Abstract
Complex interactions among genes and environmental factors are known to play a role in common human disease aetiology. There is a growing body of evidence to suggest that complex interactions are ‘the norm’ and, rather than amounting to a small perturbation to classical Mendelian genetics, interactions may be the predominant effect. Traditional statistical methods are not well suited for detecting such interactions, especially when the data are high dimensional (many attributes or independent variables) or when interactions occur between more than two polymorphisms. In this review, we discuss machine-learning models and algorithms for identifying and characterising susceptibility genes in common, complex, multifactorial human diseases. We focus on the following machine-learning methods that have been used to detect gene-gene interactions: neural networks, cellular automata, random forests, and multifactor dimensionality reduction. We conclude with some ideas about how these methods and others can be integrated into a comprehensive and flexible framework for data mining and knowledge discovery in human genetics.
An important goal of human genetics is to identify DNA sequence variations that increase or decrease susceptibility to disease. Unlike rare Mendelian diseases, which may be characterised by a single gene, susceptibility to common diseases is influenced by the nonlinear interactions of multiple genes and environmental factors. Biomolecular interactions that occur at the level of the individual can be referred to as ‘biological epistasis’[1,2] and this is what Bateson[3] had in mind when he coined the term epistasis. Deviations from additivity in a statistical model that summarises a population of individuals can be referred to as ‘statistical epistasis’;[1,2] this is what Fisher[4] had in mind when used the term epistacy. The language of gene interactions has been discussed in more detail by Phillips.[5]
Detecting and characterising attribute interactions, as statistical epistasis can be generically called, is a well known challenge in data mining[6] and is becoming increasingly recognised as a problem that needs to be addressed in human genetics and genetic epidemiology. In fact, there are reasons to believe that the effect of gene-gene interactions, or epistasis, plays a more important role than the independent main effect of any one gene in the susceptibility to common human diseases.[7] These kinds of complexities have implications that reach beyond study design and genetic analysis. For example, it has been suggested that we need to prepare the next generation of investigators for addressing these challenges.[8]
Given the complexity of biomolecular interactions (i.e. biological epistasis) in gene regulation and metabolic systems, the relationship between DNA sequence variations and clinical endpoints (i.e. statistical epistasis) is likely to involve gene-gene interactions.[7,9,10] For example, an expressed gene produces a protein, which in turn may activate or suppress another gene, and so on. Complex gene interaction networks have been described in simpler organisms such as bacteria[11] and yeast.[12] The ubiquity of gene-gene interactions in the genetic architecture of human diseases is also a plausible explanation for the fact that positive results from studies of single polymorphisms and single genes are typically not reproducible across independent samples,[13] further suggesting that important effects due to gene-gene interactions will be overlooked if not properly investigated.[7,10,14] Although the definitive methods for detecting gene-gene interactions are not known, traditional statistical methods, as discussed in more detail later in this section, are not well suited for the task, and new approaches are needed that are designed with interactions in mind. It is time for an analytical retooling that embraces, rather than ignores, the complexity of genetic architecture.[15] Only after this analytical retooling will it be possible to routinely detect and characterise statistical patterns of epistasis in human populations. The ultimate challenge will be making accurate biological inferences from computational and statistical models.[1,2]
Attribute or variable interactions (i.e. statistical epistasis) may be defined as follows.[6] Consider independent variables X1 and X2 and a class variable Y. Attributes X1 and X2 are said to interact when the relationship between X1 and Y depends on X2. A simple example illustrating nonlinear attribute interaction is an XOR (exclusive OR) model, shown in table I. Only considering one attribute at a time would result in the conclusion that neither of the independent variables correlate with Y. Looking at only X1, for example, would result in this incorrect conclusion because given a state of X1 there is a 50/50 chance of yielding either state of Y. To accurately predict the classifier variable in this interaction model, one must consider both independent variables simultaneously.
Table I.
Exclusive OR (XOR) interaction model for two binary variables
| X1 | X2 | Y |
|---|---|---|
| 0 | 0 | 0 |
| 0 | 1 | 1 |
| 1 | 0 | 1 |
| 1 | 1 | 0 |
Analogous epistatic models exist in genetics, such as the following two-locus XOR genetic model, first described by Li and Reich[16] and later by Culverhouse et al.[17] and Moore et al.[18] Consider two single nucleotide polymorphisms (SNPs): locus A with alleles ‘A’ and ‘a’ and locus B with alleles ‘B’ and ‘b’. For each SNP, there are three states or genotypes since allele order does not matter. For SNP A, the genotypes are AA, Aa and aa, and likewise for SNP B. In the XOR model shown in table II, high disease risk is dependent on inheriting a heterozygous genotype (Aa) from one SNP or a heterozygous genotype from a second SNP (Bb), but not both. The probabilities that an individual will have the disease for each of the nine possible genotype combinations are given in a penetrance table (e.g. table II). In this example, each pair of alleles has a biological population frequency of p = q = 0.5 with genotype frequency p2 for AA and BB, 2pq for Aa and Bb, and q2 for aa and bb, consistent with Hardy-Weinberg equilibrium.
Table II.
Penetrance values for combinations of genotypes from two single nucleotide polymorphisms exhibiting interactions in the absence of independent main effects
| Genotypea | Genotypea |
Marginal penetranceb | ||
|---|---|---|---|---|
| AA (0.25) | Aa (0.50) | aa (0.25) | ||
| BB (0.25) | 0 | 1 | 0 | 0.5 |
| Bb (0.50) | 1 | 0 | 1 | 0.5 |
| bb (0.25) | 0 | 1 | 0 | 0.5 |
| Marginal penetranceb | 0.5 | 0.5 | 0.5 | |
Genotype frequencies are given in parentheses.
Marginal penetrance values for the A (B) genotypes.
The marginal penetrance of a genotype is the probability of that genotype being associated with disease risk, regardless of the genotype at the other variables. It is calculated by taking the dot product of the row (or column) of penetrance values associated with the genotype of interest and the column (or row) of genotype frequencies associated with the genotypes of the other genetic variables. For example, the marginal penetrance of BB is (0, 1, 0) (0.25, 0.50, 0.25)T = 0.5. Notice in table II that the marginal penetrance values for all six genotypes are equal, indicating the absence of a main effect, or, put another way, the genetic variables do not independently affect disease risk. The genotype of one SNP only affects disease risk in the presence of the other SNP, and this despite the table penetrance values being unequal. This hypothetical example illustrates epistasis in the absence of marginal effects and represents one possible worst-case scenario in genetic analysis.
For a two-locus model, it is a simple matter to consider the effect on disease risk of all possible genotype combinations, but this quickly becomes a combinatorial challenge as the number of loci (i.e. the dimensionality of the dataset) increases. Traditional parametric statistical methods are limited in their ability to identify interacting susceptibility genes in small sample sizes because of the sparseness of the data in high dimensions. This phenomenon is referred to as the curse of dimensionality:[19] as the number of interacting genes increases, the number of genotype combinations (i.e. the dimensionality) increases exponentially, leading to the need for commensurately larger sample sizes. Unfortunately, collecting genetic datasets with such a large number of observations is prohibitively expensive. Thus, new statistical and computational methods are needed to improve the power to identify epistasis in genetic and epidemiologic samples.
Another drawback of traditional statistical methods for identifying interactions is the need to specify a model for the interaction. The problem is particularly acute when, again, the dimensionality, and hence, the number of possible interactions, is large. Logistic regression, for example, models the probability of disease (p) as a linear function of the independent variables. A logit transformation of p, ln[p/(1 − p)], is used to prevent p from taking on values outside the interval (0, 1). The independent main effects of two SNPs on the disease probability (Pind) can be modelled as (equation 1):
| (Eq. 1) |
where the independent variables are the polymorphisms A and B, which take on discrete genotype values corresponding to the three genotypes; e is the exponential function; and α and β are regression parameters. To model an interaction between A and B, the form of the interaction must be specified. For example, the interaction (Pint) can be modelled by inserting a product term of the form β3AB into equation 1 (equation 2):
| (Eq. 2) |
A test of the null hypothesis of no interaction can be carried out by testing whether β3 = 0. Rejection of this null hypothesis provides evidence for an interaction on a multiplicative scale, but the inability to reject the null hypothesis could mean that the form of the interaction requires operations more complex than simple multiplication. Using logistic regression to detect interactions when main effects are present has been investigated.[20]
One of the advantages of logistic regression is the simple physical interpretation of the model and its parameters as they relate genotypes to probability of disease. However, the advantage of interpretability is nullified if the method is unable to determine which variables interact. A framework for understanding interactions is necessary when analysing genetic data, otherwise useful knowledge (e.g. gene-gene interactions) will go undetected. Machine learning offers a powerful alternative to traditional statistical methods, an alternative that is generally model-free and able to detect nonlinear interactions in high-dimensional datasets.
Machine learning refers to algorithms that allow a computer to learn from experience. In this review, we focus on supervised machine learning, where the output variable guides the learning process. The goal of (supervised) machine learning is to predict the output variable given some input variables. The performance measure of the machine is defined as how well the machine can predict outcomes from independent test data, based on the rules it has learned from the training data. This two-step process of training and testing is referred to as cross-validation and is important for studies to find gene-gene interactions.[21,22] In unsupervised learning, the output variables are unavailable and the goal of the machine is to find patterns in the data. Classic applications of machine learning include speech and handwriting recognition, game playing and data mining.[23] Our primary interest in machine learning for this review is to extract interesting knowledge from the overabundance of genetic data. Specifically, we are interested in variable selection, that is, determining which genes and/or environmental factors lead to an individual’s increased susceptibility to disease.
Machine learning is a more sophisticated descendant of traditional statistical models such as logistic regression. Although the goal of both sets of methods is to make sense of data, machine-learning methods have more flexibility to describe complex datasets. Rather than attempt to give an exhaustive review of all machine-learning models, for simplicity we focus on four models that have been used to detect and characterise gene-gene interactions: neural networks (NNs), cellular automata (CAs), random forests (RFs) and multifactor dimensionality reduction (MDR). For more information on other relevant methods, we direct the reader to the reference list. Many machine-learning methods rely on a search or optimisation algorithm for selecting attributes and/or determining the functional form of the model. Thus, we begin the next section with a brief description of a particular set of stochastic search methods known as evolutionary algorithms.
Optimisation and Evolution
Besides the model itself, the most important part of a machine-learning method is the optimisation procedure that is used to select attributes and tune parameters. In recent decades, some researchers have looked to nature for inspiration when developing new and novel optimisation algorithms. Most of these methods are stochastic in nature. Simulated annealing, for example, is based on the thermodynamic process of cooling a molten material to find the lowest energy (i.e. minimum) solution of a problem.[24] In keeping with the biological and genetic theme, we focus on evolutionary algorithms for optimisation. Evolutionary algorithms, such as genetic algorithms[25] and genetic programming (GP),[26] are based on the mechanics of Darwinian evolution by natural selection. Although we discuss them in terms of optimisation, evolutionary algorithms also fall into the general category of machine learning. These approaches have been widely applied in the domain of bioinformatics.[27]
One goal of an optimisation procedure is to find a set of parameters that allows the machine-learning model to most accurately predict class membership (e.g. affected vs unaffected). A genetic algorithm randomly generates a population of solutions to the problem and evaluates their fitness by applying a fitness function. The fitness function is the environment in which the population must adapt. Candidate solutions are encoded as binary arrays or chromosomes. The chromosomes compete in the population and those with the highest fitness are selected to undergo exchange of random model pieces, a process referred to as recombination. Recombination generates variability among the solutions and is essential to the success of the search routine. By analogy with natural selection, less fit candidate solutions are replaced in the population with higher fitness candidates. Following recombination, the models are re-evaluated and the cycle of selection, recombination, and fitness evaluation continues until an optimum solution is identified. GP, applied in the following section to NNs, is similar to genetic algorithms, except the evolutionary operators act on binary expression trees instead of binary arrays or chromosomes.
Evolutionary algorithms, like other stochastic methods, may be well suited for optimising machine-learning models when the search space is extremely large and rugged (i.e. many local minima). Such machine-learning models often do not lend themselves well to traditional methods, such as calculus-based methods, which need an analytical or differentiable function to optimise. Furthermore, calculus-based methods begin their search for an optimum solution at a single point, making these methods susceptible to falling into local minima. The recombination operator makes evolutionary algorithms more robust against succumbing to local minima.[25]
Neural Networks
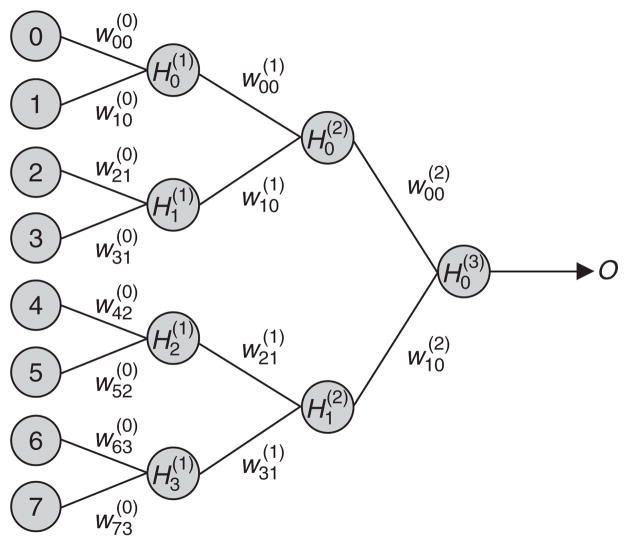
NNs are a popular machine-learning model based on the brain’s ability to solve problems. The terminology of machine learning itself is inspired by this model, which is why the term ‘learn’, as opposed to ‘fit’, is used to describe the process of finding a model that describes some data. An NN can be thought of as a directed graph composed of nodes that represent the processing elements (or neurons), arcs that represent the connections of the nodes (or synaptic connections), and directionality on the arcs that represent the flow of information, as illustrated in figure 1. The processing elements, or nodes, are arranged in layers. The input layer receives an external pattern vector for processing. Each node (Xi) in the input layer is then connected to one or more nodes in the first hidden layer (Hj(1)). The nodes in the first hidden layer are in turn connected to nodes in additional hidden layers or to each output node (O). The number of hidden layers can range from zero to as many as is computationally feasible. Each network connection has an associated weight, or coefficient, (wji(k)). The signal is conducted from the input layer through the hidden layers to the output layer. The output layer, which often consists of a single node, generates an output signal that is used to classify the input pattern.[28]
Fig. 1.
Example of a backpropagation neural network with eight input nodes (Xi) and three hidden layers with four nodes in the first layer, two nodes in the second layer and one node in the third layer. The signal is propagated through the network to yield an output signal (O), which can be put to a threshold to yield an output for affected (case) or unaffected (control). Hj(k) = value of node j in layer k; wji(k) = weight of the connection between node j in layer k with node i in layer k+1.
While NNs are often considered to be a mysterious black box, they are really a series of nonlinear statistical models, similar to regression models. NNs can be expressed as a weighted linear combination of inputs. Each hidden node can be represented as a weighted sum of its inputs. For example, the output from the input nodes to the nodes in the first hidden layer can be written as (equation 3):
| (Eq. 3) |
where σ is a nonlinear activation function, usually chosen to be sigmoid 1/(1 + e−x), and wji(0) are the weights for the connections between input nodes Xi and nodes Hj(1) in hidden layer 1. The output for nodes in subsequent hidden layers (k) can be written as a recurrence relation between the previous hidden layer nodes (equation 4):
| (Eq. 4) |
and the target output can then be modelled as a linear combination of the hidden layers (equation 5):
| (Eq. 5) |
The input pattern vector that is propagated through the network can consist of continuous or discrete values. This is also true of the output signal. Designing the network architecture must take into account the representation of the input pattern vector and how it will interact with the network while propagating information through the network.[28] Thus, the data representation scheme must be suitable to detect the features of the input pattern vector such that it produces the correct output signal. A large field of neural network design has been devoted to the question of proper data representation. More detail regarding the caveats and considerations in this task can be found in Skapura.[28]
Since learning and memory are thought to be associated with the strength of the synapse, setting the strength of the NN connections (or synaptic weights) is the mechanism that allows the network to learn.[29] The connection strengths, together with their inputs, lead to an activity level, which is then used as input for the next layer of the NN.[30] NNs often function with backpropagation types of error minimisation functions, also called gradient descent. Since learning is associated with the synaptic weights, backpropagation algorithms minimise the error by changing the weights following each pass through the network. This ‘hill-climbing’ algorithm makes small changes to the weights until it reaches a value to which any change makes the error higher, indicating that the error has been minimised.[28]
Several research groups have used NNs for genetic studies because of their potential for detecting gene-gene or gene-environment interactions in addition to main effects.[31–43] These studies had varying levels of success because of the challenges associated with designing the appropriate NN architecture. Ritchie et al.[44] proposed a novel NN technique that uses evolutionary algorithms (see section 1) to optimise both the inputs and the architecture of NNs. GP was used to optimise the NN architectures, where each GP binary expression tree represents an NN. The GP-optimised NN (GPNN) optimises the inputs from a larger pool of variables, the weights, and the connectivity of the network, including the number of hidden layers and the number of nodes in the hidden layer. Thus, the algorithm attempts to generate the appropriate network architecture for a given dataset.
Optimisation of NN architecture using GP was first proposed by Koza[26] and Koza and Rice.[45] To evaluate the ability to detect gene-gene interactions, Ritchie et al.[44] performed simulation studies in which data were simulated from a set of different models exhibiting gene-gene interactions (epistasis). Five different epista-sis models were simulated, each exhibiting interactions between two genes. To determine if the addition of the evolutionary algorithm increased the power of the NN for detecting interactions, GPNN was compared with a traditional backpropagation NN (BPNN). Two different analyses were conducted to compare the performance of the BPNN and the GPNN. First, the ability to model gene-gene interactions was determined by comparing the classification and prediction error of the two methods using data containing only the two interacting genes. Second, the ability to detect and model gene-gene interactions was investigated for both NN approaches. This was determined by comparing the classification and prediction errors of the two methods using data containing the interacting genes and a set of other nonfunctional genes.[44] GPNN was able to model nonlinear interactions as well as a traditional BPNN based on the analyses of only the interacting genes. In addition, GPNN had improved power and predictive ability compared with BPNN when applied to data containing both functional and non-functional genes. These results provide evidence that GPNN is able to detect the functional SNPs and model the interactions for the epistasis models described.[44]
Additional studies have been conducted to compare the power of GPNN with stepwise logistic regression[46] and an unconstrained GP.[47] For the logistic regression study, epistasis models were simulated that exhibited interactions between two or three genes. These genes were embedded in a total dataset of ten genes, with 200 affected individuals (cases) and 200 unaffected individuals (controls). In this study, GPNN had substantially higher power than stepwise logistic regression for all models.[46] This is further evidence that machine-learning approaches, such as GPNN, may provide more power to detect epistasis over traditional statistical methods. For the GP study, epistasis models of low heritability (between 0.5% and 3%) were simulated in datasets of 400 cases and 400 controls. It was found that GPNN had higher power and lower false-positive rates to detect gene-gene interactions than an unconstrained GP alone.[47] This demonstrates that NNs are a useful analytical representation for detecting epistatic genetic models in simulated data. Motsinger et al.[48] tested the limits of GPNN to detect simulated epistasis with extremely small genetic effects and an increasing number of interacting loci. The results indicate that GPNN has sufficient power to detect two-locus models in case-control samples of 400, 800 and 1600 individuals for all but the lowest heritability (<1%), although this limitation is overcome as the sample size increases. A similar trend is seen for higher-order models, though the decline in power happens at slightly higher heritabilities as the number of interacting loci increases. The authors successfully applied the GPNN method developed on simulated data to SNPs and environmental factors collected in a study of Parkinson’s disease. GPNN identified a gene-environment interaction, showing the method to be a useful analytical approach for genetic data.[48]
Cellular Automata
CAs are discrete, dynamical systems capable of performing computations on a lattice of cells. The state of each cell is updated synchronously at each time step according to a rule that depends on the state of the cell and its local neighbours at the previous time step. CAs were first introduced by von Neumann[49] for modelling biological self-reproduction and have since been used as models to simulate physical systems that interact locally.[50,51]
A 1-dimensional (1-D) CA is composed of a 1-D lattice of cells. The state of each cell, ai(t), at lattice site i and time step t of a k-state CA, is taken from the alphabet S = {0, 1, …, k −1}. At each time step, the state of each site is updated according to a local rule Φ: S2r+1 → S, where the rule depends on the site’s state at the previous time step as well as the states of its neighbours at the previous time step in a neighbourhood of radius r (equation 6):
| (Eq. 6) |
In this review we only consider spatially cyclic boundary conditions, so that the configuration at each time step forms a circular lattice, or, as the CA propagates in time, a circular cylinder. We focus further on nearest-neighbour (r = 1) CA interactions.
Despite the local nature of the interactions, CAs have been evolved by genetic algorithms to perform certain global computational tasks, meaning the computation of a global property of a dataset. For example, genetic algorithms have been used to evolve 1-D, two-state (k = 2), nearest-neighbour CA rules to classify based on whether or not the density of an arbitrary, fixed-length string of bits exceeds 0.5.[52,53] Usually the rules for this global computation are evolved so that the CA configuration at the final time step is all 1’s (0’s) for initial configurations that have density greater (less) than 0.5. The intuitive goal of a classifier is for the CA output at the final time step to have a reduced complexity over the input configuration, resulting in an irreversible CA, one that does not obey the second law of thermodynamics. For the output of all 1’s or 0’s, the complexity of the output configuration is minimal. CA rules with this final output have been evolved that attain very high, yet not perfect, misclassification error. However, it has been shown that perfect classification can be attained if this restriction on the final configuration is loosened (i.e. affected or unaffected).[54]
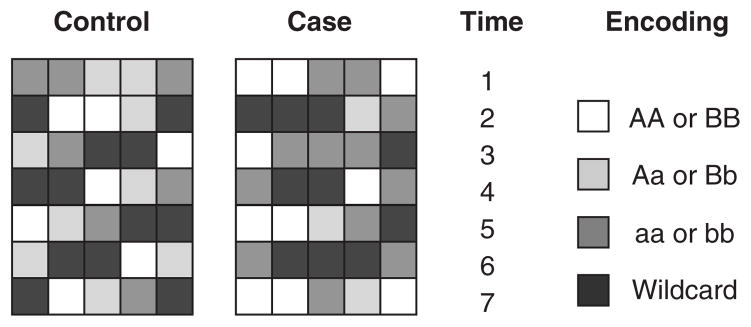
Analysing case-control SNP data is also a global computational task, where the global property is disease status, which is computed from an individual’s pattern of SNPs. Moore and Hahn used the computational ability of CAs to identify combinations of SNPs that interact to influence disease risk.[55,56] The idea behind their approach was similar to that used in the density classification problem above, with a few exceptions. Both CAs had one spatial dimension and used nearest-neighbour interactions, but, unlike the density classification problem which naturally uses binary state CAs, multi-state cells were used in the SNP analysis problem to reflect the multi-state nature of genotypic data. For example, a CA cell can be in the states 0, 1 and 2, assuming each genetic locus has only three possible genotypes. The spatial width of the CA was fixed, but part of the genetic algorithm optimisation involved selection of which SNPs to use as the CA cells. A looser restriction than all 1’s or 0’s was used on the CA output array for the classification of individuals into disease affection status (see Moore and Hahn[55,56] for more details). Figure 2 illustrates an example of CA output for simulated case-control data for two individuals (one case and one control). In this illustration, the CA is allowed to propagate for a fixed number of time steps (e.g. t = 7 units) but only the output configuration at the final time step is used to classify an individual as a case or control.
Fig. 2.
Example of cellular automata (CAs) output for simulated control and case individuals. In the first time step of both CAs, each cell represents a colour-encoded genotype for a particular genetic variation. The cells in the subsequent time steps are the result of the CA computation using a particular rule table (not shown) evolved by the genetic algorithm. The information from the cells in the final time step is used to discriminate between affected (case) and unaffected (control) disease status.
Using simulated data, Moore and Hahn demonstrated that CAs, combined with parallel genetic algorithms (see section 1) for optimisation, are capable of identifying nonlinear interactions among multiple SNPs in the absence of an independent main effect. It was shown that CAs have very good power for identifying gene-gene interactions even in the presence of real-world sources of noise such as genotyping error and phenocopy. Simulation studies indicate that an approach that uses the simplest CA features may be less able to model genetic heterogeneity, which is a well known complicating factor in genetic studies of common diseases such as type 2 diabetes mellitus;[57] however, proposals have been suggested to make CAs more robust in the face of such complicating factors.[55]
Random Forests
An RF is a collection of individual decision-tree classifiers, where each tree in the forest has been trained using a bootstrap sample of instances from the data, and each split attribute in the tree is chosen from among a random subset of attributes.[58] Classification of instances is based upon aggregate voting over all trees in the forest.
Individual trees are constructed as follows from data having N samples and M explanatory attributes:
Choose a training set by selecting N samples, with replacement, from the data.
At each node in the tree, randomly select m attributes from the entire set of M attributes in the data (the magnitude of m is constant throughout the forest building).
Choose the best split at that node from among the m attributes.
Iterate the second and third steps until the tree is fully grown (no pruning).
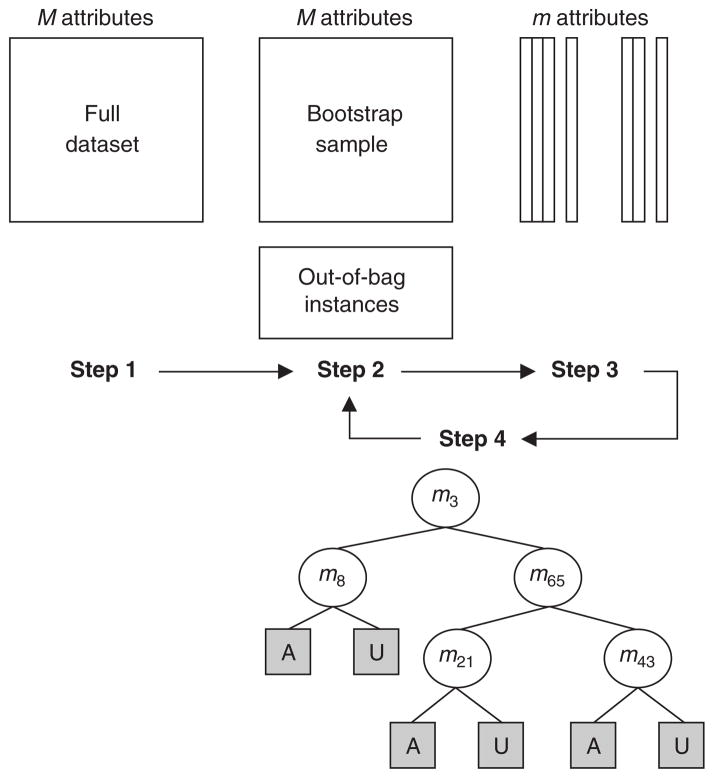
Repetition of this algorithm yields a forest of trees, each of which have been trained on bootstrap samples of instances (see figure 3). Thus, for a given tree, certain instances will have been left out during training. Prediction error is estimated from these ‘out-of-bag’ instances. The out-of-bag instances are also used to estimate the importance of particular attributes via permutation testing. If randomly permuting values of a particular attribute does not affect the predictive ability of trees on out-of-bag samples, that attribute is assigned a low importance score.[59]
Fig. 3.
Construction of individual trees using the random forest method from a full dataset of N instances and M attributes. Step 1: choose a bootstrap training sample by selecting N instances, with replacement, from the full dataset. Step 2: for a new node in the tree, randomly select m attributes from the entire set of M attributes in the data. Step 3: from among the m attributes selected in the previous step, choose the attribute that best splits the training sample at that node. Step 4: iterate the second and third steps until the tree is fully grown (no pruning). This entire algorithm is repeated to yield a forest of arbitrary size, where each individual tree has been trained on a different bootstrap sample of the data. A = affected; U = unaffected.
The decision trees comprising an RF provide an explicit representation of attribute interaction[60] that is readily applicable to the study of gene-gene or gene-environment interactions. These models may uncover interactions among genes and/or environmental factors that do not exhibit strong marginal effects.[61] Additionally, tree methods are suited to dealing with certain types of genetic heterogeneity, since early splits in the tree define separate model subsets in the data.[62] RFs capitalise on the benefits of decision trees and have demonstrated excellent predictive performance when the forest is diverse (i.e. trees are not highly correlated with each other) and composed of individually strong classifier trees.[58] The RF method is a natural approach for studying gene-gene or gene-environment interactions because importance scores for particular attributes take interactions into account without demanding a pre-specified model.[62] However, most current implementations of the importance score are calculated in the context of all other attributes in the model. Therefore, assessing the interactions between particular sets of attributes must be done through careful model interpretation, although there has been preliminary success in jointly permuting explicit sets of attributes to capture their interactive effects.[59]
In selecting functional SNP attributes from simulated case-control data, RFs outperform traditional methods such as the Fisher’s Exact test when the ‘risk’ SNPs interact, and the relative superiority of the RF method increases as more interacting SNPs are added to the model.[62] RFs have also shown to be more robust in the presence of noise SNPs relative to methods that rely on main effects, such as the Fisher’s Exact test.[59] Initial results of RF applications to genetic data in studies of asthma[59] and breast cancer[63] are encouraging, and it is anticipated that RFs will prove a useful tool for detecting gene-gene interactions.
Multifactor Dimensionality Reduction
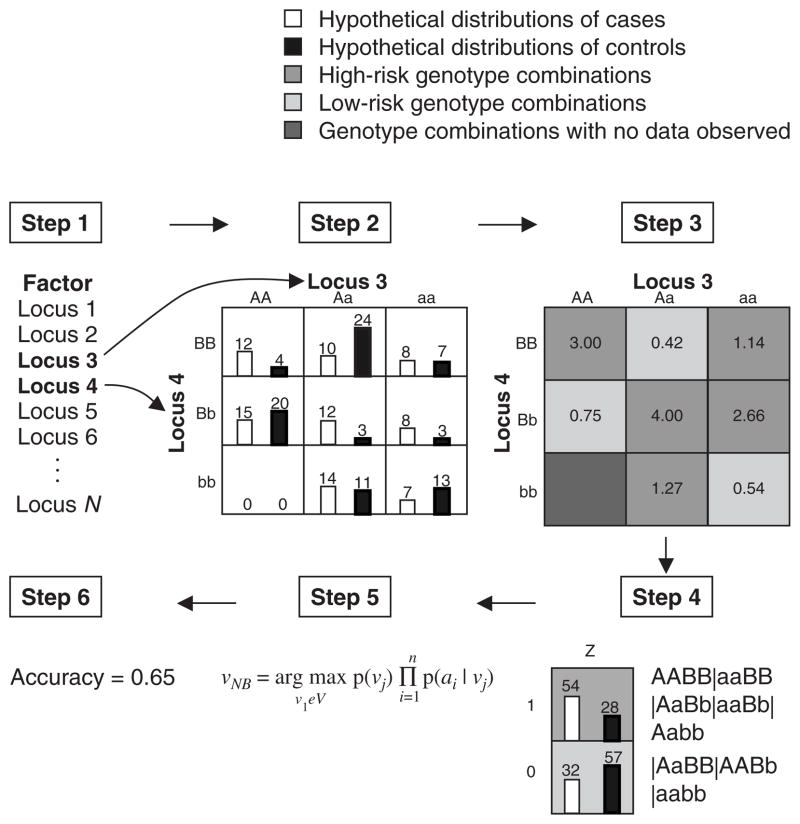
MDR is a machine-learning method specifically designed to identify interacting combinations of genetic variations associated with increased risk of common, complex, multifactorial human diseases.[64–68] This method was developed in response to the limitations posed by logistic regression in detecting gene-gene interactions in epidemiological studies (see introductory section). MDR is nonparametric in that no parameters are estimated, and it is free of any assumed genetic model. The goal of MDR is to find a combination of attributes associated with disease outcome by minimising the number of misclassified individuals. MDR pools multi-locus genotypes or environmental factors into high-risk and low-risk groups, effectively reducing the dimensionality of the predictors (i.e. attributes) from N dimensions to one dimension. This process, wherein a new attribute is defined as a function of two or more other attributes, is termed constructive induction.[69] The newly constructed attribute can be evaluated for its ability to classify and predict disease status.[70] MDR forms a hypothesis by counting the frequency of various gene combinations within the training sample, which is analogous to a naive Bayes classifier.[64] Figure 4 illustrates the general procedure for implementing the MDR method.
Fig. 4.
Summary of the general steps involved in implementing the multifactor dimensionality reduction (MDR) method. Step 1: a set of N genetic and/or discrete environmental factors is selected from the pool of all factors. Step 2: the N factors and their possible multifactor cells are represented in N-dimensional space. Step 3: each multifactor cell in the N-dimensional space is labelled as ‘high risk’ if the ratio of affected individuals to unaffected individuals (the number in the cell) exceeds some threshold T (e.g. T = 1.0) and ‘low risk’ if the threshold is not exceeded. Step 4: the collection of newly constructed multifactor attributes comprising the MDR model for the particular combination of factors. Step 5: the model is evaluated using a naive Bayes classifier. Step 6: the accuracy with which the model separates cases from controls is reported. An optimal predictive model is chosen using cross-validation and permutation testing.
In order to provide a tool that encourages interaction between the biologist and the machine-learning method, a graphical user interface (GUI) for MDR has been implemented using open-source, freely available software packages available from http://www.epistasis.org/mdr.html. The MDR software is designed in accordance with the philosophy advocated by Langley that computational methods should provide explicit support for human intervention in scientific discovery.[71–73] Taken together, the component packages of the MDR toolkit (Data Tool, Analysis Tool, and Permutation Testing Tool) encourage human intervention in all stages the discovery process, including data preprocessing, application of the learning algorithm, and evaluation and interpretation of the resulting model. Indeed, Langley stresses that “appropriate decisions about these issues are more crucial to success than decisions about which learning algorithm to use.”[73] The first step in an effective analysis involves an iterative process of careful data manipulation and filtering. Once the data have been thoroughly examined from a variety of perspectives, the biologist applies his or her specialised knowledge to the application of the machine-learning algorithm. Expert knowledge guides the analysis by assuring that candidate loci appear in the model, manipulating parameters (e.g. setting limits on the number of loci hypothesised to interact in a particular disease) and accounting for peculiarities in the data. As an aid to the evaluation and interpretation of results, the MDR software produces relevant performance statistics as well as various interactive visualisations of MDR models generated, and permutation testing is used to estimate the statistical significance of selected models.
Application of MDR to case-control datasets has routinely yielded evidence of epistasis in the absence of main effects. For example, Ritchie et al.[67] identified a statistically significant interaction among four SNPs from three estrogen metabolism genes for sporadic breast cancer. Again, this interaction was detected in the absence of independent main effects for any of the four SNPs. Evidence for epistasis has also been detected for other common diseases such as essential hypertension,[14,74] type 2 diabetes,[75] atrial fibrillation,[76,77] amyloid polyneuropathy,[78] autism,[79,80] myocardial infarction,[22] coronary calcification,[81] pharmacogenetics,[82,83] bladder cancer,[84] prostate cancer[85] and schizophrenia.[86]
A Flexible Strategy for Data Mining and Knowledge Discovery
Moore et al.[70] have proposed a four-step framework for data mining and knowledge discovery that can integrate constructive induction algorithms such as MDR with other machine-learning methods such as NNs, CAs and RFs. The first step of this flexible framework is to select interesting subsets of SNPs when the total number of SNPs is too large to exhaustively evaluate for gene-gene interactions. The key here is to identify filter strategies such as ReliefF[87] that are capable of assigning a high quality or relevance to SNPs that might be involved in a nonlinear interaction. This first step will be important in the context of genome-wide association studies[88,89] that measure thousands of SNPs. Once interesting SNPs are selected, they can then be used in conjunction with constructive induction algorithms such as MDR to generate new attributes that capture interaction information. Once a new multi-locus attribute is constructed in step two, it can then be evaluated in step three using machine-learning methods such as those described above. The goal of step four is to facilitate statistical interpretation of machine-learning models. Moore et al.[70] suggest using measures of interaction information to construct interaction graphs and interaction dendrograms as described by Jakulin and Bratko[90] and Jakulin.[91] This approach proved useful for modelling atrial fibrillation[70] and bladder cancer,[84] as it differentiated additive from nonadditive interactions. Of course, the final goal is to make biological inferences from gene-gene interactions models. This may be the greatest challenge of all.[1,2,92]
Conclusions
There is a growing awareness that susceptibility to common, multifactorial human diseases is largely due to complex nonlinear interactions among genetic and environmental variables. Traditional statistical methods often lack the ability to identify these interactions because of the inflexibility of the models and the large sample sizes required for accurate parameter estimation. New methods are needed to analyse genetic data that not only address the usual challenges posed by real-world data, such as noise, missing data, and small sample size, but that also recognise interactions as an important effect rather than a perturbation to independent main effects.[15] Interacting variables can easily be hidden to methods that are not specifically designed to detect them. To this end, we reviewed some of the machine-learning methods that have demonstrated the power and flexibility to identify combinations of interacting genes that contribute to the disease status of individuals.
Specifically, we discussed evolution-optimised NNs and CAs, as well as MDR and RFs – machine-learning models that have been successfully used to detect gene-gene interactions. For each of these models, the machine is allowed to learn from experience to discriminate between individuals whose disease affection status is known. After this learning process, during which the model parameters are optimised, the fitted model is applied to an independent dataset to test how well the model generalises.
Although the methods reviewed in this article show promise for detecting gene-gene interactions, no single computational or statistical method will be optimal for every dataset. A successful data analysis will likely combine multiple data analysis methods, both traditional and novel, that have different strengths and weaknesses. This is an advantage of the computational framework proposed by Moore et al.[70] A logical first approach to a genotypic dataset would be to use traditional statistical methods in the hopes of detecting independent main effects. Regardless of whether independent main effects are detected, it will be important to also carry out a gene-gene interaction analysis using methods such as MDR. MDR is a deterministic and conceptually simple constructive induction method that exhaustively considers every possible combination of variables up to a given order. However, the number of combinations can become computationally intractable when many-variable interactions need to be considered as in the context of a large candidate gene study or a genome-wide association study. For example, depending on the number of attributes, it may not be feasible to consider all possible seven-way and higher interactions. For higher-order interactions, it would then be necessary to implement an RF approach or a stochastic optimisation method (e.g. genetic algorithms or simulated annealing) to attempt to traverse the vast search space. Examples discussed in this review of methods that implement such stochastic optimisation methods are GPNNs and genetic algorithm-optimised CAs. Alternatively, a filter-based approach that selects interesting SNPs prior to interaction modelling might be preferable.[70]
Further research is needed to understand the biological mechanisms of disease, but it is becoming increasingly clear that interactions among genes and environmental variables will play a prominent role. The imbalance between experimental output and theoretical understanding in favour of unexplained experimental data in genetics today is reminiscent of particle physics in the 1960s and in chemistry a century earlier, before the days of the quark model and the periodic table, respectively. Perhaps a similar underlying order waits to be discovered in genetics through the collaborative efforts of geneticists, epidemiologists, bioinformaticists, computer scientists, physicians and others.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants AI059694, LM009012, AI057661, AI064625, HL65234, RR018787, ES007373 and HD047447. This work was also supported by generous funds from the Vanderbilt Program in Biomathematics and the Norris-Cotton Cancer Center at Dartmouth Medical School.
Footnotes
The authors have no conflicts of interest that are directly relevant to the content of this review.
References
- 1.Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. Bioessays. 2005 Jun;27(6):637–46. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- 2.Moore JH. A global view of epistasis. Nat Genet. 2005 Jan;37(1):13–4. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- 3.Bateson W. Mendel’s principles of heredity. Cambridge: Cambridge University Press; 1909. [Google Scholar]
- 4.Fisher RA. The correlation between relatives on the assumption of Mendelian inheritance. Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- 5.Phillips PC. The language of gene interaction. Genetics. 1998 Jul;149(3):1167–71. doi: 10.1093/genetics/149.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freitas AA. Understanding the crucial role of attribute interaction in data mining. Artif Intell Rev. 2001;16 (3):177–99. [Google Scholar]
- 7.Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 8.Sing CF, Stengard JH, Kardia SL. Genes, environment, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003 Jul;23(7):1190–6. doi: 10.1161/01.ATV.0000075081.51227.86. [DOI] [PubMed] [Google Scholar]
- 9.Gibson G, Wagner G. Canalization in evolutionary genetics: a stabilizing theory? Bioessays. 2000 Apr;22(4):372–80. doi: 10.1002/(SICI)1521-1878(200004)22:4<372::AID-BIES7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 10.Templeton AR. Epistasis and complex traits. In: Wolf JB, Brodie ED, Wade MJ, editors. Epistasis and the evolutionary process. Oxford: Oxford University Press; 2000. pp. 41–57. [Google Scholar]
- 11.Remold SK, Lenski RE. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet. 2004 Apr;36(4):423–6. doi: 10.1038/ng1324. [DOI] [PubMed] [Google Scholar]
- 12.Segre D, Deluna A, Church GM, et al. Modular epistasis in yeast metabolism. Nat Genet. 2005 Jan;37(1):77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 13.Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- 15.Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20 (12):640–7. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Reich J. A complete enumeration and classification of two-locus disease models. Hum Hered. 2000;50:334–49. doi: 10.1159/000022939. [DOI] [PubMed] [Google Scholar]
- 17.Culverhouse R, Suarez BK, Lin J, et al. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002;70:461–71. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore JH, Hahn LW, Ritchie MD, et al. Application of genetic algorithms to the discovery of complex models for simulation studies in human genetics. In: Langden WB, Cantú-Paz E, Mathias K, et al., editors. Proceedings of the Genetic and Evolutionary Computational Conference 2002. San Francisco (CA): Morgan-Kauffman; 2002. pp. 1150–5. [PMC free article] [PubMed] [Google Scholar]
- 19.Bellman R. Adaptive control processes. Princeton (NJ): Princeton University Press; 1961. [Google Scholar]
- 20.Gauderman WJ, Faucett CL. Detection of gene-environment interactions in joint segregation and linkage analysis. Am J Hum Genet. 1997 Nov;61(5):1189–99. doi: 10.1086/301597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coffey CS, Hebert PR, Krumholz HM, et al. Reporting of model validation procedures in human studies of genetic interactions. Nutrition. 2004;20 (1):69–73. doi: 10.1016/j.nut.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Coffey CS, Hebert PR, Ritchie MD, et al. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene-gene interactions on risk of myocardial infarction: the importance of model validation. BMC Bioinformatics. 2004;5:49. doi: 10.1186/1471-2105-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell T. Machine learning. Boston (MA): McGraw Hill; 1997. [Google Scholar]
- 24.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science. 1983;220:671–80. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg DE. Genetic algorithms in search, optimization, and machine learning. Reading (MA): Addison-Wesley; 1989. [Google Scholar]
- 26.Koza JR. Genetic programming: on the programming of computers by means of natural selection. Cambridge (MA): MIT Press; 1992. [Google Scholar]
- 27.Fogel GB, Corne DW. Evolutionary computation in bioinformatics. San Francisco (CA): Morgan-Kauffman; 2003. [Google Scholar]
- 28.Skapura D. Building neural networks. New York: ACM Press; 1995. [Google Scholar]
- 29.Tarassenko L. A guide to neural computing applications. London: Arnold Publishers; 1998. [Google Scholar]
- 30.Anderson J. An introduction to neural networks. Cambridge (MA): MIT Press; 1995. [Google Scholar]
- 31.Bhat A, Lucek PR, Ott J. Analysis of complex traits using neural networks. Genet Epidemiol. 1999;17 (Suppl 1):S503–7. doi: 10.1002/gepi.1370170781. [DOI] [PubMed] [Google Scholar]
- 32.Bicciato S, Pandin M, Didone G, et al. Pattern identification and classification in gene expression data using an autoassociative neural network model. Biotechnol Bioeng. 2003 Mar;81(5):594–606. doi: 10.1002/bit.10505. [DOI] [PubMed] [Google Scholar]
- 33.Curtis D, North BV, Sham PC. Use of artificial neural network to detect association between a disease and multiple marker genotypes. Ann Hum Genet. 2001;65:95–107. doi: 10.1046/j.1469-1809.2001.6510095.x. [DOI] [PubMed] [Google Scholar]
- 34.Hsia TC, Chiang HC, Chiang D, et al. Prediction of survival in surgical unresectable lung cancer by artificial neural networks including genetic polymorphisms and clinical parameters. J Clin Lab Anal. 2003;17 (6):229–34. doi: 10.1002/jcla.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Haghighi F, Falk C. Design of artificial neural network and its applications to the analysis of alcoholism data. Genet Epidemiol. 1999;17:S223–8. doi: 10.1002/gepi.1370170738. [DOI] [PubMed] [Google Scholar]
- 36.Lucek PR, Hanke J, Reich J, et al. Multi-locus nonparametric linkage analysis of complex trait loci with neural networks. Hum Hered. 1998;48 (5):275–84. doi: 10.1159/000022816. [DOI] [PubMed] [Google Scholar]
- 37.Lucek PR, Ott J. Neural network analysis of complex traits. Genet Epidemiol. 1997;14 (6):1101–6. doi: 10.1002/(SICI)1098-2272(1997)14:6<1101::AID-GEPI90>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 38.Marinov M, Weeks D. The complexity of linkage analysis with neural networks. Hum Hered. 2001;51:169–76. doi: 10.1159/000053338. [DOI] [PubMed] [Google Scholar]
- 39.Ott J. Neural networks and disease association studies. Am J Med Genet. 2001;105:60–1. [PubMed] [Google Scholar]
- 40.Saccone NL, Downey TJ, Meyer DJ, et al. Mapping genotype to phenotype for linkage analysis. Genet Epidemiol. 1997;17:S703–8. doi: 10.1002/gepi.13701707115. [DOI] [PubMed] [Google Scholar]
- 41.Serretti A, Smeraldi E. Neural network analysis in pharmacogenetics of mood disorders. BMC Med Genet. 2004 Dec;5:27. doi: 10.1186/1471-2350-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherriff A, Ott J. Application of neural networks for gene finding. Adv Genet. 2001;42:287–97. doi: 10.1016/s0065-2660(01)42029-3. [DOI] [PubMed] [Google Scholar]
- 43.Tomita Y, Tomida S, Hasegawa Y, et al. Artificial neural network approach for selection of susceptible single nucleotide polymorphisms and construction of prediction model on childhood allergic asthma. BMC Bioinformatics. 2004 Sep;5:120. doi: 10.1186/1471-2105-5-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie MD, White BC, Parker JS, et al. Optimization of neural network architecture using genetic programming improves the detection and modeling of gene-gene interactions in studies of human diseases. BMC Bioinformatics. 2003;4:28. doi: 10.1186/1471-2105-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koza JR, Rice JP. Genetic generation of both the weights and architecture for a neural network. Piscataway (NJ): IEEE Press; 1991. [Google Scholar]
- 46.Ritchie MD, Coffey CS, Moore JH. Genetic programming neural networks as a bioinformatics tool in human genetics. In: Deb K, Poli R, Banthaf W, et al., editors. Lecture notes in computer science. Vol. 3102. New York: Springer; 2004. pp. 438–48. [Google Scholar]
- 47.Bush WS, Motsinger AA, Dudek SM, et al. Can neural network constraints in GP provide power to detect genes associated with human disease? In: Rothlauf F, Branke J, Cagnoni S, et al., editors. Lecture notes in computer science. Vol. 3449. New York: Springer; 2005. pp. 44–53. [Google Scholar]
- 48.Motsinger AA, Lee SL, Mellick G, et al. Power of genetic programming neural networks for detecting high-order gene-gene interactions in association studies of human disease and an application in Parkinson’s disease. BMC Bioinformatics. 2006;7:39. doi: 10.1186/1471-2105-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Von Neumann . The theory of self-reproducing automata. Urbana (IL): University of Illinois Press; 1966. [Google Scholar]
- 50.Spezzano G, Talia D, Gregorio SD, et al. A parallel cellular tool for interaction modeling and simulation. IEEE Computational Science and Engineering. 1996;3:33–43. [Google Scholar]
- 51.Toffoli T. Cellular automata as an alternative to (rather than approximation of) differential equations in modeling physics. Physica D. 1984;10:117–27. [Google Scholar]
- 52.Mitchell M, Crutchfield JP, Hraber PT. Evolving cellular automata to perform computations: mechanisms and impediments. Physica D. 1994;75:361–91. [Google Scholar]
- 53.Packard NH. Adaptation toward the edge of chaos. In: Kelso JAS, Mandell AJ, Shlesinger MF, editors. Dynamical patterns in complex systems. Singapore: World Scientific; 1988. pp. 293–301. [Google Scholar]
- 54.Capcarrere MS, Sipper M. Necessary conditions for density classification by cellular automata. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;64:036113. doi: 10.1103/PhysRevE.64.036113. [DOI] [PubMed] [Google Scholar]
- 55.Moore JH, Hahn LW. Cellular automata and genetic algorithms for parallel problem solving in human genetics. In: Merelo JJ, Panagiotis A, Beyer H-G, editors. Lecture notes in computer science. Vol. 2439. New York: Springer; 2002. pp. 821–30. [Google Scholar]
- 56.Moore JH, Hahn LW. A cellular automata approach to detecting interactions among single-nucleotide polymorphisms in complex multifactorial diseases. Pac Symp Biocomput. 2002:53–64. [PubMed] [Google Scholar]
- 57.Busch C, Hegele R. Genetic determinants of type 2 diabetes mellitus. Clin Genet. 2002;60:243–54. doi: 10.1034/j.1399-0004.2001.600401.x. [DOI] [PubMed] [Google Scholar]
- 58.Breiman L. Random forests. Mach Learn. 2001;45 (1):5–32. [Google Scholar]
- 59.Bureau A, Dupuis J, Falls K, et al. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005 Feb;28(2):171–82. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 60.Breiman L, Friedman JH, Olshen RA, et al. Classification and regression trees. Belmont (CA): Wadsworth International Group; 1984. [Google Scholar]
- 61.Cook NR, Zee RY, Ridker PM. Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat Med. 2004 May;23(9):1439–53. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- 62.Lunetta KL, Hayward LB, Segal J, et al. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004 Dec;5(1):32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwender H, Zucknick M, Ickstadt K, et al. A pilot study on the application of statistical classification procedures to molecular epidemiological data. Toxicol Lett. 2004 Jun;151(1):291–9. doi: 10.1016/j.toxlet.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 64.Hahn LW, Moore JH. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol. 2004;4 (2):183–94. [PubMed] [Google Scholar]
- 65.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19 (3):376–82. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 66.Moore JH. Computational analysis of gene-gene interactions in common human diseases using multifactor dimensionality reduction. Expert Rev Mol Diagn. 2004;4 (6):795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- 67.Ritchie MD, Hahn LW, Roodi N, et al. Multifactor dimensionality reduction reveals high-order interactions among estrogen metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003 Feb;24(2):150–7. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 69.Michalski RS. A theory and methodology of inductive learning. Artif Intell. 1983;20:111–61. [Google Scholar]
- 70.Moore JH, Gilbert JC, Tsai CT, et al. A flexible framework for data mining and knowledge discovery in human genetics. J Theor Biol. In press. [Google Scholar]
- 71.Langley P. The computer-aided discovery of scientific knowledge. In: Carbonell JG, Siekmann J, editors. Lecture notes in artifical intelligence. Vol. 1532. New York: Springer; 1998. pp. 25–39. [Google Scholar]
- 72.Langley P. The computational support of scientific discovery. In: Carbonell JG, Siekmann J, editors. Lecture notes in artifical intelligence. Vol. 2049. New York: Springer; 2001. pp. 230–48. [Google Scholar]
- 73.Langley P. Lessons for the computational discovery of scientific knowledge. International Conference on Machine Learning; 2002 Jul 8–12; Sydney (NSW). San Francisco (CA): Morgan-Kauffman; 2002. pp. 9–12. [Google Scholar]
- 74.Williams SM, Ritchie MD, Phillips JA, et al. Multilocus analysis of hypertension. Hum Hered. 2004;57:28–38. doi: 10.1159/000077387. [DOI] [PubMed] [Google Scholar]
- 75.Cho YM, Ritchie MD, Moore JH, et al. Multifactor-dimensionality reduction shows a two-locus interaction associated with type 2 diabetes mellitus. Diabetologia. 2004;47:549–54. doi: 10.1007/s00125-003-1321-3. [DOI] [PubMed] [Google Scholar]
- 76.Motsinger AA, Donahue BS, Brown NJ, et al. Risk factor interactions and genetic effects associated with post-operative atrial fibrillation. Pac Symp Biocomput. 2006;11:514–95. [PubMed] [Google Scholar]
- 77.Tsai CT, Lai LP, Lin JL, et al. Renin-angiotensin system gene polymorphisms and atrial fibrillation. Circulation. 2004;109:1640–6. doi: 10.1161/01.CIR.0000124487.36586.26. [DOI] [PubMed] [Google Scholar]
- 78.Soares ML, Coelho T, Sousa A, et al. Susceptibility and modifier genes in Portuguese transthyretin V30M amyloid polyneuropathy: complexity in a single-gene disease. Hum Mol Genet. 2005 Feb 15;14(4):543–53. doi: 10.1093/hmg/ddi051. [DOI] [PubMed] [Google Scholar]
- 79.Ashley-Koch AE, Mei H, Jaworski J, et al. An analysis paradigm for investigating multi-locus effects in complex disease: examination of three GABAA receptor subunit genes on 15q11-q13 as risk factors for autistic disorder. Ann Hum Genet. 2006;70:281–92. doi: 10.1111/j.1469-1809.2006.00253.x. [DOI] [PubMed] [Google Scholar]
- 80.Ma DQ, Whitehead PL, Menold MM, et al. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet. 2005 Sep;77(3):377–88. doi: 10.1086/433195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bastone L, Reilly M, Rader DJ, et al. MDR and PRP: a comparison of methods for high-order genotype-phenotype associations. Hum Hered. 2004;58 (2):82–92. doi: 10.1159/000083029. [DOI] [PubMed] [Google Scholar]
- 82.Wilke RA, Reif DM, Moore JH. Combinatorial pharmacogenetics. Nat Rev Drug Discov. 2005 Nov;4(11):911–8. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- 83.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005 Jun;15(6):415–21. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 84.Andrew AS, Nelson HN, Kelsey KT, et al. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking, and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–7. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 85.Xu J, Lowey J, Wiklund F, et al. The interaction of four genes in the inflammation pathway significantly predicts prostate cancer risk. Cancer Epidemiol Bi-omarkers Prev. 2005 Nov;14(11):2563–8. doi: 10.1158/1055-9965.EPI-05-0356. [DOI] [PubMed] [Google Scholar]
- 86.Qin S, Zhao X, Pan Y, et al. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. Eur J Hum Genet. 2005 Jul;13(7):807–14. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- 87.Robnik-Sikonja M, Kononenko I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach Learn. 2003;53 (1):23–69. [Google Scholar]
- 88.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005 Feb;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 89.Wang WY, Barratt BJ, Clayton DG, et al. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005 Feb;6(2):109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 90.Jakulin A, Bratko I. Analyzing attribute dependencies. In: Lavrac N, Gamberger D, Todorovski L, et al., editors. Lecture notes in artificial intelligence. Berlin: Springer-Verlag; 2003. pp. 229–40. [Google Scholar]
- 91.Jakulin A. PhD thesis. Ljubljana, Slovenia: University of Ljubljana; 2003. Attribute interactions in machine learning. [Google Scholar]
- 92.Moore JH, Ritchie MD. The challenges of whole-genome approaches to common diseases. JAMA. 2004 Apr;291(13):1642–3. doi: 10.1001/jama.291.13.1642. [DOI] [PubMed] [Google Scholar]