Abstract
The Gloeobacter ligand-gated ion channel (GLIC) is a bacterial homolog of vertebrate Cys-loop ligand-gated ion channels. Its pore-lining region in particular has a high sequence homology to these related proteins. Here we use electrophysiology to examine a range of compounds that block the channels of Cys-loop receptors to probe their pharmacological similarity with GLIC. The data reveal that a number of these compounds also block GLIC, although the pharmacological profile is distinct from these other proteins. The most potent compound was lindane, a GABAA receptor antagonist, with an IC50 of 0.2 μM. Docking studies indicated two potential binding sites for this ligand in the pore, at the 9′ or between the 0′ and 2′ residues. Similar experiments with picrotoxinin (IC50 = 2.6 μM) and rimantadine (IC50 = 2.6 μM) reveal interactions with 2′Thr residues in the GLIC pore. These locations are strongly supported by mutagenesis data for picrotoxinin and lindane, which are less potent in a T2′S version of GLIC. Overall, our data show that the inhibitory profile of the GLIC pore has considerable overlap with those of Cys-loop receptors, but the GLIC pore has a unique pharmacology.
Introduction
The Gloeobacter ligand-gated ion channel (GLIC) is a bacterial homolog of vertebrate Cys-loop ligand-gated ion channels found in Gloeobacter violaceus, a unicellular cyanobacterium (1). The presence of GLIC in G. violaceus may contribute to the pH adaptation of this cyanobacterium that does not contain thylakoids; photosynthesis and H+ transport occur in its cell membrane. GLIC does not have a Cys-loop, and is therefore a member of the pentameric family of ligand-gated ion channels but not a Cys-loop receptor. GLIC is activated by protons and has a single channel conductance of 8 pS (2,3). GLIC has been crystallized at high (up to 2.9 Å) resolution (3,4). The crystal structures reveal an extracellular and a transmembrane domain with similar structures to Cys-loop receptors, but, unlike these proteins, GLIC lacks an intracellular domain. The structure of GLIC, determined at low pH, was originally proposed to reveal the channel in an open state, but more recent data show the receptor does slowly desensitize (5,6), and thus the structure may in fact show a desensitized, closed state.
GLIC has low overall sequence similarity to Cys-loop receptors, but many functionally important residues and structural features are conserved between these proteins. Of particular interest is the pore region of GLIC, which has high sequence similarity to that of the nicotinic acetylcholine (nACh) receptor pore. In particular GLIC has a Glu at the intracellular end, and similar or identical residues at the pore lining 2′, 6′, and 9′ positions (Fig. 1). GLIC, like the nACh receptor, is cation-selective, and, as it has been resolved to considerably higher resolution than the nACh receptor, the GLIC pore may be an appropriate model to examine the molecular details of nACh receptor pores, and interactions with pore-blocking compounds. Recently the structure of an invertebrate anion-selective Cys-loop receptor, the glutamate-gated chloride channel (GluCl), was determined, the first Cys-loop receptor whose pore region has been resolved at <4 Å (7). Nevertheless, the sequence similarity between GluCl and the nACh receptor is lower than that between GLIC and the nACh receptor, and GluCl selects for anions and not cations; thus, GLIC may be a more appropriate structural template for studying cation-selective Cys-loop receptor pores. However, it is not clear if the characteristics of the GLIC pore are similar to those of Cys-loop receptors, and so here we report the effects of a range of Cys-loop receptor ligands on GLIC responses. The aim was to probe the pharmacology of the GLIC pore to determine its functional similarity with the pores of Cys-loop receptors.
Figure 1.

Alignment of the pore lining regions of GLIC and a selection of related proteins. The residues that line the pore are highlighted. Comparison of the sequences of GLIC and nACh α1 from 0′ to 18′ reveals 28% identity and 61% similarity. A/M2 = Influenza A M2 Channel.
Materials and Methods
Cell culture and oocyte maintenance
Xenopus laevis oocyte-positive females were purchased from NASCO (Fort Atkinson, WI) and maintained according to standard methods. Harvested stage V-VI Xenopus oocytes were washed in four changes of ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, pH 7.5), defolliculated in 1.5 mg ml−1 collagenase Type 1A for ∼2 h, washed again in four changes of ND96, and stored in ND96 containing 2.5 mM sodium pyruvate, 0.7 mM theophylline, and 50 mM gentamicin.
Receptor expression
A codon-optimized version of GLIC, fused to the signal sequence of the α7 nACh receptor subunit and kindly gifted from C. Ulens (Katholieke Universiteit Leuven, Leuven, Belgium), was cloned into pGEMHE for oocyte expression. Mutant receptors were created using QuikChange mutagenesis (Agilent, Santa Clara, CA). cRNA was transcribed in vitro from linearized cDNA using the mMessage mMachine T7 kit (Ambion, Austin, TX). Stage V and VI oocytes were injected with 50 nL of ∼400 ng μL−1 cRNA, and currents were recorded 1–4 days postinjection.
Electrophysiology
Using two-electrode voltage-clamp, Xenopus oocytes were clamped at −60 mV using an OC-725 amplifier (Warner Instruments, Hamden, CT), Digidata 1322A (Axon Instruments, Union City, CA), and the Strathclyde Electrophysiology Software Package (Department of Physiology and Pharmacology, University of Strathclyde, UK; http://www.strath.ac.uk/Departments/PhysPharm/). Currents were filtered at a frequency of 1 kHz. Microelectrodes were fabricated from borosilicate glass (GC120TF-10; Harvard Apparatus, Kent, UK) using a one-stage horizontal pull (P-87; Sutter Instrument, Novato, CA) and filled with 3M KCl. Pipette resistances ranged from 1.0 to 2.0 MΩ. Oocytes were perfused with saline containing 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 10 mM MES (adjusted to the desired pH) at a constant rate of 12–15 ml min−1. Drug application was via a simple gravity-fed system calibrated to run at the same rate as the saline perfusion.
Analysis and curve fitting were performed using Prism v4.03 (GraphPad Software, La Jolla, CA). Concentration-response data for each oocyte were normalized to the maximum current for that oocyte. The mean and mean ± SE for a series of oocytes were plotted against agonist or antagonist concentration and iteratively fitted to
| (1) |
where A is the concentration of ligand present; IA is the current in the presence of ligand concentration A; Imin is the current when A = 0; Imax is the current when A = ∞; A50 is the concentration of A that evokes a current equal to (Imax + Imin)/2; and nH is the Hill coefficient.
Docking
Docking was performed using the GLIC crystal structure (PDB ID: 3EAM). Three-dimensional structures of each ligand were extracted from the Cambridge Structural Database (Ref. codes: lindane = HCCYHG02, tetracaine = XISVOK01, and picrotoxinin = CIBCUL10), and protonated forms were constructed using Chem3D Ultra 7.0 (CambridgeSoft, PerkinElmer, Waltham, MA) and energy-minimized using the MM2 force field. Rimantadine was created and minimized de novo using the same software.
Docking of the protonated ligands into GLIC was carried out using GOLD 3.0 (The Cambridge Crystallographic Data Centre, Cambridge, UK). The binding site was constrained as a docking sphere with a 20 Å radius surrounding the Cα of 6′ residues in chains A and C. These amino acids were chosen based on the binding locations of the ligands in other Cys-loop receptor pores, but the docking sphere covers the full length of the receptor pore (i.e., the region bounded by the M2 helices from −2′ to 20′). Ten genetic algorithm runs were performed on each docking exercise using default parameters. The structures were visualized using the softwares PyMOL v 1.3 (DeLano Scientific, Palo Alto, CA) and ViewerLite v 5.0 (Accelrys, San Diego, CA).
Results
GLIC activation
Current amplitude was measured at a range of external H+ concentrations, yielding a pH50 of 5.5 ± 0.1 (Fig. 2), that is comparable to previous reports (3,4).
Figure 2.
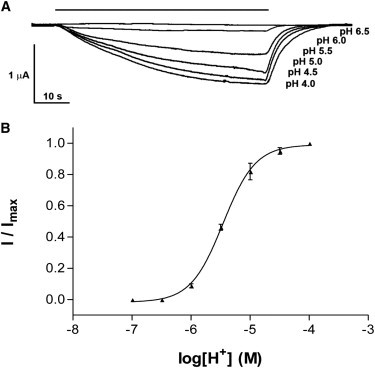
Activation of GLIC. (A) pH dependence of GLIC activation (typical of seven similar experiments) with concentration response curve shown in panel B. (Data = mean ± SE, n = 7.)
GLIC antagonists/modulators
We examined 20 compounds that act at a range of Cys-loop receptors (Table 1); some have similar effects on several different Cys-loop receptors (e.g., picrotoxin is an inhibitor of γ-aminobutyric acid (GABAA), glycine, 5-HT3, and nACh receptors) but others have different effects on different receptors and may act at multiple and/or distinct sites in the different proteins. Many (in this case, 13) of the compounds inhibited GLIC function. Lindane, a GABAA receptor inhibitor, was the most potent and the only compound tested that had an IC50 <1 μM. Three compounds (picrotoxinin, rimantadine, and tetracaine) had IC50s <10 μM; seven had IC50s between 10 and 100 μM, and two had IC50s >100 μM. Example concentration inhibition curves are shown in Fig. 3. The remaining compounds showed <5% block at 100 μM. We also examined a selection of compounds that modulate the function of various Cys-loop receptors. Dexamethasone and 5-hydroxyindole enhance 5-HT3 receptor function, whereas ivermectin is an agonist at glycine and GluCl receptors, acting at a site remote from the orthosteric binding site. None of these modulatory compounds had an effect on GLIC function.
Table 1.
Potential GLIC antagonists/modulators
| Ligand | Known Cys-loop receptor targets | pIC50 (mean ± SE) | IC50 (μM) | Hill slope | n |
|---|---|---|---|---|---|
| 5-Hydroxyindolem | 5-HT3 | NI | — | — | 3 |
| Amantadine | nACh | 4.83 ± 0.04 | 14.8 | 1.4 ± 0.2 | 4 |
| Bilobalide | GABAA, Gly, 5-HT3 | 3.94 ± 0.04 | 114 | 1.2 ± 0.1 | 4 |
| Chlorpromazine | nACh, 5-HT3 | 4.74 ± 0.06 | 18.4 | 2.2 ± 0.5 | 4 |
| Dexamethasonem | 5-HT3 | NI | — | — | 4 |
| Dieldrin | GABAA | NI | — | — | 4 |
| Diltiazem | 5-HT3 | 4.31 ± 0.04 | 48.8 | 2.2 ± 0.4 | 4 |
| Estrone | GABAA, 5-HT3 | 3.65 ± 0.20 | 224 | 0.5 ± 0.2 | 5 |
| Fipronil | GluCl, GABAA | 4.70 ± 0.03 | 20.0 | 1.1 ± 0.07 | 4 |
| Imidaclopride | nACh | NI | — | — | 4 |
| Ivermectin | GluCl, Gly, GABAA | NI | — | — | 4 |
| Lindane | GABAA, Gly | 6.64 ± 0.04 | 0.23 | 1.1 ± 0.1 | 4 |
| Mefloquine | 5-HT3, nACh | 4.66 ± 0.02 | 21.8 | 2.7 ± 0.3 | 5 |
| Pancuronium | nACh | NI | — | — | 5 |
| Picrotoxinin | GABAA, Gly, 5-HT3, GABAC | 5.59 ± 0.04 | 2.57 | 1.8 ± 0.3 | 4 |
| Quinacrine | nACh | NI | — | — | 5 |
| QX-222 | nACh, 5-HT3 | NI | — | — | 5 |
| Rimantadine | nACh | 5.59 ± 0.05 | 2.58 | 1.1 ± 0.2 | 5 |
| Tetracaine | nACh | 5.65 ± 0.06 | 2.25 | 0.8 ± 0.1 | 5 |
| α-Endosulfan | GABAA | 4.77 ± 0.15 | 17.0 | 0.7 ± 0.2 | 5 |
Most of these compounds inhibit the function of at least one Cys-loop receptor, although their sites of action are not all known and may be multiple. Ivermectin acts as an allosteric agonist at some receptors. See text for more details. Key: m, positive modulator at its known receptor; NI, no inhibition at 100 μM.
Figure 3.
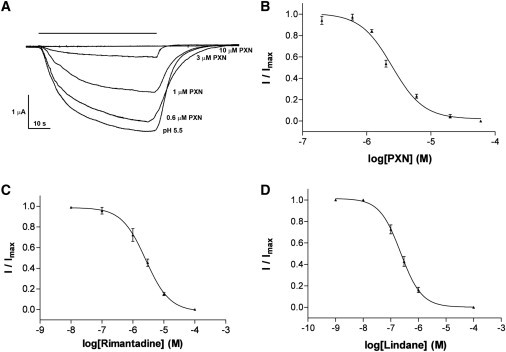
GLIC antagonists. (A) Example traces showing inhibition by picrotoxinin (PXN) and concentration-inhibition curves for PXN (B), rimantadine (C), and lindane (D). Inhibition was measured at the pH50. (Data = mean ± SE, n = 3–5.)
Ligand docking
To examine the possible location of the most potent inhibitors, we docked the ligands with highest potency (lindane, picrotoxinin, rimantadine, and tetracaine) into the GLIC channel. Ten ligand poses were generated for each compound. In Fig. 4, these poses are superimposed to show the range of positions of the docked ligands. Lindane, which is incapable of making H-bonds, showed the most varied localization. The docking data suggest it has the ability to bind at two distinct locations in the channel: at the intracellular end of the channel between the 0′ and 2′ residues (Fig. 5), and in the middle of the channel close to the 9′ Leu. Tetracaine is a relatively long molecule and the docking procedure located it between the 2′ and 9′ residues, either vertically or horizontally with respect to the channel axis; there were potential H-bond interactions with the 6′ Ser. Picrotoxinin and rimantadine were located in overlapping positions toward the intracellular side of the channel, and both were stabilized by a series of H-bonds with the 2′ Thr residues (Figs. 4 and 5).
Figure 4.
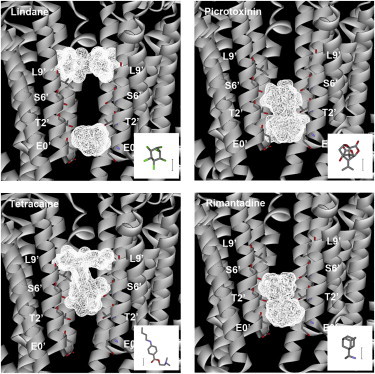
An overlay of the 10 most energetically favorable docked poses for lindane, picrotoxinin, tetracaine, and rimantadine. For lindane, the poses are almost equally distributed between two quite distinct binding locations, whereas the locations of the docked poses of the other compounds are broadly similar. The channel volume occupied is calculated from the van der Waals radii and shown in wireframe. (Inset) Structures of the docked ligands; scale bar = 2.5 Å.
Figure 5.
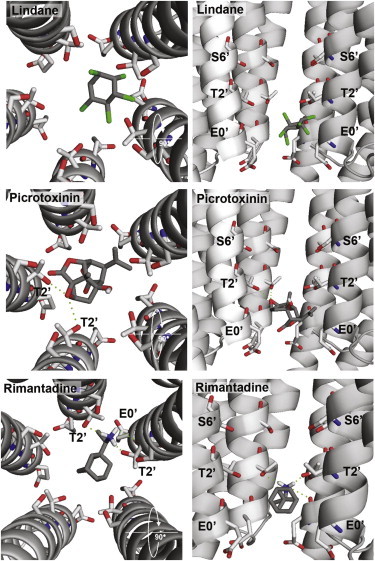
Examples of orientations of lindane, picrotoxinin, and rimantadine docked into the GLIC channel near the 2′ residue, the location supported by our mutagenesis data. There are no predicted H-bonds with lindane, but several with 2′Thr residues for both picrotoxinin and rimantadine (dashed lines).
Characterization of mutant receptors
To probe the accuracy of the docking, a GLIC receptor containing a T2′S mutation was created. This receptor had a similar pH50 to wild-type receptors: 5.2 ± 0.1 (n = 4). However, inhibition of pH50 responses in this mutant receptor revealed large changes in IC50s for picrotoxinin and lindane, although there was no significant change for rimantadine (Table 2).
Table 2.
Potencies of ligands at GLIC T2′S mutant receptors
| Ligand | pIC50 (mean± SE) | IC50 (μM) | Hill slope | Fold change in IC50 |
|---|---|---|---|---|
| Picrotoxinin | 3.776 ± 0.103∗ | 167 | 1.0 ± 0.2 | 65 |
| Rimantadine | 5.454 ± 0.088 | 3.5 | 1.1 ± 0.2 | 1.4 |
| Lindane | 4.862 ± 0.136∗ | 13.7 | 1.3 ± 0.4 | 59 |
Significantly different to wild-type; n = 3–5.
Discussion
GLIC is a proton-activated prokaryotic ligand-gated ion channel whose structure has been extensively investigated, but whose pharmacology has been less well explored. To date, only protons are known to activate GLIC, although a range of inhibitors have been identified: these include quaternary ammonium compounds, anesthetics, and divalent cations (2,8,9). These compounds are of low potency, with IC50s in the high μM or mM range, and structural and other data have revealed specific binding sites in or close to the pore, although some compounds, e.g., halothane and thiopental, also act in the extracellular domain (9–11). A range of alcohols have also been shown to inhibit or enhance GLIC function, and data have shown these again act in or close to the pore, with the short-chain (potentiatory) alcohols occupying a pocket facing the pore in the intersubunit interface (12). Quaternary ammonium compounds, anesthetics, cations, and alcohols also act at a range of Cys-loop receptors, and here we show that many other ligands that antagonize the function of these related receptors can also block GLIC responses; thus, although the pore-lining residues in this receptor show some differences to its relatives, GLIC appears to share considerable pharmacological overlap with them. Docking of the most potent of these antagonists into the pore of GLIC indicates that they interact with residues that have been established as important for interacting with channel blockers in Cys-loop receptors. It also places them at positions similar to those of other channel binding ligands that have been previously identified by cocrystallization with GLIC (9). Inhibition of GLIC responses in a T2′S mutant receptor supports an interaction at the 2′ location with picrotoxinin and lindane.
These data show that the pharmacology of the GLIC channel is distinct from that of related proteins, but the overlap in activity of compounds that block Cys-loop receptors suggests the structure of the GLIC M2 region provides a suitable template for understanding interactions with a range of channel blocking compounds in a range of receptors.
Lindane was the most potent of the compounds that we tested. Lindane is a neurotoxic organochlorine pesticide that inhibits GABAA receptor function; its binding site overlaps with the picrotoxin recognition site in the pore, and blocks Cl− flux (13,14). It also inhibits Cl− flux through glycine receptors and voltage-dependent chloride channels (15). Docking studies in GABAA receptors indicate that lindane interacts with 2′Ala and 6′Thr residues (16). We observed two possible docking locations in the GLIC pore: between the 0′ and 2′ residues or close to the 9′ residue. Data from the T2′S mutant receptor, which revealed a 50-fold change in potency, support an interaction at the lower position. Nevertheless, lindane could potentially bind at both these positions; the 9′ location is similar to that identified for binding bromo-lidocaine, which has been cocrystallized with GLIC (9), whereas the 0-2′ location is equivalent to the picrotoxin binding site in GluCl (7) and other Cys-loop receptors (16–22). It is also possible that the 9′ binding site may be a position at which lindane interacts as it descends into the channel on its way to the 0′-2′ binding site. Some support for multiple binding sites comes from another GLIC docking study that showed that the thiopental docked at three possible positions within the GLIC channel: close to the 2′, between the 2′ and 9′ residues, and between the 9′ and 16′ residues, in addition to sites within the extracellular domain and at the extracellular-transmembrane domain interface (11).
Other compounds that were relatively potent (IC50s = 1–10 μM) at inhibiting GLIC responses were picrotoxinin, tetracaine, and rimantadine. Picrotoxinin, the most active component of picrotoxin, is a classic GABAA receptor channel blocker, and also inhibits a range of other Cys-loop receptors (23–26); thus, an inhibitory effect on GLIC was not unexpected. In GABAA, glycine, and 5-HT3 receptors, mutagenesis and docking data suggest picrotoxinin interacts with the 2′ and 6′ channel residues, whereas in the GluCl receptor, structural data show the 2′ and -2′ residues are the most critical (16–22). In GLIC, our docking data show picrotoxinin is located between the 0′ and 6′ residues, with important interactions with the 2′Thr residues from several subunits; this location was supported by data from the T2′S GLIC receptor, whose picrotoxinin IC50 was reduced 65-fold (Table 2). Thus, we suggest that picrotoxin binds similarly in GluCl and GLIC receptors at the intracellular end of the pore.
Tetracaine has a number of potential binding sites in both Cys-loop receptors and other proteins, but one site is the pore of nACh receptors (27,28). Our data show it is almost equipotent with picrotoxinin at inhibiting GLIC function, and the docking data suggest it binds to a site in the channel that overlaps with that of picrotoxinin, but is located slightly higher in the pore. This is a similar location to that identified for bromo-lidocaine: the crystal structure data reveal density thought to correspond to the bromine atom near 9′Ile, which places the positively charged portion of the molecule near 6′Ser (9). This location of the positively charged portion of lidocaine is similar to the location of the charged quaternary ammonium analogs tetrabutylantimony and tetraethylarsonium shown by anomalous difference densities (9), and is consistent with the placement of the quaternary ammonium of tetracaine in our docking study.
The primary pharmacological target of rimantadine is the proton channel of the influenza A virus (A/M2), but it also blocks nACh receptor function (29,30). Structures have revealed the pore residues involved in binding rimantadine in the A/M2 pore are Val27, Ala30, Ser31, and Gly34 (31). The most critical of these may be Ser31, as 99.9% of the admantane-resistant viruses collected worldwide have an S31N mutation (32). The pore structures of Cys-loop receptors and A/M2 are quite distinct, but both are α-helical, and it may be that the role of Ser31 in A/M2 is performed by the 2′Thr residues in GLIC and Cys-loop receptor pores. Some support for this hypothesis comes from data from the T2′S GLIC receptor, which showed no change in rimantadine potency. This could indicate that the conserved residues Ser and Thr are able to interact equally efficiently with rimantadine. Alternatively, the docking data may not indicate the correct location of rimantadine. Further experiments are required to distinguish between these possibilities.
Compounds that blocked GLIC responses less potently (IC50s 100–1000 μM) were chlorpromazine, dieldrin, fibronil, α-endosulfan, mefloquine, diltiazem, estrone, bilobalide, and amantadine. These are compounds that block a range of Cys-loop receptors: nACh (chlorpromazine and admantane); 5-HT3 (mefloquine, dlitiazem, bilobalide, and estrone); GABAA (dieldrin, fibronil, and α-endosulfan) and glycine receptors (bilobalide) (13,16,18,20,30,33–41). Thus, the blocking actions of these compounds in GLIC show that the GLIC channel has pharmacological similarity to those of vertebrate Cys-loop receptors.
The pore-lining M2 region of GLIC has considerable sequence similarity with some nACh receptor subunits (e.g., >60% for α1, Fig. 1), consistent with GLIC being a cation-selective channel, although the intracellular Glu is in the −2′ rather than the −1′ position (Fig. 1). The 2′, 6′, and 9′ residues of GLIC are conserved with those of the nACh receptor, and also many other Cys-loop receptor subunits. As it is these residues that are involved in the binding of many channel blocking ligands, it explains why a number of Cys-loop channel blockers also block the GLIC pore. Our data therefore suggest that GLIC is a good template for modeling not only the nACh receptor pore, but also those of other Cys-loop receptors. There is some controversy as to whether the structure of GLIC, which was obtained at acidic pH, represents an open or a closed, desensitized state. Early studies (2–4) suggested that the channel did not desensitize at low pH, but more-recent data indicate that it does (5,6). Nevertheless, a Brownian dynamics simulation study has indicated that the GLIC crystal structure we used for docking (3EAM) is conductive (42). Interestingly, the 3EAM structure reveals a bundle of detergent molecules in the pore. It is unlikely that these significantly distort the structure, as bulkier detergent analogs appear to leave the pore structure unchanged, and molecular dynamics simulations in the absence of detergent show the structure to be stable (4). It is possible, however, that these molecules help stabilize the pore in a more open-like conformation.
In conclusion, we have identified a range of compounds that inhibit the function of GLIC. Many of these compounds also inhibit the function of Cys-loop receptors, and have been shown by mutagenesis to bind in the pore. Thus, although the pharmacological profile of GLIC is quite distinct from any of these receptors, we consider that the pore of GLIC has many similarities to its vertebrate counterparts, and therefore provides a useful model for examining structural interactions.
Acknowledgments
We thank David Weston for preliminary experiments and Lu Zhou for expert technical assistance.
S.C.R.L. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Studies. A.J.T., K.L.P., and S.C.R.L. are funded by the Wellcome Trust. M.A. is funded by a Yousef Jameel Scholarship. H.G.B. is supported by the Deutsche Forschungsgemeinschaft (grant BR 1507/4).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Tasneem A., Iyer L.M., Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bocquet N., Prado de Carvalho L., Corringer P.J. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007;445:116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- 3.Hilf R.J., Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 4.Bocquet N., Nury H., Corringer P.J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 5.Parikh R.B., Bali M., Akabas M.H. Structure of the M2 transmembrane segment of GLIC, a prokaryotic Cys loop receptor homologue from Gloeobacter violaceus, probed by substituted cysteine accessibility. J. Biol. Chem. 2011;286:14098–14109. doi: 10.1074/jbc.M111.221895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Gutierrez G., Grosman C. Bridging the gap between structural models of nicotinic receptor superfamily ion channels and their corresponding functional states. J. Mol. Biol. 2010;403:693–705. doi: 10.1016/j.jmb.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hibbs R.E., Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng Y., Yang L., Sonner J.M. Anesthetic sensitivity of the Gloeobacter violaceus proton-gated ion channel. Anesth. Analg. 2010;110:59–63. doi: 10.1213/ANE.0b013e3181c4bc69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilf R.J., Bertozzi C., Dutzler R. Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat. Struct. Mol. Biol. 2010;17:1330–1336. doi: 10.1038/nsmb.1933. [DOI] [PubMed] [Google Scholar]
- 10.Nury H., Van Renterghem C., Corringer P.J. X-ray structures of general anesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q., Cheng M.H., Tang P. Anesthetic binding in a pentameric ligand-gated ion channel: GLIC. Biophys. J. 2010;99:1801–1809. doi: 10.1016/j.bpj.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard R.J., Murail S., Harris R.A. Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. USA. 2011;108:12149–12154. doi: 10.1073/pnas.1104480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maskell P.D., Wafford K.A., Bermudez I. Effects of γ-HCH and δ-HCH on human recombinant GABAA receptors: dependence on GABAA receptor subunit combination. Br. J. Pharmacol. 2001;132:205–212. doi: 10.1038/sj.bjp.0703824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vale C., Damgaard I., Schousboe A. Cytotoxic action of lindane in neocortical GABAergic neurons is primarily mediated by interaction with flunitrazepam-sensitive GABAA receptors. J. Neurosci. Res. 1998;52:276–285. doi: 10.1002/(SICI)1097-4547(19980501)52:3<276::AID-JNR4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 15.Suñol C., Vale C., Rodríguez-Farré E. Polychlorocycloalkane insecticide action on GABA-and glycine-dependent chloride flux. Neurotoxicology. 1998;19:573–580. [PubMed] [Google Scholar]
- 16.Chen L., Durkin K.A., Casida J.E. Structural model for γ-aminobutyric acid receptor noncompetitive antagonist binding: widely diverse structures fit the same site. Proc. Natl. Acad. Sci. USA. 2006;103:5185–5190. doi: 10.1073/pnas.0600370103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reference deleted in proof.
- 18.Thompson A.J., Duke R.K., Lummis S.C. Binding sites for bilobalide, diltiazem, ginkgolide, and picrotoxinin at the 5-HT3 receptor. Mol. Pharmacol. 2011;80:183–190. doi: 10.1124/mol.111.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z., Cromer B.A., Lynch J.W. A proposed structural basis for picrotoxinin and picrotin binding in the glycine receptor pore. J. Neurochem. 2007;103:580–589. doi: 10.1111/j.1471-4159.2007.04850.x. [DOI] [PubMed] [Google Scholar]
- 20.Ffrench-Constant R.H., Rocheleau T.A., Chalmers A.E. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature. 1993;363:449–451. doi: 10.1038/363449a0. [DOI] [PubMed] [Google Scholar]
- 21.Buhr A., Wagner C., Sigel E. Two novel residues in M2 of the γ-aminobutyric acid type A receptor affecting gating by GABA and picrotoxin affinity. J. Biol. Chem. 2001;276:7775–7781. doi: 10.1074/jbc.M008907200. [DOI] [PubMed] [Google Scholar]
- 22.Xu M., Covey D.F., Akabas M.H. Interaction of picrotoxin with GABAA receptor channel-lining residues probed in cysteine mutants. Biophys. J. 1995;69:1858–1867. doi: 10.1016/S0006-3495(95)80056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erkkila B.E., Weiss D.S., Wotring V.E. Picrotoxin-mediated antagonism of α3β4 and α7 acetylcholine receptors. Neuroreport. 2004;15:1969–1973. doi: 10.1097/00001756-200408260-00027. [DOI] [PubMed] [Google Scholar]
- 24.Erkkila B.E., Sedelnikova A.V., Weiss D.S. Stoichiometric pore mutations of the GABAAR reveal a pattern of hydrogen bonding with picrotoxin. Biophys. J. 2008;94:4299–4306. doi: 10.1529/biophysj.107.118455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen R.W. Drug interactions at the GABA receptor-ionophore complex. Annu. Rev. Pharmacol. Toxicol. 1982;22:245–277. doi: 10.1146/annurev.pa.22.040182.001333. [DOI] [PubMed] [Google Scholar]
- 26.Thompson A.J., Lester H.A., Lummis S.C. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 27.Gallagher M.J., Cohen J.B. Identification of amino acids of the torpedo nicotinic acetylcholine receptor contributing to the binding site for the noncompetitive antagonist [3H]tetracaine. Mol. Pharmacol. 1999;56:300–307. doi: 10.1124/mol.56.2.300. [DOI] [PubMed] [Google Scholar]
- 28.Middleton R.E., Strnad N.P., Cohen J.B. Photoaffinity labeling the torpedo nicotinic acetylcholine receptor with [3H]tetracaine, a nondesensitizing noncompetitive antagonist. Mol. Pharmacol. 1999;56:290–299. doi: 10.1124/mol.56.2.290. [DOI] [PubMed] [Google Scholar]
- 29.Buisson B., Bertrand D. Open-channel blockers at the human α4β2 neuronal nicotinic acetylcholine receptor. Mol. Pharmacol. 1998;53:555–563. doi: 10.1124/mol.53.3.555. [DOI] [PubMed] [Google Scholar]
- 30.Matsubayashi H., Swanson K.L., Albuquerque E.X. Amantadine inhibits nicotinic acetylcholine receptor function in hippocampal neurons. J. Pharmacol. Exp. Ther. 1997;281:834–844. [PubMed] [Google Scholar]
- 31.Rosenberg M.R., Casarotto M.G. Coexistence of two adamantane binding sites in the influenza A M2 ion channel. Proc. Natl. Acad. Sci. USA. 2010;107:13866–13871. doi: 10.1073/pnas.1002051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bright R.A., Medina M.J., Klimov A.I. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 33.Arias H.R., Bhumireddy P., Bouzat C. Molecular mechanisms and binding site locations for noncompetitive antagonists of nicotinic acetylcholine receptors. Int. J. Biochem. Cell Biol. 2006;38:1254–1276. doi: 10.1016/j.biocel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Chiara D.C., Hamouda A.K., Cohen J.B. [3H]chlorpromazine photolabeling of the torpedo nicotinic acetylcholine receptor identifies two state-dependent binding sites in the ion channel. Biochemistry. 2009;48:10066–10077. doi: 10.1021/bi901271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giraudat J., Dennis M., Changeux J.P. Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: serine-262 of the δ subunit is labeled by [3H]chlorpromazine. Proc. Natl. Acad. Sci. USA. 1986;83:2719–2723. doi: 10.1073/pnas.83.8.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawthorne R., Cromer B.A., Lynch J.W. Molecular determinants of ginkgolide binding in the glycine receptor pore. J. Neurochem. 2006;98:395–407. doi: 10.1111/j.1471-4159.2006.03875.x. [DOI] [PubMed] [Google Scholar]
- 37.Huang S.H., Duke R.K., Johnston G.A. Bilobalide, a sesquiterpene trilactone from Ginkgo biloba, is an antagonist at recombinant α1β2γ2L GABAA receptors. Eur. J. Pharmacol. 2003;464:1–8. doi: 10.1016/s0014-2999(03)01344-x. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda T., Nagata K., Narahashi T. Dieldrin and picrotoxinin modulation of GABAA receptor single channels. Neuroreport. 1998;9:3189–3195. [PubMed] [Google Scholar]
- 39.Li P., Akk G. The insecticide fipronil and its metabolite fipronil sulphone inhibit the rat α1β2γ2L GABAA receptor. Br. J. Pharmacol. 2008;155:783–794. doi: 10.1038/bjp.2008.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revah F., Galzi J.L., Changeux J.P. The noncompetitive blocker [3H]chlorpromazine labels three amino acids of the acetylcholine receptor γ-subunit: implications for the α-helical organization of regions MII and for the structure of the ion channel. Proc. Natl. Acad. Sci. USA. 1990;87:4675–4679. doi: 10.1073/pnas.87.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson A.J., Lummis S.C. Antimalarial drugs inhibit human 5-HT3 and GABAA but not GABAC receptors. Br. J. Pharmacol. 2008;153:1686–1696. doi: 10.1038/bjp.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song C., Corry B. Ion conduction in ligand-gated ion channels: Brownian dynamics studies of four recent crystal structures. Biophys. J. 2010;98:404–411. doi: 10.1016/j.bpj.2009.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]


