Summary
RNA interference is a powerful tool for studying gene function, however, the reproducible generation of RNAi transgenic mice remains a significant limitation. By combining optimized fluorescence-coupled miR30-based shRNAs with high efficiency ES cell targeting, we developed a fast, scalable pipeline for the production of shRNA transgenic mice. Using this system, we generated eight tet-regulated shRNA transgenic lines targeting Firefly and Renilla luciferases, Oct4 and tumor suppressors p53, p16INK4a, p19ARF and APC and demonstrate potent gene silencing and GFP-tracked knockdown in a broad range of tissues in vivo. Further, using an shRNA targeting APC, we illustrate how this approach can identify predicted phenotypes and also unknown functions for a well-studied gene. In addition, through regulated gene silencing we validate APC/Wnt and p19ARF as potential therapeutic targets in T cell acute lymphoblastic leukemia/lymphoma and lung adenocarcinoma, respectively. This system provides a cost-effective and scalable platform for the production of RNAi transgenic mice targeting any mammalian gene.
Introduction
Genetically engineered mice are invaluable tools for understanding biology and disease. Traditionally, manipulation of gene expression in the mouse has relied on either the overexpression of cDNAs or modification (deletion/knock-in) of endogenous genes using homologous recombination (Capecchi, 2005). However, these approaches require a high investment to produce functional genetically-engineered mice. The development of site-specific recombination systems has added versatility to these mouse-modeling strategies, allowing for spatial and temporal control of gene activation or inactivation. Still, such approaches are not reversible and remain time-consuming and expensive.
RNA interference (RNAi) exploits an endogenous mechanism of gene regulation that can be adapted to suppress gene expression in vivo (Hemann et al., 2003), and in principle provides a fast and inexpensive alternative to conventional knockout approaches. Potent RNAi-mediated gene silencing can be induced by short hairpin RNAs (shRNAs) embedded within a natural microRNA (miRNA) backbone (Dickins et al., 2005). Importantly, in contrast to traditional gene targeting, shRNA expression systems act without modifying the genomic locus of the target and are, as such, reversible.
Several reports document that shRNA transgenic mice can be used to study gene function in vivo (Dickins et al., 2007; Seibler et al., 2007), although currently this approach is not widely used because the reliable generation of such animals remains a significant technical challenge. Traditional transgenesis by pronuclear DNA injection or viral transduction of zygotes or embryonic stem (ES) cells has been used (Dickins et al., 2007; Rubinson et al., 2003; Ventura et al., 2004), but generally give rise to animals with highly variable knockdown. Gene targeting by homologous recombination overcomes these drawbacks but relies on rare events.
Here, we set out to address and optimize virtually all aspects of producing shRNA-transgenic mice with the purpose of creating a rapid, flexible and scalable platform to reduce the time, cost and variability associated with developing such strains. We reasoned that an ideal system would provide potent and regulatable gene silencing, trackable shRNA expression, be economical, high throughput and adaptable to allow constitutive and tissue-specific gene knockdown. To achieve these goals, we combined optimized fluorescence-coupled shRNA technology with an Flp/FRT recombinase-mediated cassette exchange (RMCE) strategy (Hochedlinger et al., 2005) to generate a system for the production of single-copy, tet-regulatable shRNA transgenic ES cells and mice. This system is modular, flexible and includes: (i) a targeting construct developed as a recipient vector for any miR30-based shRNA and (ii) an expression cassette whereby shRNA expression is linked to a fluorescent reporter, enabling easy tracking and isolation of cells. We show that this system can be used to efficiently induce gene silencing in ES cells, ex vivo cultured cells and embryonic and adult tissues of the mouse, as well as reveal known and novel phenotypes and validate therapeutic targets in vivo. Together, this platform provides both a high throughput and cost effective way to spatially, temporally and reversibly control endogenous gene expression in mice.
Results
Generation of a targeting construct
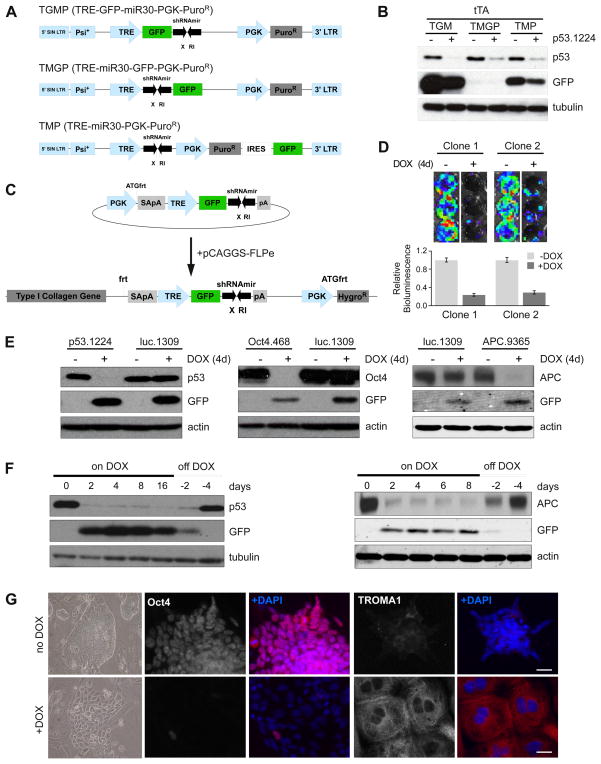
To develop a versatile shRNA targeting construct, we sought to construct an inducible, miR30-based expression cassette containing a fluorescent reporter of shRNA expression. We modified the retroviral doxycycline (DOX) inducible shRNA vector TMP, (Dickins et al., 2007) targeting the tumor suppressor Trp53 by inserting GFP either upstream (TRE-GFP-miR30-pgkPuro – TGMP) or downstream (TRE-miR30-GFP-pgkPuro – TMGP) of the miR30 cassette (Figure 1A). The presence of a GFP ‘spacer’ between the TRE promoter and the shRNA (TGM) not only allowed robust expression of GFP to serve as a reporter for shRNA expression but also enhanced p53 knockdown in comparison to the TMP and TMGP vectors (Figure 1B) (Stegmeier et al., 2005).
Figure 1. TRE-driven shRNAmirs targeted to the ColA1 locus can drive robust DOX-dependent gene knockdown in ES cells.
(A) Schematic diagram comparing the TGMP, TMGP and TMP retroviral vectors.
(B) Western blot analysis of p53.1224 infected tTA-expressing MEFs using TGMP, TMGP and TMP retroviral vectors.
(C) Schematic diagram of the pColTGM vector. Coelectroporation of pColTGM and pCAGS-Flpe recombinase promotes integration and confers hygromycin resistance.
(D) (Top) Representative luminescent images of two independent clones containing luc.1309/R26-rtTA. Cells were infected with MSCV-Luciferase and 48h post- infection cultured with or without DOX for 4 days. (Bottom) Quantification of luciferase activity. Error bars are SEM, n=2.
(E) Western blot analyses of DOX-treated ES cell clones containing R26-rtTA and luc.1309, p53.1224, Oct4.468 or APC.9365.
(F) Western blot analyses of p53.1224 and APC.9365 clones treated with DOX as indicated and then shifted to normal media for 2 or 4 days prior to harvest.
(G) Brightfield and immunofluorescence images of Oct4.468/R26-rtTA cells treated with DOX for 7d. Cells were stained with antibodies as indicated.
See also Figure S1
To enable efficient generation of targeted ES cells harboring a single DOX-regulatable GFP-shRNA, we adapted the ColA1 Flp/FRT recombinase-mediated targeting system previously described by Jaenisch and colleagues (Beard et al., 2006). Pre-engineered ‘KH2’ ES cells contain an FRT-hygro-pA “homing” cassette downstream of the ColA1 gene (hereafter referred to as CHC), as well as a reverse tet-transactivator (rtTA) targeted to the Rosa26 locus (referred to as R26-rtTA). Following coelectroporation of pCAGs-FLPe and an appropriate targeting vector, FLPe-mediated recombination between the FRT site at the ColA1 locus and those present on the targeting vector (Beard et al., 2006) (outlined in Figure S1A) confers hygromycin resistance only if correctly integrated. The highly stringent combination of site directed recombination and drug selection selects for precise targeting and from our experience, reduces the number of clones that need to be screened by 50-100 fold.
To generate the targeting construct, we modified the pBS31-ColA1 vector (Beard et al., 2006) to facilitate cloning of shRNAs into the TRE-GFP-miR30 (TGM) design (Figure 1C). Using this targeting vector, ColA1-TGM Flp-In (hereafter referred to as pCol-TGM), nearly every shRNA from the Hannon-Elledge genome-wide library (Silva et al., 2005) or RNAi codex library (Olson et al., 2006) can be inserted into the miR30 backbone in a single cloning step. Following ES cell electroporation, hygro selection and expansion of individual clones, cells were screened for DOX-dependent GFP expression (outlined in Table S1). Approximately 100% of dual-selected, hygromycin-resistant-GFP positive clones showed correct targeting by Southern blot (Figure S1B). By contrast, transgenic cells derived by pronuclear injection typically showed several integrations (Figure S1C, data not shown). We determined it was necessary to screen only 4 clones for each transfected shRNA, allowing targeting of many different shRNAs simultaneously with minimal effort.
ColA1 targeted shRNAs enable potent and reversible knockdown in ES cells
Using our workflow (Table S1), we generated several ES cell lines harboring an shRNA targeting the firefly bioluminescence reporter gene, luciferase (TG-luc.1309), to serve as a control. We also produced ES clones with shRNAs targeting a number of endogenous genes, including the pluripotency transcriptional regulator, Pou5f1 (TG-Oct4.468) and the tumor suppressor genes Trp53 (TG-p53.1224), Adenomatous Polyposis Coli (TG-APC.9365 and TG-APC.3374), p19ARF (TG-p19.157), and the transcript that encodes both p16INK4a and p19ARF (TG-p16/p19.478).
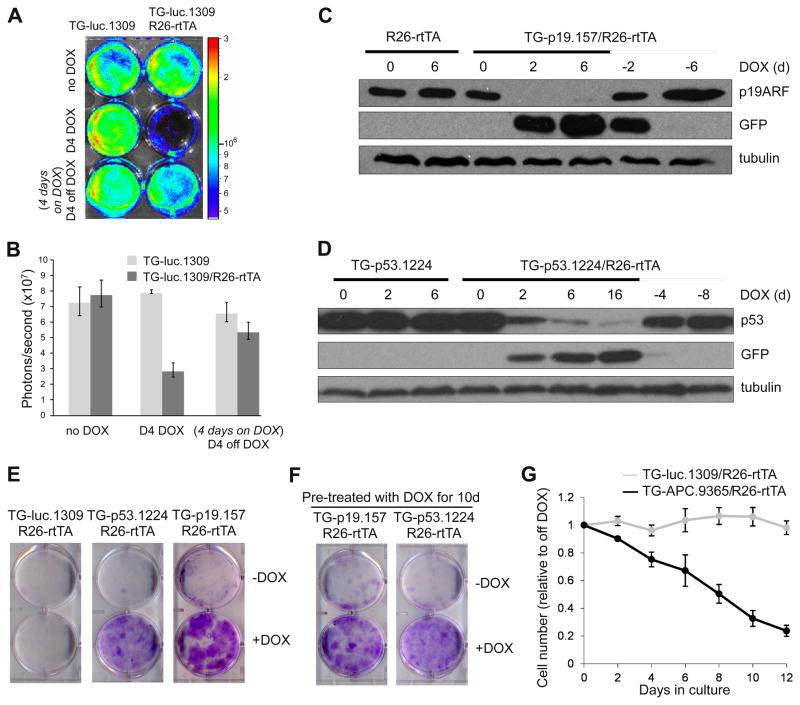
To validate the effectiveness of single-copy shRNAs targeted to the ColA1 locus, we confirmed knockdown in 5 independently selected TG-luc.1309/R26-rtTA ES cell clones transduced with a luciferase-expressing retrovirus by bioluminescence imaging (two representative clones shown in Figure 1D). In addition, we measured strong suppression of endogenous protein expression in selected TG-p53.1224/R26-rtTA, TG-Oct4.468/R26-rtTA and TG-APC.9365/R26-rtTA ES cell clones after DOX treatment and this knockdown was fully reversible (Figure 1E,F). GFP expression was not detected in untreated samples but strongly correlated with gene knockdown (Figure 1E, S1D) and thus provides a visual biomarker of shRNA expression.
Knockdown of endogenous gene expression using RNAi was associated with dramatic phenotypes in ES cells. Specifically, TG-p53.1224/R26-rtTA cells showed marked resistance to the DNA damaging agent Adriamycin following p53 suppression by DOX addition (Figure S1E), whereas, TG-Oct4.468/R26-rtTA ES cells treated with DOX underwent differentiation toward a trophoblast lineage (Figure 1G and Figure S1F). These phenotypes precisely reflect what has been observed in cells harboring the respective gene deletions (Lowe et al., 1993; Nichols et al., 1998) and illustrate that expression of a single shRNA transgene at the ColA1 locus can induce significant gene knockdown and mimic loss of function phenotypes.
ColA1 targeted shRNAs enable potent and reversible gene suppression in vitro
We next generated fully ES cell-derived transgenic mice using tetraploid embryo complementation, an approach that reduces time and cost by eliminating the need to screen founder lines for germline transmission. To test the potency and reversibility of knockdown in each transgenic line, we isolated MEFs carrying the TG-shRNA and R26-rtTA alleles and measured target protein expression. As seen in ES cell clones, DOX treatment induced significant depletion of luciferase activity in double transgenic (TG-luc.1309/R26-rtTA), but not single transgenic (TG-luc.1309) MEFs (Figure 2A,B). Moreover, luciferase activity was recovered upon DOX withdrawal, indicating the knockdown was fully reversible. Similarly, in MEFs derived from each of the TG-p53.1224, TG-APC.9365, TG-p19.157 and TG-p16/p19.478 shRNA founder lines we observed substantial and reversible DOX-dependent protein knockdown (Figure 2C,D and Figure S2A,B). In TG-APC.9365 MEFs, gene silencing led to a significant and reversible induction of downstream signaling, as measured by expression of the Wnt target gene Axin2 (Figure S2C).
Figure 2. Reversible knockdown of gene targets in primary mouse embryonic fibroblasts derived from ColA1-TGM ES-cell derived mice.
(A) Bioluminescence imaging of luc.1309 and luc.1309;R26-rtTA MEFs infected with MSCV-luciferase. MEFs were treated with DOX, then shifted into DOX-free media as indicated.
(B) Quantification of the bioluminescence signal to assess luciferase knockdown. Error bars represent SEM, n=2.
(C, D) Western blot analysis of MEFs harvested from a cross between C57BL/6 WT mice and p19.157;R26-rtTA or p53.1224;R26-rtTA founder mice, treated with DOX as indicated.
(E) Colony formation assay of R26-rtTA MEFs containing either luc.1309, p53.1224 or p19.157 plated at low density and cultured with or without DOX for 12 days.
(F) MEFs pre-treated with DOX for 10d were plated at low density and cultured with or without DOX for 12 days.
(G) Proliferation of DOX-treated TG-luc.1309;R26-rtTA and TG-APC.9365;R26-rtTA MEFs relative to off-DOX populations. Error bars represent SEM, n=4.
See also Figure S2.
Deletion of several of the genes targeted using transgenic shRNAs affects proliferation in MEFs. Like the corresponding null mutants, suppression of either p53 or p19ARF promoted immortalization as demonstrated by their ability to form colonies when plated at low density (Figure 2E) (Lundberg et al., 2000). Additionally, TG-p53.1224/R26-rtTA and TG-p19.157/R26-rtTA MEFs pretreated with DOX lost their ability to form colonies when removed from DOX-containing media, indicating that DOX withdrawal restores tumor suppressor function and reverts the immortalized phenotype (Figure 2F). Conversely, induction of APC.9365 in MEFs caused a progressive proliferation arrest (Figure 2G) consistent with what has been described following activation of Wnt signaling in MEFs via overexpression of a stable mutant β-catenin (Damalas et al., 2001). As has been noted for cells harboring gene deletions, suppression of p19ARF by RNAi enabled the rapid expansion of isolated pre-B cells in vitro (data not shown), and suppression of p53 dramatically enhanced colony formation of isolated hepatic stellate cells (V. Krizhanovsky, P.P., and S.W.L., unpublished observations). Thus, cells derived from distinct tissues show consistent, efficient and reversible gene knockdown that is associated with predicted phenotypic changes.
Transgenic shRNAs do not impair normal microRNA processing
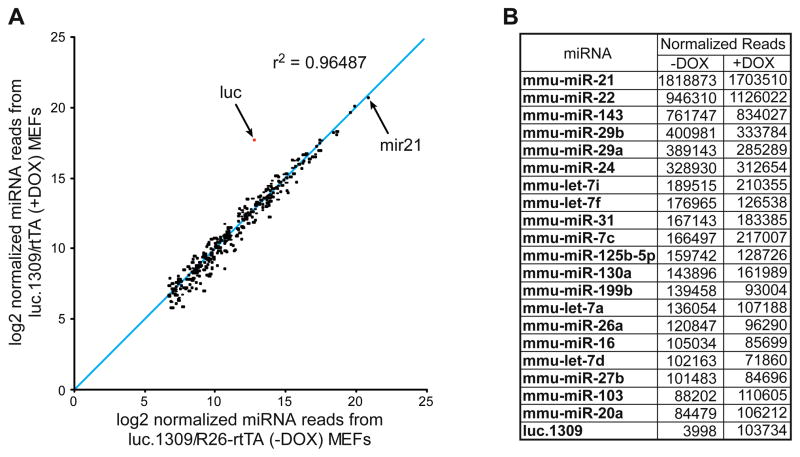
Studies suggest that high levels of shRNA expression can cause saturation of the miRNA processing machinery (Grimm et al., 2006). To determine whether shRNA expression in our system disrupts endogenous miRNA processing, we measured the abundance of all mature miRNA species using high-throughput sequencing. Small RNA libraries (Malone et al., 2009) prepared from single (TG-luc.1309) and bi-transgenic (TG-luc.1309/R26-rtTA) MEFs showed few or no changes in the expression or relative proportions of endogenous miRNAs. (Figure 3 and Figure S3). As expected, we observed a substantial (25-fold) DOX-dependent induction of the synthetic luc.1309 mature miRNA species (Figure 3). We also noted a basal level of luc.1309 expression in the absence of DOX and rtTA, reflecting low-level DOX- and rtTA-independent leakiness from the TRE promoter in MEFs that is cell type specific (data not shown). Importantly, this potential leakiness does not cause significant gene knockdown in TG-p53.1224, TG-p19.157, TG-p16/p19.478 or TG-APC.9365 MEFs (Figure 2C,D and Figure S2). These results are consistent with reports that miR30 based shRNAs show reduced general toxicity compared to other RNAi triggers (McBride et al., 2008) and demonstrate that DOX-regulated shRNAs expressed from the ColA1 locus allow reversible silencing of gene expression without disrupting endogenous miRNA processing.
Figure 3. Endogenous miRNA levels are unaffected by exogenous shRNA induction.
(A) Scatter plot representing the normalized expression of the 319 most abundant miRNAs from DOX-treated and untreated TG-luc.1309/R26-rtTA MEFs. R2 correlation coefficient excludes TG-luc.1309 sequence reads.
(B) Number of normalized reads obtained for the top 20 miRNAs and luc.1309 expressed in MEFs.
See also Figure S3.
ColA1 targeted shRNAs enable potent and reversible gene suppression in vivo
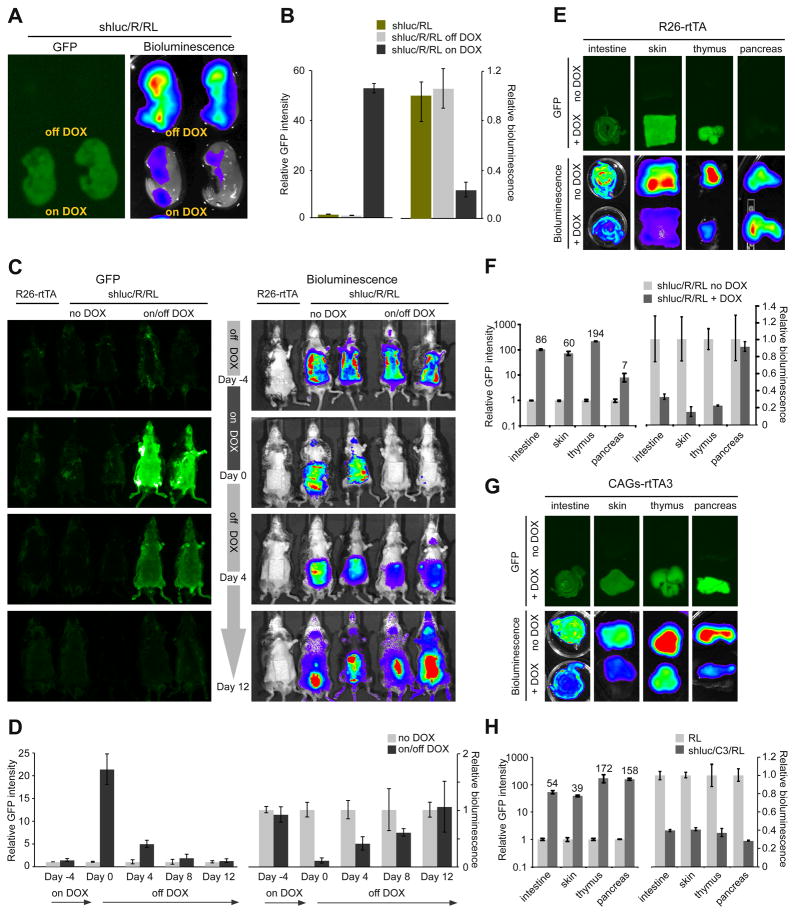
To evaluate the dynamics of reversible gene silencing in vivo, we bred mice that express luciferase from the endogenous Rosa26 promoter to TG-luc.1309/R26-rtTA mice to produce triple transgenics (TG-luc.1309/R26-rtTA/R26-luciferase, or shluc/R/RL) and littermate controls (shluc/RL). Rosa26 expression is reportedly high in embryonic tissues (Soriano, 1999), therefore, we first tested knockdown in E17.5 embryos. Triple transgenic embryos showed marked induction of GFP expression and ~75% knockdown of luciferase relative to controls (Figure 4A,B), consistent with the maximal level of knockdown we observed in ES cells and transgenic MEFs with this shRNA (Figure 1D, 2A,B).
Figure 4. Reversible GFP-marked DOX-dependent knockdown in live animals.
(A) GFP and bioluminescent images of E17.5 TG-luc.1309;R26-rtTA;Rosa-Luciferase (shluc/R/RL) embryos sacrificed from pregnant females treated with or without DOX.
(B) Relative GFP intensity and (C) bioluminescence between shluc/RL and shluc/R/RL embryos. Error bars represent SEM, n=7.
(D) In vivo bioluminescent time course imaging of TG-luc.1309;R26-rtTA;Rosa-Luciferase (shluc/R/RL) triple transgenics and controls. Animals were treated with or without DOX for 4 days, then removed from treatment.
See also Figure S4
We next measured the dynamics of luciferase silencing longitudinally in the skin of triple transgenic mice. After 4 days of DOX-treatment, shluc/R/RL animals showed significant Luciferase knockdown (and high GFP expression) relative to untreated triple transgenic mice (Figure 4C,D). This suppression was reversible, with loss of GFP and increased Luciferase expression 4 days following DOX withdrawal and after 12 days Luciferase activity returned to basal levels. To evaluate knockdown in additional tissues we imaged individual organs for GFP and bioluminescence. Consistent with previous reports (Beard et al., 2006), we observed significant variation in the level of R26-rtTA-mediated GFP induction between different organs (Figure 4E,F; S4A,B). Importantly, tissues that displayed strong induction (>20 fold) of GFP expression (skin, intestine, thymus and liver) showed Luciferase knockdown approaching the maximal potency of this shRNA in vitro (Figure 4, Figure S4C).
A tet-transactivator line that enables efficient knockdown in a broad range of adult tissues
During the course of our in vivo analyses we noted that the presence of two R26-rtTA transgenic alleles increased the DOX-dependent expression of GFP-shRNAs relative to a single allele (not shown). This indicated that, not only was rtTA expression from the Rosa26 promoter tissue restricted (Figure S4), but it was limiting for maximal expression from the TRE promoter in many adult tissues. To address this limitation, we sought to develop a stronger, more ubiquitous, strain expressing a highly sensitive reverse tet-transactivator (rtTA3) downstream of the strong CAG promoter (Niwa et al., 1991). After characterizing a strain that contained a single genomic insertion of CAG-rtTA3 (data not shown), we generated triple transgenic mice carrying TG-luc.1309/CAG-rtTA3/R26-luciferase (shluc/C3/RL) and imaged individual organs for knockdown (Figure 4G; S4D). In contrast to shluc/R/RL mice, shluc/C3/RL triple transgenics showed robust induction of GFP in almost all tissues examined and, accordingly, tissues that expressed high levels of GFP displayed knockdown of Luciferase to levels consistent with the intrinsic potency of this shRNA (Figure 4G,H; S4D-F). These results establish CAG-rtTA3 as a broadly useful tet-transactivator strain and demonstrate that strong expression from the TRE promoter at the ColA1 locus is possible in almost all adult mouse tissues.
Studying gene function during embryonic and post-natal development: Reversible and irreversible consequences of hyperactive Wnt signaling
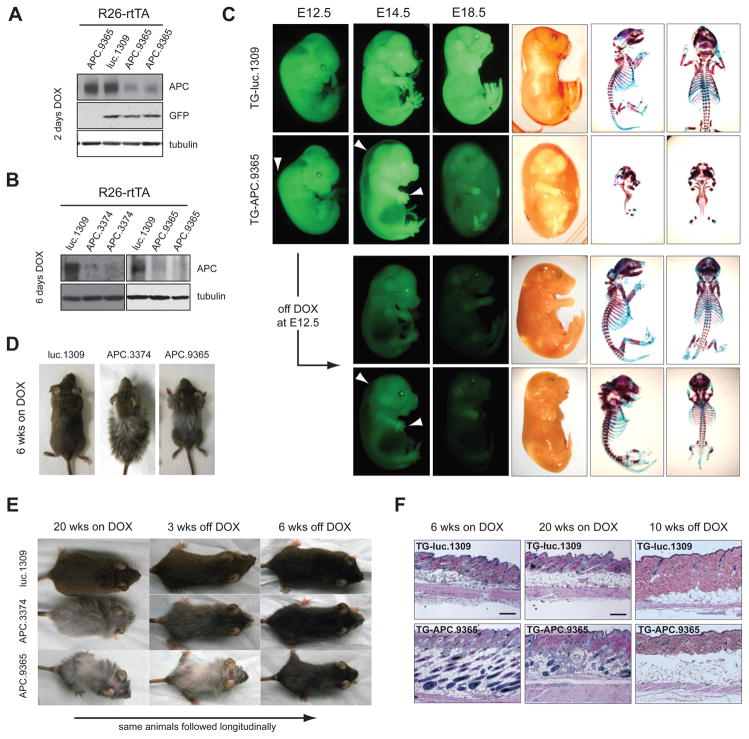
We reasoned that the inducibility and reversibility of gene silencing enabled by our system would facilitate the study of gene function during defined windows of development and disease, particularly for those genes whose inactivation results in embryonic lethality, such as APC (Moser et al., 1995). Similarly we observed embryonic lethality following early (E0.5) depletion of APC (data not shown). While conditional deletion strategies with various tissue specific Cre recombinases have enabled conditional deletion at later stages of development in specific tissues [see, for example, (Colnot et al., 2004; Kuraguchi et al., 2006; Sansom et al., 2004)] we examined the consequences of global APC knockdown at different developmental stages by simply varying the timing of DOX treatment (Figure 5A,B). Although untreated and TG-luc.1309/R26-rtTA animals developed normally (Figure 5C, not shown), TG-APC.9365/R26-rtTA bi-transgenic embryos DOX-treated at E8.5 displayed prominent hydrops fetalis (pronounced fetal edema) (Figure 5C, Figure S5A) and died prior to birth (n=14). At E14.5 and E18.5, DOX-treated embryos revealed a severe delay or absence in growth and endochondral ossification of multiple skeletal elements (spine, ribs and limbs) (Figure 5C, Figure S5B) identical to the phenotype recently reported using skeletal specific mutation of APC (Miclea et al., 2009).
Figure 5. APC loss disrupts embryonic and postnatal development.
(A) Western blot of whole protein lysates from E10.5 embryos on DOX for 2 days and(B) E14.5 embryos on DOX for 6 days.
(C) GFP and brightfield images of embryos treated with DOX from E8.5 or pulsed with DOX from E8.5–E12.5. Arrows indicate fluid accumulation along the dorsal ridge and defects in limb and digit development at E12.5 and E14.5. Alcian blue and Alizarin red stained skeletons from E18.5 embryos.
(D) Representative photographs of TG-luc.1309/R26-rtTA, TG-APC.3374/R26-rtTA and TG-APC.9365/R26-rtTA double transgenic mice on DOX for 6 weeks.
(E). Representative photographs of Luc.1309, APC.3374 and APC.9365 treated with DOX for 20 weeks and then removed from DOX-treatment for 6 weeks.
(F) H&E sections of skin taken from Luc.1309/R26-rtTA and TG-APC.9365/R26-rtTA double transgenic mice treated with DOX for 6 weeks, 20 weeks and 20 weeks on DOX/6 weeks off DOX as indicated. Scale bars are 100μm.
See also Figure S5.
A theoretical advantage of our system is the potential for interrogating gene/pathway function during defined developmental windows by controlling gene repression and reactivation. To test the feasibility of this approach, we studied the impact of transient APC suppression during embyogenesis. shAPC embryos pulse-treated with DOX for 4 days (E8.5 to E12.5) showed no edema and marked recovery of some bone growth (ribs, spine, skull), although both hind and forelimb development remained severely stunted (Figure 5C), especially when knockdown was driven by an additional R26-rtTA allele (Figure S5C). Interestingly, DOX-pulsed embryos also developed extra digits (total 5–9) that were largely unsegmented and appeared duplicated along their length (Figure S5D). These data imply that regulation of Wnt signaling by APC dictates the timed development and differentiation of different skeletal elements during embryogenesis and that even transient disruption to APC function can cause irreversible defects in tissue patterning.
To examine whether our system could be used to study post-natal development, we avoided the consequences of APC suppression in utero by delaying DOX treatment until near birth. Although shAPC (9365 or 3374) induction 1–2 days prior to birth was tolerated, it resulted in post-natal defects including stunted development of the snout and runting (Figure S5E, not shown), consistent with tissue restricted APC excision during mid-embryogenesis (Kuraguchi et al., 2006). Continued APC silencing for 6 weeks (initiated at birth or 4–6 weeks of age) promoted excessive hair growth (Figure 5D) while long-term DOX treatment beyond 20 weeks resulted in progressive hair loss and significant disruption to the normal structure of the hair follicles (Figure 5E,F). Interestingly, reactivation of APC after 4 weeks of DOX-treatment prevented eventual hair loss but could not revert the development of a stunted snout (data not shown). In contrast, dysregulation of hair follicle function was reversible, as re-expression of APC in largely hairless mice promoted rapid recovery of pelage (Figure 5E, S5F). In all, these data demonstrate that APC and the precise control of Wnt is crucial at many stages of mammalian embryonic and post-natal development and define both permanent and reversible consequences of APC loss. Such insights go beyond what could be obtained through conditional knockout approaches and, in principle, conceptually similar approaches could be applied to study any endogenous gene.
Studying gene function in disease initiation and maintenance: APC suppression is required for the initiation and maintenance of T-ALL
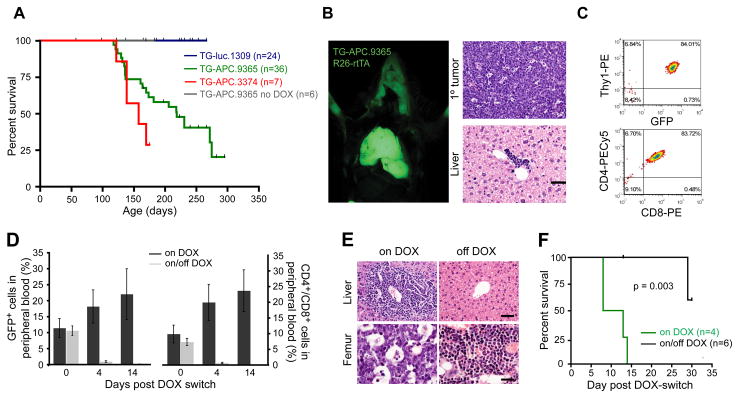
Perhaps one of the more interesting potential applications of inducible shRNA transgenics is to study disease associated with loss of gene function, where reversibility of gene suppression would determine whether restoring gene function would ameliorate disease symptoms and help pinpoint therapeutic targets. Activation of the Wnt pathway, often through deregulation of APC, is one of the most frequent events in human cancers (Clevers, 2006) and represents a potential target (Gunther et al., 2003). To explore this possibility, shAPC/R26-rtTA mice were maintained on DOX from 4–6 weeks of age, until they showed signs of disease. All moribund mice presented with lymphoma with a median survival of 7.4 (TG-APC.9365) and 5.3 (TG-APC.3374) months (Figure 6A,B). Primary leukemia/lymphomas were detected in the thymus and lymph nodes (Figure 6B, not shown). Additionally, secondary organs (e.g. liver) showed minor infiltration of lymphoblastic disease (Figure 6B) and were transplantable into secondary recipients (Figure 6 and S6). In all cases examined (7/7) leukemia/lymphomas expressed both CD4 and CD8 on their surface (Figure 6C), indicative of uncommitted T-lymphoid progenitors. Hence, the disease had many features of human T cell acute lymphoblastic leukemia/lymphoma (T-ALL), a disease in which Wnt signaling has recently been associated with (Yang et al., 2005).
Figure 6. APC loss is required for the maintenance of T lymphoblastic leukemia/lymphoma.
(A) Kaplan Meier analysis of R26-rtTA/TG-shRNA double transgenic mice treated continually with DOX from 4–6 weeks of age.
(B) Fluorescent image of TG-APC.9365/R26-rtTA animals with GFP positive tumor in the thymus. All analyzed animals that became moribund developed T lymphoblastic leukemia/lymphoma apparent in the thymus, lymph nodes (not shown) and liver. Scale bars: 50μm
(C) Representative flow cytometry plots of cells isolated from primary lymphomas and stained for surface markers: Thy1, CD4 and CD8
(D) GFP positive (left) and CD4/CD8 double positive cells in the peripheral blood of SCID mice transplanted with TG-APC.3374 lymphoma following the DOX-switch. Error bars = SEM.
(E) H&E stained sections of the liver (upper) and bone marrow (femur – lower) in mice on DOX or removed from DOX for 2 weeks (off DOX). Scale bars: 50μm (liver) and 20μm (femur).
(F) Kaplan Meier analysis of SCID mice transplanted with TG-APC.3374 primary lymphoma. Graph is plotted from the time of DOX-switch (5 weeks after transplant).
See also Figure S6
Previous studies using overexpressed transgenes suggest that hyperactive Wnt signaling is required for the maintenance of some murine cancers (Gunther et al., 2003). However, our shAPC transgenic enabled us, for the first time, to explore the role of APC suppression and physiological Wnt pathway deregulation in tumor maintenance. Five weeks post-transplant, when mice showed ~10% GFP-positive leukemia cells in the peripheral blood, animals were randomized into two cohorts and monitored for disease progression in the presence and absence of DOX (Figure 6D, Figure S6A). Animals that remained on DOX became moribund within 1–2 weeks, showing massive disease infiltration into secondary organs (Figure 6E, Figure S6B, not shown). In contrast, off-DOX animals showed a rapid clearance of GFP+ and CD4+/CD8+ cells from the peripheral blood and secondary organs (Figure 6D and E), leading to a significant survival benefit (Figure 6F, p=0.003). Strikingly, one animal that presented with lymphoma associated hind limb paralysis at 4 weeks, regained normal limb function only 3 days following DOX withdrawal (Movies S6A,B). Importantly, 2/5 animals showed relapse of lymphoblastic disease 4 weeks after DOX withdrawal. Such relapsed cancers were entirely GFP negative and showed strong re-expression of APC (Figure S6C), suggesting that the disease can eventually become independent of APC suppression. These data demonstrate that APC suppression is required for tumor maintenance, validate the Wnt pathway it controls as a therapeutic target in T-ALL and provide a system for studying resistance mechanisms to Wnt antagonists. More generally, they illustrate that the shRNA approach can effectively recapitulate disease etiology predicted from genetic mouse models and rapidly determine whether the pathology depends on this initiating lesion.
Studying genetic modifiers of disease: p19ARF suppression promotes the initiation and maintenance of KrasG12D-induced lung cancer
We envisioned that our shRNA transgenic platform would facilitate the analysis of genes that modulate disease onset and progression in a variety of pre-existing mouse models of human disease. As a proof-of-principle, we took advantage of a pre-existing mouse strain harboring a latent oncogene Kras (G12D) allele, in which the ras oncogene can be activated by intranasal injection of adenoviral Cre (adeno-Cre) leading to the development of lung adenocarcinoma (Jackson et al., 2001). Studies suggest that hypermethylation of the ARF tumor suppressor occurs in approximately 30% of human non-small lung cancers (Ozenne et al., 2010) and that gene deletions targeting ARF can cooperate with oncogenic Kras to induce lung adenocarcinoma in mice (Fisher et al., 2001). To test whether gene suppression of ARF using RNAi could produce similar results, we produced mice carrying 4 alleles: 1) TG-p19.157, 2) LSL-KrasG12D, 3) rCCSP-rtTA (rat Clara cell secretory protein) and 4) R26-LSL-luciferase. We also used an additional shRNA transgenic line targeting Renilla luciferase (TG-Ren.713) (Zuber et al., 2010), a control shRNA that does not repress the luciferase reporter.
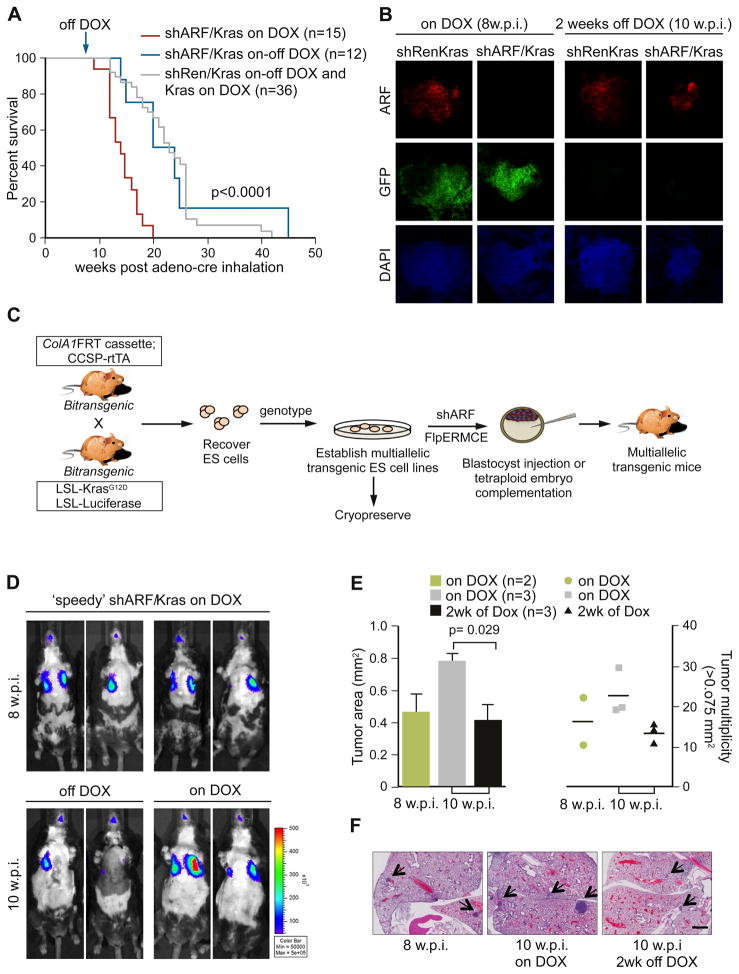
Transgenic mice harboring either TG-p19.157 or TG-Ren.713 and rCCSP-rtTA/LSL-KrasG12D/R26-LSL-luciferase (referred to as shARF/Kras or shRen/Kras respectively), were inoculated with adenoviral Cre and simultaneously treated with DOX to induce the p19ARF shRNA. As expected, luciferase imaging showed that shARF/Kras animals exhibited markedly increased tumor burden compared to shRen/Kras and Kras alone controls (Figure S7A) and, by 12–14 weeks post infection, shARF/Kras DOX-treated mice displayed rapid shallow breathing, cachexia and an increase in both size and number of tumor nodules in the lung (Figure S7B,C,D). Histopathological analyses revealed pulmonary neoplasias (Figure 7E) similar to those seen in p19ARF knockout models combined with KrasG12D activation (Fisher et al., 2001), while controls showed predominantly hyperplastic growth with rare non-invasive lung adenomas (data not shown). The increased tumor burden in shARF/KRas mice resulted in significantly reduced survival (p<0.0001) compared to shRen/Kras and Kras controls (Figure 7A). Thus, gene suppression using transgenic RNAi can recapitulate a null phenotype even in a complex multi-allelic system.
Figure 7. Reversible suppression of ARF in a multi-allele disease model documents a contribution to tumor maintenance.
(A) Kaplan Meier analysis of Kras, shRen/Kras and shARF/Kras treated with Adeno-Cre and DOX at 4-6 weeks. Cohorts were randomized and followed with or without DOX treatment, 8 w.p.i.
(B) Confocal microscopy images of 150μm sections from lungs of shRen/Kras and shARF/Kras animals on DOX at 8 w.p.i. or off DOX for 2 weeks (10 w.p.i.). Sections were stained with an antibody to detect ARF and DAPI. Scale bar = 100μm.
(C) The ‘speedy’ mouse workflow to derive and retarget multiallelic ES cell clones from intercrossed mice.
(D) Bioluminescence images of shARF/Kras mice produced by the ‘speedy’ approach (C) and treated with Adeno-Cre and DOX at 5 weeks of age. At 8 w.p.i., cohorts were randomized and followed with or without DOX treatment for 2 weeks.
(E) Quantification of lesions in the lungs of ‘speedy’ shARF/Kras mice at 8 and 10 w.p.i. For each animal, area and multiplicity represent the mean scored across 10 H&E stained sections taken at 100μm apart. Error bars represent SEM (left).
(F) Representative H&E stained sections of the lungs from mice in (D,E). Arrows mark lesions scored. Scale bar = 1mm.
See also Figure S7
As described above, a unique feature of our platform is the ability to manipulate gene expression after disease onset. In the case of shARF/Kras lung carcinomas, this feature allowed us to explore the potential of an experimental strategy currently undergoing clinical development. Drugs targeting DNA methyltransferases (DNMTI) and histone deacetylase inhibitors (HDACI) have been put forward as a therapeutic strategy to re-activate tumor suppressor genes in established tumors, and we reasoned the effects of ARF reactivation in lung carcinomas would point towards a subset of expected “on-target” effects of such agents. Therefore, shARF/Kras animals with detectable disease were withdrawn from DOX treatment. After two weeks, ARF reactivation coincided with a massive induction of cleaved-caspase 3 (Figure 7B; S7F) and measurable decrease in bioluminescent signal (Figure S7A), suggesting that a p19ARF-induced apoptotic response can promote tumor regression in KRasG12D driven lung cancer. Histological analyses showed clear tumor regression within DOX-removed animals (data not shown), which likely contributed to a significant survival benefit following ARF re-expression (Figure 7A, p<0.0001) compared to those animals continually treated with DOX (Figure 7A). However, DOX-removed shARF/Kras mice died at a similar time as Kras controls (Figure 7A), perhaps owing to a subset of mutant tumors that failed to reactivate ARF or acquired mutations in the rtTA allele. Consistent with the latter, upon sacrifice, some lung tumors were GFP positive, indicating continued expression of the shRNA. Nonetheless, these results are consistent with recent reports describing the effects of p53 reactivation in lung carcinomas (Feldser et al., 2010; Junttila et al., 2010), and imply that reversing ARF silencing in lung adenocarcinomas will have a therapeutic benefit. More broadly, they illustrate how transgenic RNAi can be effectively employed to control gene regulation in complex genetic models.
Accelerating the analysis of complex models of disease using conditional RNAi
The large volume of information emerging through genomic and genetic screens has implicated many new genes in human disease. While our shRNA transgenic platform provides one strategy to rapidly explore the biology of such genes in vivo, the time and cost associated with introducing each new shRNA transgenic to more complex genetic backgrounds remains a limitation. In an attempt to rapidly produce mice with multiple alleles, recent work documents the utility of producing mice with complex genotypes from ES cells engineered to harbor multiple genetically engineered alleles (Zhou et al., 2010). We sought to synthesize this strategy with our shRNA platform by exploring the feasibility of rederiving ES cells from mice harboring the shRNA targeting cassette together with other disease promoting alleles. We reasoned that this ‘speedy’ strategy (Figure 7C) would enable investigators to rapidly produce mice harboring any naturally-occurring or engineered mouse alleles with any inducible shRNA, thus bypassing slow and expensive breeding regimes, particularly when interested in the effects of more than one shRNA.
As a proof-of-principle using the lung adenocarcinoma model, we re-derived ES cell clones containing the four relevant alleles: LSL-KrasG12D, rCCSP-rtTA, R26-LSL-luciferase and the ColA1 FRT ‘homing cassette’ and targeted 2 clones with the p19.157 shRNA as previously described (Figure 1A). ES cell-derived mice and chimeras were successfully generated by tetraploid embryo complementation or injection into C57BL/6 albino host blastocysts and ‘speedy’ shARF/Kras mice were treated with adeno-Cre. All ‘speedy’ mice developed tumors detectable by bioluminescence imaging 8 w.p.i. (n=8) as anticipated and showed rapid tumor regression in response to ARF reactivation (Figure 7D). Additionally, histological examination revealed that tumor multiplicity and size was significantly attenuated following DOX removal (Figure 7E,F). These data precisely recapitulate the results produced through intercrossing but at a fraction of the time and cost. With the system now in place it can be used to examine the impact of virtually any gene on Kras-induced lung cancer development and regression without additional intercrossing. More generally, this approach can be adapted to study any gene in any disease state that can be modeled in the mouse, and is amenable to the simultaneous testing of many candidate disease-modulating shRNAs.
Discussion
Transgenic RNAi offers a flexible and systematic alternative to homologous recombination for studying gene function in vivo. While RNAi cannot replace gene targeting in creating precise genetic lesions, it enables fast, reproducible, and cost-effective generation of transgenic mouse models with unique capabilities for spatial, temporal, and reversible suppression of endogenous genes. Here we set out to optimize every step in the production and analysis of shRNA-transgenic mice by developing: 1) a system that enables efficient ES cell targeting, enhanced gene silencing, and fluorescence-coupled tracking of shRNA expression in vivo and 2) an approach to rapidly combine shRNAs with existing disease alleles. Importantly, the system is modular such that it can be modified to incorporate alternate reporters (eg. turboRFP, mCherry), tandem shRNA cassettes, or be adapted for controlled expression of additional non-coding RNA species (unpublished data). Additionally, we have recently developed a systematic and high-throughput screening methodology to identify the most efficient shRNAs targeting any given transcript (Fellmann et al., in press); these shRNAs can then be seamlessly incorporated into the production of shRNA transgenic mice. Together with the availability of optimized RNAi triggers, the system provides a high throughput platform that streamlines production of ES cells and transgenic mice harboring conditional shRNAs, enabling exquisite control of endogenous gene expression in vivo.
RNAi-mediated gene silencing does not produce a complete null situation and hence shRNA transgenics will not replace the need for conditional knockout mice, at least in some settings. Rather, we envision that this technology will provide a complimentary approach when speed and scale are desirable – a situation that may be increasing as biologists try to understand the avalanche of information emerging from various genome projects. We also envision that our system will enable the evaluation of hypomorphic phenotypes reflective of many human conditions and disease states and may be applicable to the examination of potential drug targets and in this context, the level and duration of gene suppression that can be tolerated in vivo. Here we demonstrate the strength of our system for investigating gene function in vitro and in vivo during multiple stages of embryonic and postnatal development and disease progression. Importantly, the ability to reversibly regulate gene expression allowed us to explore the consequences of transient APC loss during development and define a small window of development where APC is essential for regulating skeletal morphogenesis.
Using single and multi-allelic approaches, we also showed how shRNA transgenic mice can be used to accurately model human disease and, through gene reactivation, assess the potential of therapeutic targeting strategies. Indeed, the system provides the unique capability of genetically mimicking the action of potential drugs whose action would likely be transient and incomplete, enabling validation of drug targets and characterization of drug toxicities in a manner that has not previously been possible. Finally, further application of the multi-allelic ‘speedy’ approach should provide a relatively high throughput platform for in vivo characterization of candidate genes identified using shRNA-based screens or genome-wide association studies and will facilitate triaging and prioritizing drug targets in expensive drug development programs.
Despite the advances shown here, the limited scope and effectiveness of available tet-transactivator strains and the paucity of shRNAs that act efficiently at single copy represent two remaining barriers to achieving an ideal system. As a first step towards addressing the former issue, we developed a tet-transactivator strain (CAG-rtTA3) that provides stronger and more widespread expression of TRE-regulated shRNAs than any other strain we have tested, including R26-rtTA, Actin-rtTA and CMV-rtTA (data not shown). Although some organs, such as the brain, spleen and lung showed lower GFP expression and reduced luciferase knockdown (see Figure S4), we believe this is likely due to limited delivery of DOX to these cells (in the case of brain) or sub-optimal rtTA expression rather than inaccessibility of the targeted ColA1 locus. In fact, tissue-specific expression of tet-transactivators in CCSP-rtTA and Vav-tTA strains can promote effective ColA1-targeted shRNA expression in lung epithelium (Figure 7) and B-cells (data not shown), respectively. It will be important to determine which of the >100 existing tet-transactivator strains (http://www.zmg.uni-mainz.de) will provide sufficient tTA/rtTA expression for effective knockdown and to develop additional robust and versatile strains.
We believe that transgenic systems such as the one described here have the potential to transform mammalian genetics. Indeed, in lower organisms such as D. melanogaster and C. elegans, the application of in vivo RNAi on a broad scale has had an enormous impact on the ability of these models to provide insights into the physiology of whole organisms in isolation or in the context of other disease associated alleles. With similar capabilities now achievable in mice, it should be possible to rapidly explore all aspects of mammalian physiology and provide clues to the molecular control of normal development, aging, and disease. Accordingly, we have now implemented a 96-well targeting format to enable paralleled production of hundreds of ES cells and transgenic mice with inducible and reversible, RNAi-mediated gene silencing. With this pipeline it is now possible and indeed feasible, to develop single and multi-allelic shRNA transgenic animals to evaluate in parallel the function of many mammalian genes.
Experimental Procedures
Targeting Constructs and ES cell targeting
Details of targeting construct and shRNA cloning can be found in Supplementary information. Coelectroporation of 50 μg pCol-TGM with 25 μg pCAGs-FLPe at 400 V and 125 μF was performed using two pulses in a Gene PulserII (Bio-Rad, Hercules, CA). 36–48 h post-electroporation, GFP was measured by flow cytometry (Guava EasyCyte) to assess the electroporation efficiency. Only cultures with >10% GFP expression were maintained and selected in 140 μg/ml hygromycin. Clones were screened as indicated (Table S1).
Transgenic and ‘speedy’ mice
Rosa26-LSL-Luciferase females and Stra8-Cre males (Sadate-Ngatchou et al., 2008) were crossed to produce RL mice. DOX was administered to mice via food pellets (625mg/kg) from Harlan Teklad. The Cold Spring Harbor Animal Care and Use Committee approved all procedures described in this work. TG-shRNA transgenic mice were genotyped by genomic PCR using shRNA-specific forward primers and a common reverse primer (Table S2), yielding an ~250-bp product. CAG-rtTA3 mice were generated using pronuclear injection and genotyped by PCR. rtTA3 Southern blots were performed on genomic DNA digested with BamHI, using a probe covering the coding region of rtTA3. ‘Speedy’ mice were produced using ES cells rederived from E3.5 blastocysts (Bryja et al., 2006).
Alcian Blue and Alizarin Red skeletal staining
E14.5-E18.5 embryos were euthanized and all skin was removed. Animals were fixed in 95% Ethanol for 24hrs, transferred to 100% Acetone for 24hrs and stained in 0.015% Alcian Blue/0.005% Alizarin Red in 0.8M Acetic acid and 70% Ethanol for 24 hrs. Remaining soft tissue was digested in 1% KOH over 3 days, changing the solution daily and skeletons were gradually transferred into 100% glycerol.
Lymphoma transplants and monitoring
Primary lymphomas were mechanically disrupted and passed through a 40μM filter to a single cell suspension. 2 × 105 viable cells were injected intravenously into SCID mice pretreated with DOX for 24hrs. Peripheral blood was stained with antibodies to CD45 (CD45-APC (30-F11, BD Pharmigen), CD4-PECy5 (GK1.5, Biolegend) and CD8-PE (53-6.7, Biolegend) and flow cytometry was performed on a Guava HT EasyCyte and analyzed using InCyte software (Guava Technologies). At necropsy, single cell suspensions from spleen, bone marrow and peripheral LNs were analyzed by flow cytometry as described above.
Cell culture and expression analysis
ES cell cultures were selected and maintained on irradiated (40 Gy) MEFs derived from the DR4 mouse strain (Tucker et al., 1997). To test ES cells for luciferase knockdown, TG-luc.1309 ES cells were infected with MSCV-IRES-Luciferase and treated cells with 1μg/ml DOX-hyclate (Sigma). For Trp53 protein blots in Figures 1, 2 and Figure S1E, cells were treated with 0.5μg/ml Adriamycin (Bedford Laboratories, OH) for 4h to induce Trp53. To assess survival of ES cells following DNA damage, cells cultured with and without DOX for 4 days were treated with Adriamycin (0.4–256 ng/ml) for 12 h, stained with propidium iodide (1μg/ml) and counted by flow cytometry (Guava EasyCyte). MEF cultures and protein blotting were performed as described previously (Dickins et al., 2005).
RNA extraction and quantitative real-time PCR
For quantitative analysis of mRNA levels, cDNA was generated from total RNA using random primers and MultiScribe reverse transcriptase (Applied Biosystems) and amplified by PCR with SYBR Green® dye detection (Applied Biosystems, Warrington UK). Primers were described previously (Kado et al., 2008; Niwa et al., 2000; Takahashi and Yamanaka, 2006) and PCR product detection was performed on the IQ5 iCycler (BioRad) under standard conditions. For details of small RNA cloning see Supplementary information.
Lung histopathology and immunofluorescence
Adenoviral Cre was purchased from the University of Iowa Gene Transfer Vector Core and 2.5 × 107 PFU was administered to mice 4–6 wk of age (Jackson et al., 2001). Lung plucks were inflated with 10% buffered formalin and embedded and stained according to standard protocols. For tumor burden analyses (Fig. 7E, S7B), all lung lobes were orientated in the same manner and 10 cross sections 100μm apart were quantified using NDP view software (Hamamatsu). For immunofluorescence, lungs were flushed with ice cold PBS and filled with agarose via tracheal intubation. Cooled, rigid tissue was sectioned on a Leica vibratome at 150μm. Sections were heated to remove agarose and fixed in 3.7% formaldehyde at RT. Lung slices were permeabilized with 0.5% triton x-100, blocked with 1% BSA and stained overnight at 4C. Specimens were counterstained with DAPI.
Supplementary Material
Acknowledgments
We thank C. Beard and R. Jaenisch (Whitehead Institute) for pBS31 flp-in and pCAGs-FLPe-Puro vectors, ColA1 3′ probe and KH2 ES cells. We thank X. Guo, Y. Park and P.W.J. Russell for discussions and assistance in procedures. We thank E. McGlinn and C. Tabin (Harvard Medical School) for advice with the skeletal preparations of embryos. Thanks to L. Bianco, J. Coblentz, E. Earl and the Cold Spring Harbor Laboratory animal house staff. We gratefully acknowledge Janelle Simon, Danielle Grace, and Jacqueline Cappellani for technical assistance and members of the Lowe laboratory for advice and discussions. This study was supported by a Mouse Models of Human Cancer Consortium grant and a program project grant from the National Cancer Institute. P.K.P is MSTP Fellow of Stony Brook University, L.E.D. is supported by a National Health & Medical Research Council of Australia overseas Biomedical Training Fellowship, C.M. was supported by a fellowship from the Deutsche Forschungsgemeinschaft and a AACRAstellas USA Foundation Fellowship in Basic Cancer Research, J.Z. is the Andrew Seligson Memorial Fellow, R.A.D is a VESKI Fellow and G.J.H and S.W.L. are Howard Hughes Medical Institute investigators. P.K.P, G.J.H and S.W.L. are founders on the scientific advisory board of Mirimus, Inc., a company that has licensed technology related to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Bryja V, Bonilla S, Arenas E. Derivation of mouse embryonic stem cells. Nature protocols. 2006;1:2082–2087. doi: 10.1038/nprot.2006.355. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, Giovannini M, Perret C. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc Natl Acad Sci U S A. 2004;101:17216–17221. doi: 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damalas A, Kahan S, Shtutman M, Ben-Ze’ev A, Oren M. Deregulated beta-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. EMBO J. 2001;20:4912–4922. doi: 10.1093/emboj/20.17.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, Sanchez-Rivera FJ, Resnick R, Bronson R, Hemann MT, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunckin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel Sensor assay. Mol Cell. doi: 10.1016/j.molcel.2011.02.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, Cardiff RD, Chodosh LA. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, Brown Swigart L, Pham DM, Seo Y, Evan GI, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468:567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado M, Lee JK, Hidaka K, Miwa K, Murohara T, Kasai K, Saga S, Morisaki T, Ueda Y, Kodama I. Paracrine factors of vascular endothelial cells facilitate cardiomyocyte differentiation of mouse embryonic stem cells. Biochemical and biophysical research communications. 2008;377:413–418. doi: 10.1016/j.bbrc.2008.09.160. [DOI] [PubMed] [Google Scholar]
- Kuraguchi M, Wang XP, Bronson RT, Rothenberg R, Ohene-Baah NY, Lund JJ, Kucherlapati M, Maas RL, Kucherlapati R. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Hahn WC, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Current opinion in cell biology. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclea RL, Karperien M, Bosch CA, van der Horst G, van der Valk MA, Kobayashi T, Kronenberg HM, Rawadi G, Akcakaya P, Lowik CW, et al. Adenomatous polyposis coli-mediated control of beta-catenin is essential for both chondrogenic and osteogenic differentiation of skeletal precursors. BMC Dev Biol. 2009;9:26. doi: 10.1186/1471-213X-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AR, Shoemaker AR, Connelly CS, Clipson L, Gould KA, Luongo C, Dove WF, Siggers PH, Gardner RL. Homozygosity for the Min allele of Apc results in disruption of mouse development prior to gastrulation. Dev Dyn. 1995;203:422–433. doi: 10.1002/aja.1002030405. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Olson A, Sheth N, Lee JS, Hannon G, Sachidanandam R. RNAi Codex: a portal/database for short-hairpin RNA (shRNA) gene-silencing constructs. Nucleic Acids Res. 2006;34:D153–157. doi: 10.1093/nar/gkj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. International journal of cancer. 2010;127:2239–2247. doi: 10.1002/ijc.25511. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Wang Y, Dausman J, Jaenisch R. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25:3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Takeuchi S, Tsukasaki K, Yamada Y, Hata T, Mori N, Fukushima A, Seo H, Koeffler HP, Taguchi H. Methylation analysis of the adenomatous polyposis coli (APC) gene in adult T-cell leukemia/lymphoma. Leukemia research. 2005;29:47–51. doi: 10.1016/j.leukres.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rideout WM, Zi T, Bressel A, Reddypalli S, Rancourt R, Woo JK, Horner JW, Chin L, Chiu MI, et al. Chimeric mouse tumor models reveal differences in pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. Nat Biotechnol. 2010;28:71–78. doi: 10.1038/nbt.1595. [DOI] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2010;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.