 100.00 | 0.27 | 0 |
| IFNA5 | 1300 | 443 | 4 | 3 | 1 | 0.54 | 0.54 | 0.27 | 2.96 | 1.08 | 0.27 |
| IFNA6 | 1464 | 441 | 3 | 1 | 0 | 0.54 | 0.27 | 0 | 0.81 | 0.27 | 0 |
| IFNA7 | 1285 | 441 | 5 | 0 | 0 | 99.46 | 0 | 0 | 99.46 | 0 | 0 |
| IFNA8 | 1889 | 440 | 1 | 0 | 0 | 7.26 | 0 | 0 | 7.26 | 0 | 0 |
| IFNA10 | 1644 | 438 | 13 | 0 | 3 | 98.12 | 0 | 34.14 | 100.00 | 0 | 34.68 |
| IFNA13 | 1461 | 440 | 3 | 0 | 0 | 5.11 | 0 | 0 | 6.99 | 0 | 0 |
| IFNA14 | 1216 | 448 | 2 | 0 | 1 | 0.54 | 0 | 0.27 | 0.81 | 0 | 0 |
| IFNA16 | 1700 | 439 | 7 | 1 | 0 | 97.58 | 97.04 | 0 | 100.00 | 97.04 | 0 |
| IFNA17 | 1710 | 438 | 6 | 1 | 0 | 65.86 | 65.86 | 0 | 98.39 | 65.86 | 0 |
| IFNA21 | 1801 | 440 | 9 | 1 | 1 | 4.03 | 0.27 | 0.27 | 8.87 | 0.27 | 0.27 |
| IFNB1 | 1520 | 439 | 5 | 1 | 0 | 1.61 | 0.27 | 0 | 3.23 | 0.27 | 0 |
| IFNE | 2156 | 490 | 3 | 1 | 1 | 2.15 | 0.27 | 7.26 | 3.49 | 0.27 | 7.26 |
| IFNK | 2186 | 496 | 4 | 3 | 0 | 6.99 | 2.69 | 0 | 10.75 | 3.76 | 0 |
| IFNW1 | 2151 | 451 | 6 | 5 | 0 | 4.30 | 4.30 | 0 | 5.91 | 5.65 | 0 |
| IFNAR1 | 5250 | 1303 | 9 | 1 | 0 | 20.43 | 0.27 | 0 | 29.30 | 0.27 | 0 |
| IFNAR2 | 5737 | 1201 | 10 | 5 | 0 | 58.87 | 0.81 | 0 | 74.19 | 2.42 | 0 |
| IFNG | 4172 | 395 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IFNGR1 | 4345 | 1130 | 10 | 2 | 0 | 1.87 | 0.27 | 0 | 5.65 | 0.54 | 0 |
| IFNGR2 | 4015 | 715 | 6 | 1 | 0 | 76.33 | 0.27 | 0 | 77.15 | 0.27 | 0 |
| IL28A | 1325 | 303 | 8 | 1 | 1 | 92.74 | 80.11 | 0.27 | 100.00 | 80.11 | 0.27 |
| IL28B | 3031 | 434 | 12 | 3 | 0 | 62.63 | 3.49 | 0 | 65.59 | 3.76 | 0 |
| IL29 | 1558 | 443 | 5 | 1 | 0 | 70.16 | 70.16 | 0 | 73.12 | 70.16 | 0 |
| IL28RA | 3295 | 1177 | 10 | 4 | 0 | 3.76 | 3.76 | 0 | 11.29 | 4.84 | 0 |
| IL10RB | 3696 | 721 | 5 | 1 | 0 | 32.53 | 0.54 | 0 | 34.41 | 0.54 | 0 |
Absolute numbers of polymorphisms observed in our entire dataset
Among the different variants observed in each category, the value (in %) of the most frequent polymorphism is shown
Proportion of chromosomes (in %) in the general population carrying at least one nonsynonymous or nonsense variant. NS stands for nonsynonymous. “PRD” refers to “probably damaging” mutations as predicted by PolyPhen v2 HumDiv.
Purifying selection has operated differently among IFN family members
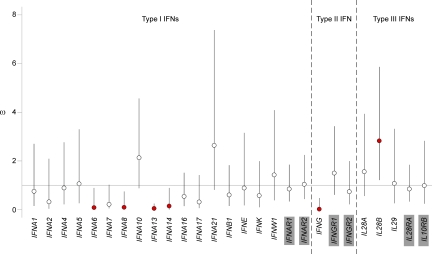
We investigated whether and how natural selection has driven the observed heterogeneous patterns of diversity of the various IFNs and their receptors by first estimating the direction and strength of selection within the human species as a whole. To this end, we measured dS and dN, i.e., the number of silent and nonsynonymous fixed differences between humans and chimpanzees, together with pS and pN, i.e., the number of silent and nonsynonymous polymorphic sites observed within humans. We used the McDonald-Kreitman Poisson random field method (Sawyer and Hartl, 1992; Bustamante et al., 2005) to estimate ω (i.e., ω α θN/θS, where θN and θS are estimates of the rate of nonsynonymous and silent mutations) and to assess the selection pressure driving amino acid substitutions. Under neutrality, ω is not significantly different from 1. Values <1 indicate a deficit of nonsynonymous variants, whereas values >1 reflect an excess of amino acid changes. We found that only IFNA6, IFNA8, IFNA13 and IFNA14, and IFNG had ω values significantly <1, consistent with their evolution under the strongest purifying selection (Fig. 3). Among type I IFNs, we removed from our analyses a few low-frequency nonsynonymous mutations that were found to result from gene conversion at IFNA6, IFNA13, and IFNA14, whereas no gene conversion events were detected at IFNA8 (Table S5). Our simulation analyses showed that the removal of gene conversion–derived events cannot produce spurious signals of purifying selection (Fig. S1). However, because of the minimal, but nonnull, uncertainty in our procedure for gene conversion detection, IFNA8 represents the most robust target of purifying selection among type I IFNs. At the other extreme, IL28B was the only gene that had a ω value significantly greater than 1, consistent with the action of positive selection.
Figure 3.
Estimation of the intensity of natural selection acting on the various IFN families and their receptors. The strength of natural selection was assessed by estimating ω values. Under neutrality, ω is not significantly different from 1. Values below 1 are consistent with selection against nonsynonymous variants, whereas values greater than 1 indicate an excess of amino acid changes. Bars indicate 95% Bayesian confidence intervals, and red circles indicate genes with ω estimates significantly above or below 1. Genes shaded in gray correspond to those encoding the receptor subunits of each IFN family.
Positive selection has targeted type III IFNs in non-African populations
We next investigated the ways in which positive selection has affected IFN genes in a population-specific manner, as populations from different continents have clearly been historically exposed to different selection pressures (Novembre and Di Rienzo, 2009). We performed various intraspecies neutrality tests on various aspects of the data, including the allele frequency spectrum (i.e., Tajima’s D, Fu and Li’s D* and F*, and Fay and Wu’s H tests), levels of population differentiation (i.e., FST), and haplotype-based tests (i.e., derived intrallelic nucleotide diversity [DIND] and integrated haplotype score [iHS] tests; Kreitman, 2000; Nielsen et al., 2007). As most of these tests are known to be sensitive to the effects of demography and selection, we used simulation-based or empirical procedures to correct for the influence of demography on the patterns of population genetic diversity. For the allele frequency spectrum and DIND tests, we incorporated into our neutral expectations two demographic models based on multiple, noncoding genomic regions sequenced in a set of populations similar to those used in this study (Voight et al., 2005; Laval et al., 2010). For the population differentiation tests, we obtained a background expectation of genome-wide FST by analyzing the publicly available HGDP-CEPH dataset (Li et al., 2008) from the same set of individuals we sequenced in this study. In addition, we complemented our analyses of recent positive selection by obtaining the iHS values for each SNP in the populations of the HapMap Phase II dataset (Frazer et al., 2007).
We defined genes under selection conservatively as those (a) for which significant results were obtained after both demographic corrections or for which results were significant at the genome-wide level, and (b) for which significant results were obtained in at least two tests of selection based on different aspects of the data (e.g., allele frequency spectrum tests and FST) in the same population. With these stringent criteria, most type I IFNs and the type II IFN did not show compelling signatures of selection in any of the continental populations here studied. In contrast, we found that positive selection had strongly affected the members of the type III IFN family in European and Asian populations (Fig. 4, Fig. 5, and Table 2). In addition, the results of the neutrality tests for type III IFNs remained significant after correction for multiple testing, emphasizing the intensity of the events of positive selection detected. The three type III IFN genes are adjacent to each other on an ∼50-kb region of chromosome 19 (Fig. 6), but they nonetheless displayed low levels of linkage disequilibrium in all populations (Fig. S2). This suggests that independent positive selection events have targeted IL28A, IL28B, and IL29.
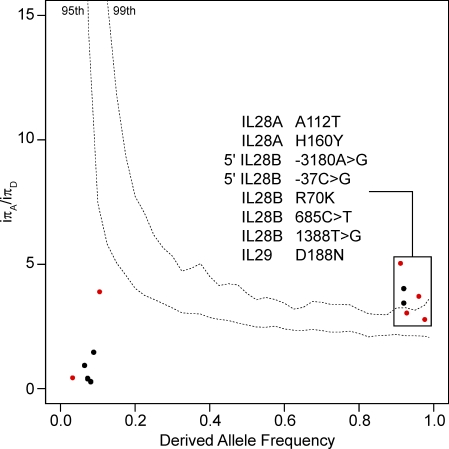
Figure 4.
Detection of positive selection in Asia using the DIND test. We plotted iπA/iπD values against derived allele frequencies (DAFs). P-values were obtained by comparing the iπA/iπD values for the three type III IFN genes against the expected iπA/iπD values obtained in 104 simulations, taking into account the most conservative demographic model (Laval et al., 2010). The top dashed line on the graph corresponds to the 99th percentile, and the bottom to the 95th percentile. Dots above the 95th percentile dashed line correspond to mutations that have increased in frequency faster than expected under neutrality in the Asian population. These mutations have been most likely targeted by positive selection, and thus conferred an advantage to the host. Red and black dots correspond to nonsynonymous and silent polymorphisms, respectively. For DIND analyses concerning all genes encoding the 27 IFNs and their receptors in all populations, see Fig. S3.
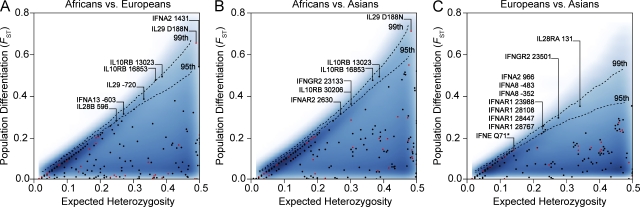
Figure 5.
Detection of positive selection on the basis of levels of population differentiation. Population pairwise FST values are plotted against expected heterozygosity for all the SNPs identified in our study for Africans versus Europeans (A), Africans versus Asians (B), and Europeans versus Asians (C). The dashed lines represent the 99th and 95th percentiles of the HGDP-CEPH genotyping dataset for the same individuals (represented by the density area in blue; Li et al., 2008). Black dots correspond to silent polymorphisms and red dots correspond to nonsynonymous polymorphisms.
Table 2.
Sequence-based neutrality tests across the 27 genes encoding the IFNs and their receptors
| AFRICA | EUROPE | ASIA | ||||||||||
| Gene | TD | D | F | Hn | TD | D | F | Hn | TD | D | F | Hn |
| TYPE I IFNs | ||||||||||||
| IFNA1 | −0.15 | 0.09 | 0.00 | −1.06 | −1.59† | −0.82 | −1.31 | −3.20* | −0.97 | −0.65 | −0.91 | −3.07* |
| IFNA2 | −1.42 | −0.01 | −0.64 | −0.94 | −0.02 | 0.41 | 0.31 | 0.90 | 0.54 | 0.12 | 0.31 | 0.20 |
| IFNA4 | −0.88 | −1.57 | −1.56 | −0.69 | 0.28 | −0.97 | −0.61 | −2.59* | 1.24 | −0.68 | 0.04 | 0.43 |
| IFNA5 | −0.93 | −0.65 | −0.89 | 0.06 | −1.09 | −2.42† | −2.33*† | −0.66 | 0.19 | −0.42 | −0.25 | 0.40 |
| IFNA6 | −0.92 | 0.28 | −0.16 | 0.60 | −0.22 | −0.31 | −0.33 | 0.67 | 0.31 | −0.07 | 0.07 | 0.45 |
| IFNA7 | −1.78 | −1.55 | −1.94 | −0.96 | −1.32 | −1.55 | −1.74 | 0.27 | −0.58 | 0.12 | −0.14 | −1.14 |
| IFNA8 | −0.47 | −0.11 | −0.29 | 0.72 | −0.72 | −2.21† | −1.99 | 0.47 | −0.33 | −0.49 | −0.52 | 0.85 |
| IFNA10 | −0.39 | 1.11 | 0.62 | −1.81* | −1.20 | −0.59 | −0.98 | −1.95 | 0.86 | −1.59 | −0.83 | −0.41 |
| IFNA13 | 0.10 | 1.36*† | 1.09*† | 0.35 | −0.63 | −0.65 | −0.77 | −1.00 | −0.17 | −3.01*† | −2.38*† | 0.09 |
| IFNA14 | −1.70 | −1.80 | −2.10 | −0.66 | −1.37 | −2.83*† | −2.77*† | −0.94 | 0.68 | −0.31 | 0.01 | 0.28 |
| IFNA16 | −1.14 | 0.33 | −0.31 | −3.40* | −0.96 | 1.28 | 0.51 | −4.75**† | 0.67 | 0.27 | 0.49 | −0.06 |
| IFNA17 | 0.21 | 0.68*† | 0.60† | −0.64 | −0.06 | −0.68 | −0.53 | −1.50 | 1.51 | −0.82 | 0.07 | 0.27 |
| IFNA21 | −1.12 | −2.37 | −2.28 | 0.16 | 0.62 | 0.28 | 0.47 | 0.42 | 0.51 | 0.41 | 0.53 | −0.30 |
| IFNB1 | −1.38 | −1.06 | −1.41 | -0.79 | −0.28 | −1.25 | −1.09 | −1.00 | −0.08 | −0.42 | −0.36 | −0.65 |
| IFNE | −0.51 | −0.35 | −0.49 | 0.69 | −0.29 | −0.42 | −0.45 | −0.03 | 0.19 | −1.02 | −0.71 | 0.08 |
| IFNK | −0.89 | −0.65 | −0.89 | 0.08 | −0.90 | −1.52 | −1.54 | −0.53 | −0.84 | −3.25*† | −2.81*† | −0.99 |
| IFNW1 | −1.07 | −1.14 | −1.33 | 0.61 | −1.36 | −3.45**† | −3.20** | 0.36 | −0.33 | −0.82 | −0.77 | 0.76 |
| IFNAR1 | −0.82 | −0.77 | −0.95 | -0.27 | 0.46 | −0.43 | −0.11 | 1.10 | −0.05 | −3.50**† | −2.66*† | 0.75 |
| IFNAR2 | −0.99 | −2.58 | −2.31 | -0.71 | 1.03 | −0.39 | 0.19 | −0.10 | −0.01 | −1.76 | −1.28 | −0.94 |
| TYPE II IFN | ||||||||||||
| IFNG | −1.60 | −1.39 | −1.76 | -2.10* | 0.34 | −1.02 | −0.65 | −0.42 | 0.70 | 0.12 | 0.37 | −0.11 |
| IFNGR1 | −1.44 | −2.13 | −2.23 | -0.26 | 0.27 | −2.02 | −1.43 | 0.44 | −0.37 | −0.95 | -0.88 | 0.55 |
| IFNGR2 | −1.13 | −3.09 | −2.77 | -0.37 | 0.39 | −1.63 | −1.05 | −0.54 | −0.73 | −4.08**† | −3.36**† | −0.92 |
| TYPE III IFNs | ||||||||||||
| IL28A | −0.95 | −1.99 | −1.90 | -2.50 | −0.93 | −1.33 | −1.42 | −0.05 | −1.92*†† | −3.39**† | −3.42**† | −0.89 |
| IL28B | −0.18 | 1.18**† | 0.75† | 0.42 | 0.66 | −1.06 | −0.48 | −1.73 | −1.57*† | −3.01*† | −2.94*† | −2.35* |
| IL29 | −1.00 | −2.00 | −1.95 | -0.33 | −1.72*† | −3.81**†† | −3.68**†† | −1.41 | −1.71*†† | −0.86 | −1.36 | −1.56 |
| IL28RA | −0.64 | −0.93 | −0.97 | -0.24 | −0.20 | −1.52 | −1.22 | −0.28 | -0.83 | −2.63*† | −2.33*† | −0.19 |
| IL10RB | −0.32 | −0.63 | −0.60 | −0.66 | 1.50 | −0.55 | 0.26 | 0.28 | 0.53 | −1.32 | −0.74 | −0.35 |
TD, Tajima’s D; D, Fu & Li’s D*; F, Fu & Li’s F*; Hn, normalized Fay && Wu’s H. **/*, P ≤ 0.01 and ≤ 0.05, respectively, according to the model of (Voight et al., 2005); ††/†, P ≤ 0.01 and ≤ 0.05, respectively, according to the model of (Laval et al., 2010).
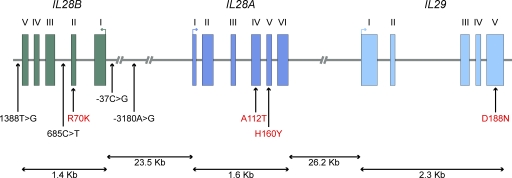
Figure 6.
Spatial distribution of positively selected variants across type III IFN genes. Genomic organization of the three members of the type III IFN family, IL28B, IL28A, and IL29. Filled boxes correspond to exonic regions, and arrows above exons indicate the direction of the open reading frame. Genetic variants displaying signatures of population-specific positive selection are shown; noncoding SNPs are indicated in black and amino acid changes in red. At the gene level, IL28A and IL28B displayed signatures of positive selection in Asia, and IL29 in both Europe and Asia (Table 2). At the SNP level, the action of positive selection targeting the five SNPs at IL28B and the two at IL28A was supported by the DIND test (Fig. 4), whereas that at the IL29 D188N variant was supported by the DIND test and iHS, as well as by the levels of population differentiation (Fig. 5).
IL28A and IL28B deviated significantly from neutral expectations in the Asian population, in allele frequency spectrum tests (Table 2). Furthermore, the derived alleles of two SNPs in IL28A and five in IL28B were found to be associated with significantly lower levels of surrounding nucleotide diversity, given their high population frequency (>90%), in Asia (see the DIND test in Fig. 4). Interestingly, the two SNPs in IL28A and one in IL28B correspond to amino acid–altering variants (IL28A SNP 983G>A, A112T, rs8103362; IL28A SNP 1227C>T, H160Y, rs61735713; IL28B SNP 502G>A, R70K, rs8103142). These amino acid changes therefore appear to have increased in frequency more rapidly than would be expected under neutrality, in the Asian population, consistent with the action of population-specific positive selection.
We also detected strong signals of positive selection at the IL29 locus. First, allele frequency spectrum tests detected a significant excess of rare variants in both Europeans and Asians (Table 2). Second, very high levels of population differentiation were observed at the IL29 locus between African and Eurasian populations (mean FST = 0.42). In particular, the nonsynonymous variant 2054G>A (D188N, rs30461, predicted to be probably damaging by PolyPhen), presented extreme levels of differentiation between Africans and Eurasians (FST Africa/Asia = 0.71, P = 0.014; FST Africa/Europe = 0.66, P = 0.025, using the HGDP-CEPH dataset; Fig. 5). Remarkably, this SNP not only presented the highest degree of population differentiation of all SNPs in our dataset but was also among the most highly differentiated SNPs at the level of the entire human genome. Indeed, the D188N variant falls into the group of 139 nonsynonymous SNPs presenting the largest allele frequency differences among populations in the 1,000 Genomes project (Durbin et al., 2010). In addition, this nonsynonymous variant gave a significant result for the DIND test in Asian populations (iπA/iπD = 3.71, P < 0.01; Fig. 4) and gave a significant iHS value of -2.285 in Europeans from the HapMap Phase II dataset (iHS was not calculated for Asian HapMap populations, because this SNP has a frequency >95%). These results suggest that IL29 variation, and the D188N variant in particular, has conferred a selective advantage to Eurasian populations.
DISCUSSION
In this study, we demonstrate that the different IFN families, and their individual members, have followed different evolutionary trajectories in humans. First, we found that type I IFN subtypes differ in their levels of evolutionary constraint. Amino acid–altering variation has been constrained for some type I IFNs, with IFNA6, IFNA8, IFNA13, and IFNA14 found to have been subject to the strongest purifying selection (Fig. 3). Low levels of amino acid–altering variation were also observed at IFNA2, IFNA5, IFNA21, IFNB1, IFNK, and IFNW1 (Fig. 2 and Table 1). Conversely, selective constraints have been relaxed for other type I IFNs, which harbor nonsynonymous variants at high population frequencies (IFNA1, IFNA4, IFNA7, IFNA10, IFNA16, and IFNA17). Furthermore, some IFNs present nonsense mutations in the homozygous state (IFNA10 and IFNE), suggesting that they might be currently undergoing pseudogenization. We also found that some nonsynonymous polymorphisms at several IFNA genes, for the most part observed at low population frequencies, appear to have been introduced by gene conversion from their paralogs (Table S5). This observation supports the notion that, besides gene duplication, gene conversion has contributed to the evolution of type I IFNs in mammals (Hughes, 1995; Woelk et al., 2007; Génin et al., 2009b). Together, the strong constraints characterizing some type I IFNs suggest that they fulfill an essential, nonredundant function in host defense. In contrast, the high population frequencies of missense or nonsense mutations, occurring through mutation or gene conversion, found in other type I IFN subtypes, suggest that these molecules are highly redundant.
Our findings provide evolutionary evidence of the complexity of the biological actions of type I IFNs. Indeed, in the mouse model, they play a key role in protective antiviral immunity to multiple experimental infections (Jouanguy et al., 2007; Vilcek, 2006). In humans, primary immunodeficiencies of the type I IFN pathway, including STAT-1 and TYK-2 deficiencies, have also shown that type I IFNs are critical for antiviral immunity (Dupuis et al., 2003; Chapgier et al., 2006, 2009; Minegishi et al., 2006; Zhang et al., 2008). The integration of our population genetics data into a clinical framework thus indicates that at least one subgroup of type I IFNs plays a critical, nonredundant role in antiviral immunity in natural conditions. A greater tolerance for the increase in frequency of missense or nonsense mutations in the general population is observed in another set of type I IFNs, suggesting that the functions they fulfill are largely overlapping with other IFN subtypes. The existence of multiple type I IFNs and differences in their degrees of diversity and redundancy may attest to the great capacity of this host defense system to evolve, to develop efficient antiviral responses.
However, there is a growing body of work showing that type I IFN activity can also be detrimental to the host (Decker et al., 2005; Vilcek, 2006; Trinchieri, 2010). Experimental data from mice and clinical observations in human patients have shown that IFN production can be harmful in the context of infection and can increase morbidity (Gresser et al., 1975; Rivière et al., 1977; Vilcek, 1984). Such adverse effects of increased IFN production, such as systemic lupus erythematosus (Banchereau et al., 2004; Crow, 2007; Le Bon et al., 2006a; Le Bon et al., 2006b), have also been observed in the context of autoimmune diseases (Crow et al., 2003; Banchereau and Pascual, 2006). In addition, there is increasing evidence to suggest that type I IFNs have opposing roles in viral and bacterial infection (Decker et al., 2005; Vilcek, 2006; Trinchieri, 2010). Such a multifaceted mechanism of host defense is illustrated by IFN-α8 and IFN-α13, which are both under strong selective constraint yet display high and low antiviral potency, respectively (Foster et al., 1996; Foster and Finter, 1998; Koyama et al., 2006; Jaks et al., 2007; Lavoie et al., 2011). However, differences in bioactivity between IFN subtypes will depend not only on their respective potencies and distinct receptor-binding chemistries, as recently shown for a subset of type I IFNs (Thomas et al., 2011), but also on their individual production. Few studies have systematically assessed the levels of expression of the multiple type I IFNs (Coccia et al., 2004; Génin et al., 2009a) and, because of the differences in experimental conditions used, there is as yet no clear consensus as to which subtypes are the most expressed in different cell types. In light of this, we hypothesize that type I IFNs presenting various antiviral potencies and/or production could have been maintained, and selected for, to regulate global type I IFN activity. To test this hypothesis, further analyses are needed to (a) characterize the expression and potency of the individual type I IFN subtypes under different conditions of infection and in different cell types, in particular for those exhibiting the strongest signatures of purifying selection; (b) define how the variation in both production and potency of the various subtypes is under genetic control (i.e., host genetic variation in both protein-coding regions and regulatory regions); and (c) evaluate how the combination of the whole set of type I IFN subtypes, with their varying functional diversity, affects downstream transcriptional programs and host responses at the organism level.
Second, we found that the type II IFNG was the only gene, across all three families of human IFNs and their receptors, to display a complete absence of amino acid–altering mutations. This gene was subject to the strongest purifying selection of all IFNs, and we previously showed that IFNG is among the ∼10% of immune-related genes subject to the most intense selective constraints on amino acid variation in humans (Manry et al., 2011). Clinical genetic studies have demonstrated that six genes involved in the IFN-γ circuit (IL-12/23–IFN-γ) play a critical role in protective immunity (Filipe-Santos et al., 2006). Specifically, disorders of IFN-γ production caused by mutations affecting IL-12B, IL-12RB1, or specific NEMO mutations, and impaired IFN-γ responses caused by IFNGR1, IFNGR2, or specific STAT1 mutations, are associated with Mendelian susceptibility to mycobacterial disease in patients resistant to most viruses (Zhang et al., 2008). Population and clinical data show that no variation with a significant impact on protein function is tolerated at loci involved in IFN-γ–mediated immunity, indicating that the IFN-γ pathway is essential and nonredundant in host survival, including host defense against mycobacteria.
Finally, our data showed that type III IFNs are the only group of IFNs where selective pressures have involved processes of geographically restricted adaptation, revealing that genetic variation at these genes has conferred a selective advantage to specific human populations (Fig. 6). There is increasing evidence from clinical genetic studies to support a major role of these molecules in antiviral immunity (Zhang et al., 2008), so the selection pressure acting on type III IFN genes may be of viral origin. Strong support for this notion has been provided by recent genome-wide association studies. Indeed, the five IL28B polymorphisms we identified as being under positive selection in Asia have been associated with the spontaneous clearance of HCV and a better response to pegylated IFNα-ribavirin treatment for chronic HCV infection in populations of African, European, and Asian ancestry (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Thomas et al., 2009; Rauch et al., 2010). These SNPs include two located in the regulatory region of IL28B (SNP -3180A>G, rs12979860; and SNP -37C>G, rs28416813), one in an intron (SNP 685C>T rs11881222), one in the 3′ region (SNP 1388T>G, rs4803217), and one nonsynonymous SNP of IL28B (SNP 502G>A, R70K, rs8103142). Interestingly, based on the odds ratios for protective alleles, it has been suggested that variation of the IL28B gene may confer a stronger protective effect in Asians than in individuals of European or African ancestry (O’Brien, 2009). However, the environmental, genetic, and evolutionary factors underlying this difference remain unknown.
Our data provide new insight into the relationship between type III IFN variation and ethnic background by showing that Asian populations have evolved the most adaptively and protective alleles have increased in frequency among Asians as a result of positive selection, rather than simple genetic drift. Given the chronic and insidious nature of HCV pathogenesis, it is unlikely that HCV, at least in its modern form, is really responsible for the selection pressure exerted on IL28B. In light of this, we hypothesize that other ancestral and more virulent flaviviruses are responsible for the selective footprints observed. To this end, it will be instructive to determine whether IL28B polymorphisms are associated with natural immunity to related viruses (e.g., hemorrhagic flaviviruses and/or encephalogenic alphaviruses).
The overlap between the IL28B variants found here to be under positive selection and those associated with the spontaneous clearance of HCV infection provides an important proof-of-concept for the value of the evolutionary approach, as a complement to epidemiological and medical genetics studies. This is particularly important for positively selected IL28A and IL29 variants, whose function is not yet fully appreciated. The strongest signature of positive selection we observed concerned a nonsynonymous SNP in IL29 (SNP 2054G>A, D188N, rs30461) in European and Asian populations. However, the way in which this variant confers a selective advantage to the host and the pathogens responsible for exerting a selective pressure on IL29 remains to be identified. Because the three type III IFNs operate as independent genetic entities, we propose that the signatures of positive selection, which appear to be independent, displayed by each type III IFN reflect their different relative contributions to human fitness and survival. Additional studies are required to unravel the immunological role and phenotypic expression of type III IFN subtypes, particularly polymorphisms shown to be under positive selection, in relation to susceptibility to, or the pathogenesis of infectious diseases or autoimmune disorders.
In conclusion, our population genetics data indicate that the various members of the human IFN families differ in biological relevance, ranging from highly constrained to redundant and expendable. The identification of individual IFN genes subject to strong constraints or to adaptive evolution, attesting to their important role in immunity to infection, paves the way for additional studies to evaluate the potential of these molecules for use in vaccination, diagnosis, and treatment. More generally, our study provides a paradigm of the use of population genetics in the context of infection, with a view to improving our understanding of the biological importance of immunity-related genes in host defense in the natural setting.
MATERIALS AND METHODS
Population samples.
Sequence variation for all human IFNs and their receptors was determined in 186 individuals from sub-Saharan Africa, Europe, and Asia (62 individuals per geographic region) from the HGDP-CEPH panel (Cann et al., 2002). Sub-Saharan African populations were composed of 19 Bantu from Kenya, 21 Mandenka from Senegal, and 22 Yoruba from Nigeria; European populations were composed of 20 French, 14 Italians, 6 Orcadians, and 22 Russians; the Asian populations were composed of 15 Han Chinese and 33 individuals from Chinese minorities, 10 Japanese, and 4 Cambodians. Population structure within continental regions has been shown to be limited (Li et al., 2008) and to have a negligible influence on the inference of natural selection (Manry et al., 2011). This study was approved by the Institut Pasteur Institutional Review Board (no. RBM 2008.06).
DNA sequencing.
We sequenced the 27 genes encoding the IFNs and their receptors using Sanger sequencing. Given the extremely high sequence identity among IFN genes (i.e., the type I IFNA genes and type III IFN genes, are organized into two distinct clusters of paralogous genes), Sanger sequencing was the most appropriate choice to differentiate with confidence regions that are highly paralogous. Another advantage of Sanger sequencing is the reliable detection of low-frequency variants, which are the substrate used to detect and estimate the intensity of purifying selection. This contrasts with publicly available whole genome sequence datasets that, even if they include IFNs (e.g., 1,000 Genomes), are depleted for low-frequency variants, particularly nonsynonymous mutations (Durbin et al., 2010), because of the low coverage at which the genomes have been sequenced. For each gene, we sequenced all the exon regions and at least as much of the nonexon regions, including intron, 5′, and 3′ regions (Table S1). All sequences were obtained with the Big Dye Terminator kit and a 3730 XL automated sequencer from Applied Biosystems. Sequence files and chromatograms were inspected with GENALYS software (Takahashi et al., 2003). All singletons or ambiguous polymorphisms were systematically reamplified and resequenced. We were unable to resequence the first exon of IFNGR2 and the first exon of IL10RB for technical reasons, probably because of the very high GC content of the region (73 and 72%, respectively). The reference sequences used are given in Table S1.
Gene conversion analyses.
Because interlocus gene conversion requires high levels of sequence identity between loci (Mansai and Innan, 2010), we sought to detect gene conversion events based on (a) the local homology observed between two paralogs and (b) the ancestral/derived state of each base pair in humans determined using the corresponding chimpanzee orthologue. For a pair of genes, X and Y, resequenced in two different individuals, each sequence was subdivided in fragments of 40 bp. Each fragment defined in gene X was compared with all the possible fragments in gene Y, irrespective of their location. This procedure was performed for all pairs of individuals in our sample (n = 186). We retained pairs of fragments with a sequence identity >90%. Within these pairs, a mutation observed at a given position in gene X was declared as being a putative gene conversion event when its derived state (fixed or polymorphic) was equal to the ancestral or derived state observed at the same position in gene Y. However, situations that could be most parsimoniously explained by a single point mutation (i.e., a mutation fixed for the derived state in gene X and fixed for the ancestral allele at the same position in gene Y) were not considered as a conversion event. For each gene, this method provided a set of mutations probably resulting from gene conversion. We declared the putative acceptor and donor genes on the basis of the frequencies of converted mutations (i.e., the donor has the highest frequency of the conversion event).
We evaluated the power of the method to detect gene conversion events and the influence of their removal on the detection of selection by means of coalescence simulations, using SIMCOAL v2 (Laval and Excoffier, 2004). We simulated two duplicated genes in the human and chimpanzee lineages, with the gene duplication predating the divergence of the two species. 20 human sequences (10 individuals) and 1 chimpanzee sequence were simulated with the following parameters: the recombination rate expected in humans (1cM/Mb), a human/chimpanzee divergence time of 5 million years, constant population sizes (n = 1,000 individuals), and a mutation rate adjusted to the number of mutations observed in our dataset. The two duplicated genes, each 2,000 bp long, were simulated using a global sequence identity between paralogs set to be equal to 90%. In addition, 30% of sites were set to be nonsynonymous, with a sequence identity set to be equal to 95% in coding regions. When gene conversion was introduced, a fixed number of conversion events were simulated using a tract length set at 100 bp (consistent with empirical estimates of mean tract length (Mansai and Innan, 2010)). We allowed conversion when sequence identity between tracts was >60%, specifying that 90% of the conversion events simulated involved a sequence identity >90%. Next, all mutations present in the tract of the donor gene (both fixed and polymorphic within the human lineage) were copied in the acceptor gene. To evaluate the impact of gene conversion in a more realistic scenario, we also introduced purifying selection on nonsynonymous sites by using a lower mutation rate at nonsynonymous sites with respect to silent sites. We then applied our method of gene conversion detection and calculated the power to detect the gene conversion events and the false discovery rate (Fig. S1).
All variants identified as resulting from gene conversion were not considered in the statistical analyses to detect selection. It should be noted that ignoring the events most parsimoniously explained by a point mutation (i.e., fixed mutations in humans) will have no effect on the detection of selection using analyses based on intraspecies polymorphism, and cannot generate false-positive signals of purifying selection using interspecies tests corrected for gene conversion (Fig. S1). In addition, we verified manually if some of the amino acid–altering polymorphisms detected as resulting from gene conversion could be ambiguously explained by gene conversion or mutation, because the erroneous removal of such sites can create spurious signals of purifying selection. Two events were identified as false positives, in IFNA10 and IFNA17, and the corresponding mutations were thus not excluded from our analyses.
Statistical analyses.
We used Haploview software (Barrett et al., 2005) to obtain and visualize levels of linkage disequilibrium in the various genomic regions. Haplotype reconstruction was performed by the Bayesian method, implemented in Phase (v.2.1.1; Stephens and Donnelly, 2003). We applied the algorithm five times, using different randomly generated seeds, and checked the consistency of the results across runs. The entire dataset was used for the calculation of sequence-based neutrality statistics, including Tajima’s D, Fu & Li’s D*, Fu & Li’s F*, Fay & Wu’s H, in DnaSP v5.1 (Rozas et al., 2003). P-values for the various neutrality tests were estimated from 104 coalescent simulations, performed with SIMCOAL 2.0 (Laval and Excoffier, 2004) under a finite-site model and using the recombination rate of the tested region reported in HapMap Phase II (Frazer et al., 2007) and the deCODE recombination rate given in the UCSC database (http://genome.ucsc.edu; Kong et al., 2002). Each of the 104 coalescent simulations was conditional on the observed sample size and the number of segregating sites observed for each gene. We corrected for the effects of demography on diversity patterns by considering two demographic models based on resequencing data for noncoding regions in a set of populations similar to those studied here (Voight et al., 2005; Laval et al., 2010). The main difference between these two demographic models is that the Laval’s model takes intercontinental population migration into account (Laval et al., 2010).
We used the McDonald-Kreitman Poisson random field method (Sawyer and Hartl, 1992; Bustamante et al., 2005) to search for the effects of natural selection, taking into account both interspecies divergence and within-species polymorphism. For the detection of recent positive selection events, we used the DIND test, based on the iπA/iπD ratio, where iπA and iπD are the levels of nucleotide diversity associated with the haplotypes carrying the ancestral and the derived alleles, respectively (Barreiro et al., 2009). This test is based on the rationale that a derived allele under positive selection present at high frequency in the population should display lower levels of nucleotide diversity at linked sites than expected, and therefore higher than expected a iπA/iπD ratio. Singletons and doubletons were excluded from this analysis. To define statistical significance, the iπA/iπD values estimated for all IFNs and their receptors were compared against a background distribution obtained by means of 104 simulations of the genomic regions concerned, conditional on the number of segregating sites and the recombination rate of the regions, and integrating the demographic models previously described (Voight et al., 2005; Laval et al., 2010). We also used tests based on levels of extended haplotype homozygosity, such as the iHS (Voight et al., 2006). These tests share a similar rationale: an allele that has a high population frequency and that is associated with an unusually long-range haplotype as compared with genome-wide expectations is likely to have been targeted by recent positive selection. This is explained by the rapid increase in allele frequency of the advantageous allele, meaning that recombination will not have enough time to substantially break down the haplotype on which the selected mutation arose (Nielsen et al., 2007). We assessed the levels of population differentiation for the entire SNP panel, using the FST statistics derived from the analysis of variance (Excoffier et al., 1992). We identified SNPs presenting extreme levels of population differentiation, a signature of positive selection (Sabeti et al., 2006; Nielsen et al., 2007; Barreiro et al., 2008) by comparing the observed FST values for individual SNPs in the genes studied here with a genome-wide FST distribution. This was calculated using ∼640,000 SNPs genotyped in the same subset of individuals from the HGDP-CEPH dataset (Li et al., 2008), with the exception of five individuals who were not genotyped. Because FST values depend on allele frequencies, FST comparisons were confined to SNPs presenting similar allele frequencies (i.e., similar expected heterozygosities). Empirical p-values for each SNP in the 27 genes were estimated as previously described (Barreiro et al., 2009). As the genome-wide FST distribution of the HGDP-CEPH dataset, used here to represent the neutral distribution, includes loci targeted by positive selection (Pickrell et al., 2009), the comparison of FST values of IFNs against this distribution represents a highly conservative approach to detecting selection. We defined genes under selection conservatively as those (a) for which significant results were obtained after both demographic corrections or for which results are significant at the genome-wide level, and (b) for which significant results were obtained in at least two tests of selection based on different aspects of the data (e.g., allele frequency spectrum tests and FST) in the same population. To test the robustness of our results, and to prevent the detection of false positive signatures of positive selection, we measured the probability, within and between the genes in our dataset, of observing neutral simulations exhibiting “significant” results. Specifically, we calculated the number of simulations that exhibit at least two significant tests among Tajima’s D, DIND test, and FST for each gene given the observed p-values, and corrected by the number of genes in our dataset (27 genes). The functional impact of all amino acid–altering mutations (benign, possibly damaging, or probably damaging) was predicted with the Polyphen algorithm v2 HumDiv (Adzhubei et al., 2010). This method, which takes into account protein structure and/or sequence conservation information for each gene, has been shown to be the best predictor of the fitness effects of amino acid substitutions (Williamson et al., 2005).
Online supplemental material.
Fig. S1 shows the simulations of our method to detect gene conversion events and the impact of their removal on the detection of selection. Fig. S2 shows the levels of linkage disequilibrium in the genomic region encompassing the three type III IFN genes. Fig. S3 shows the DIND analyses for all genes encoding the IFNs and their receptors in all populations. Table S1 provides details of the resequenced regions for the genes encoding the IFNs and their receptors. Table S2 lists all exonic SNPs detected at the IFN genes and their receptors. Table S3 shows the diversity indices across the genes encoding the IFNs and their receptors. Table S4 lists the number of gene conversion events, including silent and nonsynonymous sites, detected in type I and type III IFN families. Table S5 lists detected gene conversion events, where acceptor genes receive a derived amino acid–altering polymorphism from a putative donor gene. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111680/DC1.
Acknowledgments
We would like to thank Sandra Pellegrini, Gilles Uzé, Matthew Albert, Claire Leblond, and Katherine Siddle for discussions and critical reading of the manuscript.
This work was supported by the Institut Pasteur, the Agence Nationale de la Recerche (ANR-08-MIEN-009-01), the Fondation pour la Recherche Médicale (FRM), the Centre National de la Recherche Scientifique, Merck-Serono, and an EPFL-Debiopharm Life Sciences Award to L. Quintana-Murci. J. Manry was supported by a FRM fellowship and Y. Itan by an AXA post-doctoral fellowship.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- DAF
- derived allele frequency
- DIND
- derived intraallelic nucleotide diversity
- HCV
- hepatitis C virus
- HGDP-CEPH
- Human Genome Diversity Panel–Centre d’Etude du Polymorphisme Humain
- iHS
- integrated haplotype score
- MAF
- minor allele frequency
- SNP
- single-nucleotide polymorphism
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., et al. ; International HapMap 3 Consortium 2010. Integrating common and rare genetic variation in diverse human populations. Nature. 467:52–58 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Pascual V. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 25:383–392 10.1016/j.immuni.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Banchereau J., Pascual V., Palucka A.K. 2004. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 20:539–550 10.1016/S1074-7613(04)00108-6 [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Quintana-Murci L. 2010. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11:17–30 10.1038/nrg2698 [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Laval G., Quach H., Patin E., Quintana-Murci L. 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40:340–345 10.1038/ng.78 [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J.K., Bouchier C., Tichit M., Neyrolles O., Gicquel B., et al. 2009. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5:e1000562 10.1371/journal.pgen.1000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., et al. 2005. Natural selection on protein-coding genes in the human genome. Nature. 437:1153–1157 10.1038/nature04240 [DOI] [PubMed] [Google Scholar]
- Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., et al. 2002. A human genome diversity cell line panel. Science. 296:261–262 10.1126/science.296.5566.261b [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Abel L. 2007. Human genetics of infectious diseases: a unified theory. EMBO J. 26:915–922 10.1038/sj.emboj.7601558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Abel L., Quintana-Murci L. 2011. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu. Rev. Immunol. 29:447–491 10.1146/annurev-immunol-030409-101335 [DOI] [PubMed] [Google Scholar]
- Chapgier A., Wynn R.F., Jouanguy E., Filipe-Santos O., Zhang S., Feinberg J., Hawkins K., Casanova J.L., Arkwright P.D. 2006. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol. 176:5078–5083 [DOI] [PubMed] [Google Scholar]
- Chapgier A., Kong X.F., Boisson-Dupuis S., Jouanguy E., Averbuch D., Feinberg J., Zhang S.Y., Bustamante J., Vogt G., Lejeune J., et al. 2009. A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 119:1502–1514 10.1172/JCI37083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia E.M., Severa M., Giacomini E., Monneron D., Remoli M.E., Julkunen I., Cella M., Lande R., Uzé G. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34:796–805 10.1002/eji.200324610 [DOI] [PubMed] [Google Scholar]
- Crow M.K. 2007. Type I interferon in systemic lupus erythematosus. Curr. Top. Microbiol. Immunol. 316:359–386 10.1007/978-3-540-71329-6_17 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Black D.N., Ali M., Bond J., Jackson A.P., Lefson M., Michaud J., Roberts E., Stephenson J.B., Woods C.G., Lebon P. 2003. Cree encephalitis is allelic with Aicardi-Goutiéres syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J. Med. Genet. 40:183–187 10.1136/jmg.40.3.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C., et al. 2006a. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 38:917–920 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. 2006b. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38:910–916 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- Decker T., Müller M., Stockinger S. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675–687 10.1038/nri1684 [DOI] [PubMed] [Google Scholar]
- Díaz M.O., Pomykala H.M., Bohlander S.K., Maltepe E., Malik K., Brownstein B., Olopade O.I. 1994. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics. 22:540–552 10.1006/geno.1994.1427 [DOI] [PubMed] [Google Scholar]
- Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- Durbin R.M., Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Gibbs R.A., Hurles M.E., McVean G.A.; 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe-Santos O., Bustamante J., Chapgier A., Vogt G., de Beaucoudrey L., Feinberg J., Jouanguy E., Boisson-Dupuis S., Fieschi C., Picard C., Casanova J.L. 2006. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin. Immunol. 18:347–361 10.1016/j.smim.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Foster G.R., Finter N.B. 1998. Are all type I human interferons equivalent? J. Viral Hepat. 5:143–152 10.1046/j.1365-2893.1998.00103.x [DOI] [PubMed] [Google Scholar]
- Foster G.R., Rodrigues O., Ghouze F., Schulte-Frohlinde E., Testa D., Liao M.J., Stark G.R., Leadbeater L., Thomas H.C. 1996. Different relative activities of human cell-derived interferon-alpha subtypes: IFN-alpha 8 has very high antiviral potency. J. Interferon Cytokine Res. 16:1027–1033 [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. ; International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature. 449:851–861 10.1038/nature06258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 461:399–401 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- Génin P., Lin R., Hiscott J., Civas A. 2009a. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 29:3435–3450 10.1128/MCB.01805-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Génin P., Vaccaro A., Civas A. 2009b. The role of differential expression of human interferon—a genes in antiviral immunity. Cytokine Growth Factor Rev. 20:283–295 10.1016/j.cytogfr.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Glocker E.O., Kotlarz D., Boztug K., Gertz E.M., Schäffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., et al. 2009. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361:2033–2045 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser I., Tovey M.G., Maury C., Chouroulinkov I. 1975. Lethality of interferon preparations for newborn mice. Nature. 258:76–78 10.1038/258076a0 [DOI] [PubMed] [Google Scholar]
- Henco K., Brosius J., Fujisawa A., Fujisawa J.I., Haynes J.R., Hochstadt J., Kovacic T., Pasek M., Schamböck A., Schmid J., et al. 1985. Structural relationship of human interferon alpha genes and pseudogenes. J. Mol. Biol. 185:227–260 10.1016/0022-2836(85)90401-2 [DOI] [PubMed] [Google Scholar]
- Hughes A.L. 1995. The evolution of the type I interferon gene family in mammals. J. Mol. Evol. 41:539–548 10.1007/BF00175811 [DOI] [PubMed] [Google Scholar]
- Innan H., Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11:97–108 10.1038/nrg2689 [DOI] [PubMed] [Google Scholar]
- Jaks E., Gavutis M., Uzé G., Martal J., Piehler J. 2007. Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 366:525–539 10.1016/j.jmb.2006.11.053 [DOI] [PubMed] [Google Scholar]
- Jouanguy E., Zhang S.Y., Chapgier A., Sancho-Shimizu V., Puel A., Picard C., Boisson-Dupuis S., Abel L., Casanova J.L. 2007. Human primary immunodeficiencies of type I interferons. Biochimie. 89:878–883 10.1016/j.biochi.2007.04.016 [DOI] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kotenko S.V., Krause C.D., Izotova L.S., Pollack B.P., Wu W., Pestka S. 1997. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 16:5894–5903 10.1093/emboj/16.19.5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77 10.1038/ni875 [DOI] [PubMed] [Google Scholar]
- Koyama T., Sakamoto N., Tanabe Y., Nakagawa M., Itsui Y., Takeda Y., Kakinuma S., Sekine Y., Maekawa S., Yanai Y., et al. 2006. Divergent activities of interferon-alpha subtypes against intracellular hepatitis C virus replication. Hepatol. Res. 34:41–49 10.1016/j.hepres.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Kreitman M. 2000. Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 1:539–559 10.1146/annurev.genom.1.1.539 [DOI] [PubMed] [Google Scholar]
- Laval G., Excoffier L. 2004. SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics. 20:2485–2487 10.1093/bioinformatics/bth264 [DOI] [PubMed] [Google Scholar]
- Laval G., Patin E., Barreiro L.B., Quintana-Murci L. 2010. Formulating a historical and demographic model of recent human evolution based on resequencing data from noncoding regions. PLoS ONE. 5:e10284 10.1371/journal.pone.0010284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie T.B., Kalie E., Crisafulli-Cabatu S., Abramovich R., DiGioia G., Moolchan K., Pestka S., Schreiber G. 2011. Binding and activity of all human alpha interferon subtypes. Cytokine. 56:282–289 10.1016/j.cyto.2011.07.019 [DOI] [PubMed] [Google Scholar]
- Le Bon A., Durand V., Kamphuis E., Thompson C., Bulfone-Paus S., Rossmann C., Kalinke U., Tough D.F. 2006a. Direct stimulation of T cells by type I IFN enhances the CD8+ T cell response during cross-priming. J. Immunol. 176:4682–4689 [DOI] [PubMed] [Google Scholar]
- Le Bon A., Thompson C., Kamphuis E., Durand V., Rossmann C., Kalinke U., Tough D.F. 2006b. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J. Immunol. 176:2074–2078 [DOI] [PubMed] [Google Scholar]
- Lewin R. 1987. Africa: cradle of modern humans. Science. 237:1292–1295 10.1126/science.3114877 [DOI] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 319:1100–1104 10.1126/science.1153717 [DOI] [PubMed] [Google Scholar]
- Manry J., Laval G., Patin E., Fornarino S., Tichit M., Bouchier C., Barreiro L.B., Quintana-Murci L. 2011. Evolutionary genetics evidence of an essential, nonredundant role of the IFN-γ pathway in protective immunity. Hum. Mutat. 32:633–642 10.1002/humu.21484 [DOI] [PubMed] [Google Scholar]
- Mansai S.P., Innan H. 2010. The power of the methods for detecting interlocus gene conversion. Genetics. 184:517–527 10.1534/genetics.109.111161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Saito M., Morio T., Watanabe K., Agematsu K., Tsuchiya S., Takada H., Hara T., Kawamura N., Ariga T., et al. 2006. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 25:745–755 10.1016/j.immuni.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A.G. 2007. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 8:857–868 10.1038/nrg2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Di Rienzo A. 2009. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 10:745–755 10.1038/nrg2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T.R. 2009. Interferon-alfa, interferon-lambda and hepatitis C. Nat. Genet. 41:1048–1050 10.1038/ng.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 2000. Mechanisms of molecular evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1623–1626 10.1098/rstb.2000.0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 2010. Gene Conversion and Evolution of Gene Families: An Overview. Genes. 1:349–356 10.3390/genes1030349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Krause C.D., Walter M.R. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8–32 10.1111/j.0105-2896.2004.00204.x [DOI] [PubMed] [Google Scholar]
- Pickrell J.K., Coop G., Novembre J., Kudaravalli S., Li J.Z., Absher D., Srinivasan B.S., Barsh G.S., Myers R.M., Feldman M.W., Pritchard J.K. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19:826–837 10.1101/gr.087577.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L., Alcaïs A., Abel L., Casanova J.L. 2007. Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases. Nat. Immunol. 8:1165–1171 10.1038/ni1535 [DOI] [PubMed] [Google Scholar]
- Rauch A., Kutalik Z., Descombes P., Cai T., Di Iulio J., Mueller T., Bochud M., Battegay M., Bernasconi E., Borovicka J., et al. ; Swiss Hepatitis C Cohort Study; Swiss HIV Cohort Study 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 138:1338–1345: 1345: e1–e7 10.1053/j.gastro.2009.12.056 [DOI] [PubMed] [Google Scholar]
- Rice G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M., Fuller J.C., Jackson R.M., Lamb T., Briggs T.A., et al. 2009. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41:829–832 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière Y., Gresser I., Guillon J.C., Tovey M.G. 1977. Inhibition by anti-interferon serum of lymphocytic choriomeningitis virus disease in suckling mice. Proc. Natl. Acad. Sci. USA. 74:2135–2139 10.1073/pnas.74.5.2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J.C., Messeguer X., Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 19:2496–2497 10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. 2006. Positive natural selection in the human lineage. Science. 312:1614–1620 10.1126/science.1124309 [DOI] [PubMed] [Google Scholar]
- Sawyer S.A., Hartl D.L. 1992. Population genetics of polymorphism and divergence. Genetics. 132:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 10.1038/ni873 [DOI] [PubMed] [Google Scholar]
- Stephens M., Donnelly P. 2003. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73:1162–1169 10.1086/379378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 10.1038/ng.447 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Matsuda F., Margetic N., Lathrop M. 2003. Automated identification of single nucleotide polymorphisms from sequencing data. J. Bioinform. Comput. Biol. 1:253–265 10.1142/S021972000300006X [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 10.1038/ng.449 [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 461:798–801 10.1038/nature08463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Moraga I., Levin D., Krutzik P.O., Podoplelova Y., Trejo A., Lee C., Yarden G., Vleck S.E., Glenn J.S., et al. 2011. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 146:621–632 10.1016/j.cell.2011.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent J.M., Olson S., Lawn R.M. 1982. Chromosomal localization of human leukocyte, fibroblast, and immune interferon genes by means of in situ hybridization. Proc. Natl. Acad. Sci. USA. 79:7809–7813 10.1073/pnas.79.24.7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207:2053–2063 10.1084/jem.20101664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzé G., Schreiber G., Piehler J., Pellegrini S. 2007. The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 316:71–95 10.1007/978-3-540-71329-6_5 [DOI] [PubMed] [Google Scholar]
- Vilcek J. 1984. Adverse effects of interferon in virus infections, autoimmune diseases and acquired immunodeficiency. Prog. Med. Virol. 30:62–77 [PubMed] [Google Scholar]
- Vilcek J. 2006. Fifty years of interferon research: aiming at a moving target. Immunity. 25:343–348 10.1016/j.immuni.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Voight B.F., Adams A.M., Frisse L.A., Qian Y., Hudson R.R., Di Rienzo A. 2005. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. USA. 102:18508–18513 10.1073/pnas.0507325102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock E.F., Sibley W.A. 1965. Circulating Virus, Interferon and Antibody after Vaccination with the 17-D Strain of Yellow-Fever Virus. N. Engl. J. Med. 273:194–198 10.1056/NEJM196507222730404 [DOI] [PubMed] [Google Scholar]
- Williamson S.H., Hernandez R., Fledel-Alon A., Zhu L., Nielsen R., Bustamante C.D. 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA. 102:7882–7887 10.1073/pnas.0502300102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk C.H., Frost S.D., Richman D.D., Higley P.E., Kosakovsky Pond S.L. 2007. Evolution of the interferon alpha gene family in eutherian mammals. Gene. 397:38–50 10.1016/j.gene.2007.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M.H., Aggarwal S., Ho W.H., Foster J., Zhang Z., Stinson J., Wood W.I., Goddard A.D., Gurney A.L. 2000. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J. Biol. Chem. 275:31335–31339 10.1074/jbc.M005304200 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Boisson-Dupuis S., Chapgier A., Yang K., Bustamante J., Puel A., Picard C., Abel L., Jouanguy E., Casanova J.L. 2008. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol. Rev. 226:29–40 10.1111/j.1600-065X.2008.00698.x [DOI] [PubMed] [Google Scholar]