Abstract
The recruitment of coactivators by nuclear hormone receptors (NRs) promotes transcription by subverting chromatin-mediated repression. Although the histone methylation enzyme CARM1 and an ATP-remodeling complex have been individually implicated in nuclear receptor-dependent transcription, neither a functional nor mechanistic linkage between these systems has been identified. In the process of purifying endogenous CARM1-interacting proteins, we identified an associated complex, nucleosomal methylation activator complex (NUMAC), which includes at least eight components of SWI/SNF, including the ATPase BRG1. In the NUMAC complex, the methylase, CARM1, acquires the ability to covalently modify nucleosomal histones, and the directed nucleosome versus free core histone methylation-specificity change is increased dramatically. Reciprocally, CARM1 stimulates the ATPase activity of BRG1, a key component in nucleosome remodeling. In vivo, CARM1 and BRG1 coassemble on an estrogen receptor (ER)-target gene to cooperatively activate ER-dependent transcription. This association of ATP-remodeling factors with HMT CARM1 defines a new component of regulation in the nuclear hormone-signaling pathway.
Keywords: CARM1, SWI/SNF, chromatin remodeling, histone methyltransferase
The NR superfamily of transcription factors regulates gene expression in response to small lipophilic ligands whose binding promotes the recruitment of coregulators that reverse the block to transcription imposed by condensed chromatin (McKenna and O'Malley 2002). The coregulators typically display either histone-modifying activities or ATP-dependent chromatin remodeling activities that utilize the energy of ATP hydrolysis to promote the accessibility of transcription factors to hormone response elements (Travers 1999; Dilworth and Chambon 2001). The best-characterized NR coactivators include the p160 family proteins SRC-1, SRC-2 (TIF2/GRIP1), SRC-3 (ACTR/pCIP/AIB1/RAC3), the histone acetyltransferases (HATs) CBP/p300, ATP-dependent chromatin remodeling complexes, and Mediator complex (DRIP/TRAP) that bridges basal transcriptional machinery during transactivation (Westin et al. 2000; Aranda and Pascual 2001; Narlikar et al. 2002). Whereas these activities, which often exist as multisubunit complexes, are targeted to NR-regulated promoters in a combinatorial manner, how, and precisely which complexes are assembled remains unresolved.
ATP-remodeling complexes, consisting of from 2 to 12 subunits, can affect the position or mobility of nucleosomes in an ATP-dependent manner to either enhance or alleviate the repressive effects of chromatin (Travers 1999). ATP-remodeling complexes all contain the SWI2/SNF2 family of ATPases, which has homology to DNA helicases. They can be grouped into four subfamilies on the basis of the identity of their prototype ATPase as follows: the SWI2/SNF2 subfamily (shares the greatest homology to the yeast SWI2/SNF2 ATPase), the ISWI/SNF2H subfamily (defined by similarity to the Drosophila ISWI), the Mi-2 subfamily (defined by similarity to the CHD ATPases), and the INO80 subfamily (defined by the central INO80 ATPase; Becker 2002). Although the two mammalian homologs of SWI2/SNF2, BRG1, and Brahma (BRM) are highly related, they are apparently recruited by different classes of transcription factors and participate in distinct signaling pathways (Kadam and Emerson 2003). Among the divergent mammalian SWI/SNF complexes, BAF (also called hSWI/SNF-A) and PBAF (hSWI/SNF-B) are the two predominant forms, which possess either BRG1/BRM or BRG1, respectively (Nie et al. 2000). The SWI/SNF (BRG1) complex has been proposed to activate transcription by steroid nuclear receptors such as ER and GR in vivo (Fryer and Archer 1998; DiRenzo et al. 2000), whereas PBAF was shown to activate VDR and PPARs in a chromatin-based transcription reaction in vitro (Lemon et al. 2001). Although the mechanism of BRG-1 recruitment by ER or GR is unclear, both the ATPase and cognate ligands are required for transactivation (Ichinose et al. 1997; DiRenzo et al. 2000; Nie et al. 2000).
More recently, two histone methyltransferases (HMTs), CARM1 and PRMT1, have been shown to act as NR coactivators, but only in the presence of the p160 and p300/CBP proteins (Chen et al. 1999a; Koh et al. 2001). This activity is most effective on preacetylated histones, suggesting that CARM1 acts downstream of NR HATs (Xu et al. 2001; Daujat et al. 2002). The CARM1 methyltransferase catalytic function is required for receptor transactivation (Chen et al. 1999a) with methylation at R17 of histone H3 serving as a marker of active hormone response elements (Ma et al. 2001; Bauer et al. 2002). These lines of evidence suggest that directed methylation is a component of the histone code of NR-mediated hormone signaling (Stallcup 2001; Kouzarides 2002). The observation that recombinant CARM1 methylates histone H3 of free core histones, but much less efficiently with nucleosomes, suggests that association with other factors may facilitate its action on chromatin substrates in vivo (Xu et al. 2001). During glycerol gradient fractionation of HeLa nuclear extracts, CARM1 was found in high-molecular weight fractions (see below; data not shown), suggesting that CARM1 is a probable component of an in vivo complex.
In an attempt to dissect the molecular basis underlying transcriptional activation by the NR coactivator CARM1, we have established cell lines that express Flag-tagged CARM1 and purified the endogenous CARM1-associated complex, NUMAC. The NUMAC complex contains multiple SWI/SNF subunits and is found to have enriched activity for nucleosome histone methylation. CARM1 and BRG1 physically interact in vitro and coassemble on an estrogen receptor (ER) target gene to cooperatively activate ER-dependent transcription in vivo. The association of ATP-remodeling factors with HMT CARM1 reveals a new arm in the nuclear hormone-signaling cascade.
Results
Purification of Flag-tagged CARM1-associated complex,NUMAC
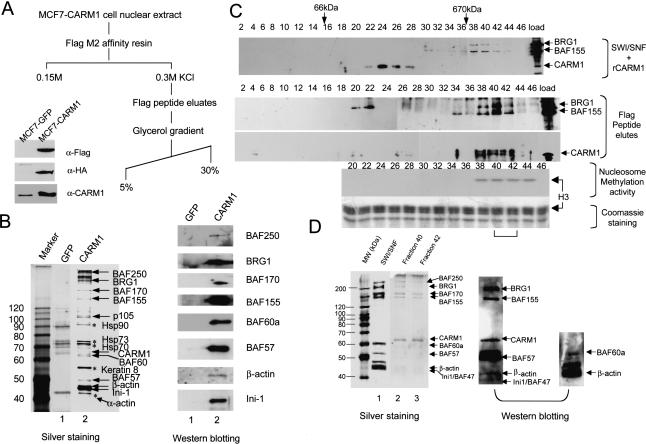
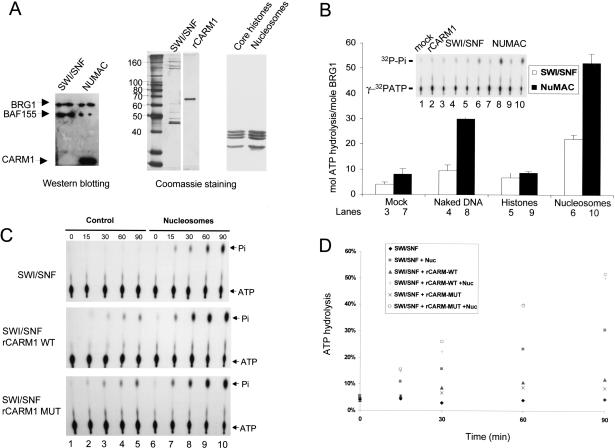
To identify proteins regulating HMT activity in vivo, we generated MCF7 cells that stably express CARM1 with an amino-terminal Flag tag and a carboxy-terminal HA tag to facilitate the purification. MCF7 cells were selected because CARM1 is expressed at its highest level in these, among 10 cell lines examined (data not shown). Western blot analysis with anti-Flag and HA antibodies showed efficient expression of CARM1 (Fig. 1A), and approximately the same amount of Flag-CARM1 is produced as the endogenous protein (Fig. 1A). Using a two-step purification scheme (Fig. 1A), CARM1 and its associated factors were purified over a Flag-M2 affinity resin in the presence of estradiol (Fig. 1B, left, lane 2). The eluted samples were trypsin digested, and the chromatographed mixture was subjected to high-throughput mass spectrometry sequencing (Washburn et al. 2001). This resulted in the identification of 15 proteins, including CARM1. After subtracting proteins present in the control MCF7-GFP cell lines (Fig. 1B, left, lane 1), the 10 remaining components of this collection included eight members of the SWI/SNF ATP-dependent chromatin remodeling complex (Fig. 1B, left, lane 2), CARM1 and p105. Western blotting using BAF-specific antibodies also reveals the specific association of SWI/SNF components in the Flag-eluate from CARM1-expressing cells (Fig. 1B, right, lane 2), as they are absent in the Flag-eluate from GFP control cells (Fig. 1B, right, lane 1).
Figure 1.
Characterization of CARM1-complex. (A) Schematic for CARM1-complex purification. Western blot showing detection of Flag-CARM1-HA in transfected MCF7 (lane 2) compared with the control GFP-stable cells (lane 1) using α-flag, HA, or CARM1 antibodies. (B) Determination of CARM1-associated polypeptides. Silver-staining (left) and Western analyses (right) of nuclear proteins from GFP-stable cells (lane 1) and CARM1-stable cells (lane 2) after Flag-peptide elution. Half of the gel was subjected to Western blotting with antibodies against SWI/SNF subunits (right) and superimposed on the silver-stained gel (left) to position each subunit. Heat-shock proteins and α-actin (asterisks) are probably contaminants, as they were also present in control MCF7-GFP cell eluate (lane 1). (C) Western analysis of CARM1, SWI/SNF, and Flag-eluted CARM1 complex by glycerol gradient. Premixed CARM1 and SWI/SNF (top) or Flag-eluted complex (bottom) was applied to a 5%-30% (v/v) gradient. A total of 100 μL of fractions were collected and immunoprecipitated with 20% (w/v) TCA, followed by Western detection with the mixture of BRG1, BAF155, and CARM1 antibodies. BRG1 and BAF155 peaks at fraction 40 are separated from CARM1 peak at fraction 24 (top). The fractions where BSA (66 kD) and Thyroglobulin (670 kD) molecular weight markers sediment are shown on the top of the blot. CARM1 in the Flag-eluted complex cosediments with BRG1 and BAF155 through glycerol gradient (bottom). A total of 10 μL of each Flag-eluted fraction was assayed for methyltransferase activity with 1.5 μg of nucleosomes and 3H-AdoMet as substrates. The autoradiography shows the methylation activity is concentrated in fractions 38-44 and occurs on histone H3 when superimposed on the Coomassie-staining gel. The bracket indicates fractions being further analyzed in D. (D) Silver-staining (left) and Western blotting (right) of NUMAC in fractions 40 and 42 from C. Fractions were TCA precipitated, SDS-PAGE resolved, and silver stained (left) or analyzed by Western blotting (right). The weak staining of small molecular-weight BAFs by silver is probably due to inefficient precipitation by TCA, as the non-TCA-precipitated samples contain small molecular weight BAFs as revealed by Western blotting (right). After Western transfer, the nitrocellulose membranes were first blotted with α-BAF60a/β-actin antibodies, then stripped and reprobed with the mixture of BRG1/BAF155/CARM1/BAF57/INI-1 antibodies.
SWI/SNF is one of the major chromatin-remodeling complexes and exists in biochemically distinct forms. For example, SWI/SNF A contains BAF250, whereas SWI/SNF Bis distinguished by the BAF180 subunit (Peterson 2002). The presence of BRG1, BAF250, BAF170, BAF155, BAF57, β-actin, and INI-1 within NUMAC were confirmed by Western blotting (Fig. 1B). β-actin and the actin-related proteins (ARPs) have been found in several SWI/SNF-like complexes, whose function is to tether remodeling machinery to the nuclear matrix (Olave et al. 2002). However, BAF180 (a PBAF-specific subunit) was not detected, suggesting close similarity to the mammalian SWI/SNF-A complex that also lacks this component. Likewise, neither the ATPase hSNF2H (a human ISWI homolog) nor the NR coactivator SRC-p160 proteins were found in the CARM1 complex (data not shown). The second step of purification was glycerol gradient sedimentation. We first combined recombinant CARM1 (rCARM1, see Material and Methods) isolated from Sf9 cells (Xu et al. 2001) and highly purified SWI/SNF complex isolated from HeLa cells stably expressing Flag-Ini-1 (Phelan et al. 1999) together onto glycerol gradients. rCARM1, peaked at fraction 24, eluted closer to the top of the gradient, whereas SWI/SNF components BRG1 and BAF155 cosedimented in fractions 38 to 44 (Fig. 1C, top). However, when the Flag-purified CARM1 complex was subjected to glycerol gradient fractionation, resulting in depletion of free CARM1, the majority of CARM1 remained associated with SWI/SNF components (fraction 38-44, cf. with SWI/SNF profile at top), which includes the nucleosome histone H3 methylation activity (activity panel, Fig 1C, cf. with the Coomassie staining). The presence of eight SWI/SNF subunits in these fractions (F40 and F42, lanes 2,3) were detected by silver staining (Fig. 1D, left) and verified by Western blotting (Fig. 1D, right).
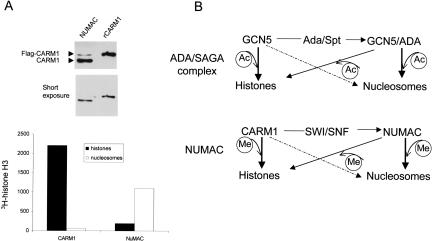
We next measured the substrate specificity of methyltransferase complex using various substrates, that is, free core histones and nucleosomal histones. Intriguingly, in contrast to rCARM1, which preferentially methylates free core histones (Fig. 2A, lanes 1,2), the Flag-peptide eluate readily methylates histone H3 present in either nucleosomes or free-core histones (Fig. 2A, lanes 3,4). Strikingly, the glycerol gradient fractions containing both CARM1 and SWI/SNF subunits, such as F40, preferentially methylate histone H3 in nucleosomes, and the ability to methylate free histones is largely compromised (Fig. 2A, lanes 5,6). We refer to this entity as the nucleosomal methylation activator complex (NUMAC). After normalizing recombinant CARM1 with the native form in NUMAC by Western blotting (Fig. 7A, below), their respective enzymatic activities toward equal amounts of free core histones versus nucleosomes were compared. Whereas rCARM methylates free core histones 36-fold higher than nucleosomes, NUMAC displays a preference for nucleosomes over histones by sixfold (Fig. 7A, below). The reversed activity ratio of rCARM1 toward free histones versus nucleosomes (36:1) versus NUMAC (1: 6) produces a net 200-fold change in specificity (Fig. 7A, below). It is worthwhile to note that although we purify NUMAC using a Flag-M2 affinity resin, the CARM1 that is present in NUMAC is mostly endogenous CARM1, which has no tag (Fig. 7A, below). This phenomenon is consistent with our observation that CARM1 can form oligomers in vivo, and supports the notion that endogenous CARM1 associates with SWI/SNF to form NUMAC.
Figure 2.
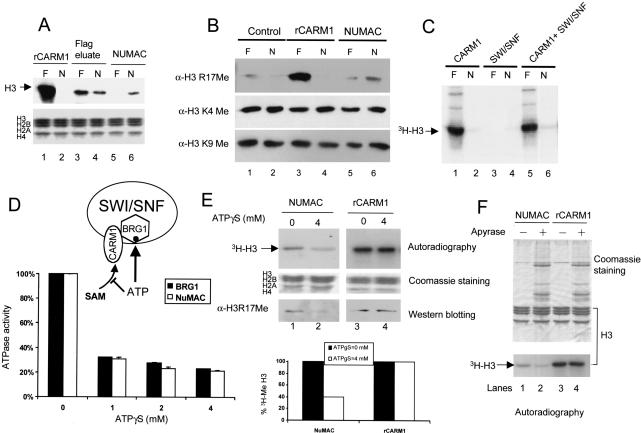
NUMAC ATPase activity is required for nucleosomal histone methylation. (A) Nucleosomal histone methylation activity of 0.2 μg of rCARM1 (lanes 1,2), 0.2 μg of Flag-peptide eluates (lanes 3,4), or 0.1 μg of NUMAC (Fraction 40, lanes 5,6) were determined with 3 μg of either free core histones (F) or nucleosomes (N) in the methylation reaction. Autoradiograph (top) shows 3H-H3 in nucleosomes after methylation, and Coomassie staining (bottom) shows total core histones. (B) Determination of methylation sites in free core histones (F) and nucleosomes (N) by rCARM1 (lanes 3,4) and NUMAC (lanes 5,6). A total of 3 μg of free core histones (lanes 1,3,5) or nucleosomes (lanes 2,4,6) were incubated with 0.1 μg of BSA (lanes 1,2), 0.1 μg of rCARM1 (lanes 3,4), or 0.1 μg of NUMAC (lanes 5,6) in the presence of 1 mM s-AdoMet. Reaction products were resolved by SDS-PAGE and analyzed using anti-H3R17Me antibodies. The blot was stripped and reprobed with anti-H3K4Me or H3K9Me antibodies to indicate equal loading and the unaltered methylation at those sites. (C) Addition of rCARM1 to SWI/SNF minimally affects nucleosomal methylation. A total of 0.1 μg of rCARM1 (lanes 1,2), 3 μg of SWI/SNF (lanes 3,4), or 0.1 μg of rCARM1 preincubated with 3 μg of SWI/SNF (lanes 5,6) at 27°C for 15 min were incubated with 3 μg of either free core histones (lanes 1,3,5) or nucleosomes (lanes 2,4,6) in the methylation reaction. 3H-H3 is shown in the autoradiograph. (D) Dual role of ATP on NUMAC and determination of ATPγS concentration required to inhibit BRG1-ATPase activity. A total of 0.05 μg of BRG1 or 0.5 μg of NUMAC were incubated with [γ-32P]ATP and 0-4 mM ATP in the ATPase assay. Free phosphate was separated from [γ-32P]ATP on TLC plates, and the percentage of hydrolyzed [γ-32P]ATP was plotted. (E) Inhibition of NUMAC nucleosome (lanes 1,2) but not rCARM1 histone (lanes 3,4) methylation by 4 mM ATPγS. A total of 0.5 μg of NUMAC or 0.1 μg of rCARM1 was incubated with 3 μg of nucleosomes or free core histones, respectively, in the presence of 0.1 mM 3H-AdoMet. Autoradiography and Coomassie staining of SDS-PAGE are shown. Half of the reactions were subjected to Western blotting using α-H3R17Me antibody. The 3H-histone H3 band was cut out and counted with scintillation solvent. The amount of 3H-histone H3 in the presence of 4 mM ATPγS (lanes 2,4) was normalized by the level without ATPγS (lanes 1,3) to show the inhibitory effect of ATPγS (bottom). (F) Either 0.5 μg of NUMAC or 0.1 μg of rCARM1 were used to methylate 3 μg of nucleosome or free core histones, respectively. A 0.2 unit of apyrase was preincubated with the reaction mixture at 27°C for 15 min to deplete ATP before addition of 3H-AdoMet substrates.
Figure 7.
ADA/SPT and SWI/SNF function as nucleosome modification adaptor for GCN5-HAT and CARM1-HMT complex, respectively. (A) NUMAC (∼0.2 μg) and rCARM1 (∼30 ng) were normalized to equal amounts of CARM1 by Western blotting (both long exposure and short exposure are shown). The methylation of free core histones (3 μg) or nucleosomes (3 μg) with 3H-AdoMet by NUMAC or rCARM1 were performed as described in the Materials and Methods. The reaction mixtures were resolved on SDS-PAGE and 3H-histone H3 was cut out and counted in the scintillation buffer. (B) The association of GCN5-HAT with different proteins in multiple complexes endows upon it nucleosomal histone acetylation activity and expanded substrate specificity (top). Similarly, the association of CARM1 with SWI/SNF subunits potentiates its nucleosome histone methylation activity (bottom). The thickness of the lines represents the more or less favorable substrates. After normalizing CARM1 and histones amounts, the specific activity of free core histones to nucleosomes switches from 36:1 to 1:6.
Methylation site-specific antibodies confirm that NUMAC preferentially methylates R17 of H3 in nucleosomes, but not free histones (Fig. 2B, cf. lanes 5,6 and 1,2), whereas rCARM1 almost exclusively methylates R17 of H3 in free core histones relative to nucleosomes (Fig. 2B, cf. lanes 3,4 and 1,2). Reprobing this blot with anti-methylated H3K4 and H3K9 antibodies revealed that methylation of these residues are unchanged (Fig. 2B, lanes 1-6) during the reaction. Thus, methylated K4 and K9 in histone H3 automatically serve as loading controls. These results suggest that the methylation activity of NUMAC is specific for R17 of H3 in nucleosomal histones, and consequently, the methylation activity of NUMAC is mediated by CARM1. This preferential nucleosome substrate specificity is reminiscent of PR-set7, which methylates H4 in nucleosomes, but reacts poorly with free histones (Nishioka et al. 2002).
We next attempted to reconstitute the nucleosome activity by mixing purified rCARM1 with the highly purified SWI/SNF complex isolated from HeLa-INI-1 cells. This reaction is very inefficient, and results in only marginal stimulation of nucleosomal histone methylation (Fig. 2C, lane 6), which suggests that either proper folding of each subunit in vivo or ordered subunit recruitment may be required for the assembly of NUMAC and its enzymatic activity. We have also attempted to reassemble NUMAC by denaturation and renaturation of the rCARM1 and SWI/SNF. However, CARM1 alone or in combination with SWI/SNF completely lost its methylase activity upon renaturation (data not shown).
It has been reported that the ATP-dependent nucleosome disruption activity of NuRD stimulates the deacetylation of nucleosomal histones (Xue et al. 1998). Therefore, we tested whether ATP-dependent remodeling mediated by NUMAC similarly facilitates nucleosomal histone methylation. First, we determined whether addition of ATP could augment NUMAC-dependent nucleosomal histone methylation. Only a modest stimulatory effect on NUMAC was observed by addition of 1 mM ATP (data not shown). Second, we measured whether there are pre-existing amounts of ATP in our methylation reaction. Using a firefly luciferase on the basis of bioluminescence assay, we found that our preparation of NUMAC and nucleosomes associate with ∼10 nM ATP. Third, to demonstrate whether NUMAC methyltransferase activity is dependent on ATP, we inhibited ATPase activity with the ATP analog ATPγS and analyzed the change in nucleosome methylation by NUMAC. As diagrammed in Figure 2D, ATP can potentially play dual roles in the NUMAC nucleosome methylation reaction by affecting CARM1 and BRG1 independently. On one hand, ATP is a substrate of the BRG1 ATPase, and is therefore essential for its remodeling activity. On the other hand, ATP can be inhibitory to CARM1 due to its similarity to S-adenosylmethionine, the substrate of CARM1. For these reactions, we first determined the amount of ATPγS that efficiently inhibits the ATPase activity of BRG1, but has no inhibitory effect on methyltransferase activity. We found that 4 mM of ATPγS efficiently inhibited the ATPase activity of either BRG1 or NUMAC (Fig. 2D) and displayed a corresponding inhibition of NUMAC methylase activity by 60% (Fig. 2E, lanes 1,2). In contrast, the methylase activity of rCARM1 was not affected (Fig. 2E, lanes 3,4), although higher concentrations of ATPγS can inhibit rCARM1 by competing with the substrate S-adenosyl methionine (SAM; data not shown). The ATPγS inhibition of nucleosome histone H3 methylation by NUMAC (Fig. 2E, autoradiograph) correlates with the reduction of R17Me in histone H3 (Fig. 2E). This data supports the notion that CARM1 is the catalytic subunit of NUMAC that is responsible for nucleosome methylation. To further demonstrate that ATP-dependent nucleosome remodeling facilitates nucleosome methylation, we used apyrase to remove ATP from the methylation reaction. Addition of 0.2 units of apyrase inhibits nucleosome histone H3 methylation by NUMAC, but has no effect on methylation of core histones by rCARM1 (Fig. 2F, lanes 3,4). The incomplete inhibition of nucleosome methylation by addition of ATPγS (Fig. 2E) and removal of ATP (Fig. 2F, lanes 1,2) implies that there is an ATP-independent nucleosome methylation event (see Discussion).
ATP-dependent remodeling activity of NUMAC and the stimulation of SWI/SNF remodeling activities by CARM1
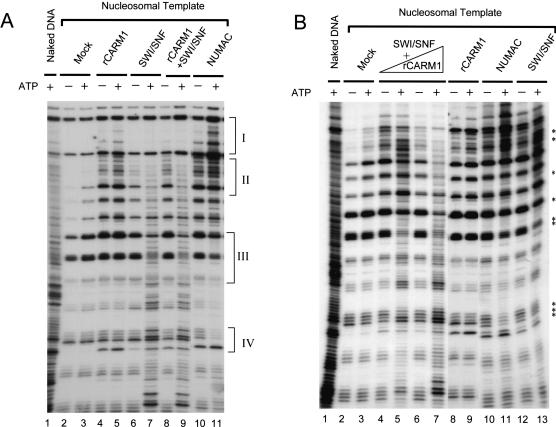
We next investigated whether NUMAC retains the ability of SWI/SNF to disrupt nucleosomes, and if so, whether the existence of CARM1 affects remodeling activity. As shown in Figure 3A (lanes 4,5) and Figure 3B (lanes 8,9), rCARM1 alone does not have ATP-dependent remodeling activity. As expected, both NUMAC and SWI/SNF exhibit ATP-dependent remodeling activity (Fig. 3A, lanes 6,7,10,11, denoted by bracket). Intriguingly, the nucleosome disruption patterns generated by NUMAC and SWI/SNF are different. NUMAC increases, but SWI/SNF decreases DNaseI digestion in regions indicated as brackets I and II in an ATP-dependent manner, whereas the opposite effect is observed in regions indicated as brackets III and IV (Fig. 3A), implying that incorporation of CARM1 may alter the enzymatic properties of the ATPase in SWI/SNF to remodel mononucleosomes. Next, we examined the stimulation of CARM1 on SWI/SNF remodeling activity. No stimulation of SWI/SNF by rCARM1 was observed at high concentrations of SWI/SNF, which probably has reached its maximal activity (Fig. 3A, cf. lanes 8,9 and 6,7). To observe the stimulatory effect of CARM1, we used limiting amounts of SWI/SNF. Both NUMAC and SWI/SNF exhibit weak ATP-dependent remodeling activity at limiting amounts of either complex (Fig. 3B, lanes 10-13, denoted by stars). Preincubation of recombinant CARM1 with SWI/SNF (1:1 molar ratio) results in a marked increase in nucleosome remodeling (Fig. 3B, cf. lanes 4,5 and 12,13), which was further increased at higher ratios of CARM1 to SWI/SNF (5:1; Fig. 3B, cf. lanes 6,7 and 12,13). As an alternative approach to investigate the mechanistic difference between NUMAC and SWI/SNF, and to quantify the change in SWI/SNF nucleosome remodeling activity induced by CARM1, we used the restriction endonuclease digestion assay (Logie and Peterson 1997). NUMAC fails to induce the exposure of a single site in the nucleosome array as SWI/SNF does in this assay (data not shown). However, we consistently observe that CARM1 stimulates SWI/SNF-dependent nucleosome disruption by 1.5 to twofold (data not shown). This result further supports the notion that SWI/SNF and NUMAC remodel nucleosomes using different mechanisms; moreover, it distinguishes NUMAC from the simple mixture of rCARM1 and SWI/SNF. Further studies to reveal their mechanistic difference in remodeling is under way. Nevertheless, the stimulation of SWI/SNF remodeling by rCARM1 suggests that CARM1 can stimulate SWI/SNF chromatin remodeling activity in vitro, possibly by enhancing BRG1-catalyzed ATP hydrolysis, increasing the effectiveness of the hydrolysis toward the remodeling event, or increasing the affinity of SWI/SNF binding to chromatin substrates, or any combination of the above.
Figure 3.
Chromatin-remodeling activity of NUMAC and stimulation of SWI/SNF-remodeling activity by CARM1. (A) NUMAC and SWI/SNF exhibit different nucleosome disruption patterns. The ATP-dependent remodeling as revealed by DNaseI digestion of mononucleosomes after incubating with mock (0.1 μg of BSA, lanes 2,3), rCARM1 (0.1 μg, lanes 4,5), SWI/SNF (0.6 μg, lanes 6,7), rCARM1+SWI/SNF (0.1 μg rCARM1 pre-incubated with 0.6 μg of SWI/SNF at 27°C for 15 min, which gave a molar ratio of 5:1 for rCARM1/SWI/SNF, lanes 8,9), NUMAC (0.6 μg, lanes 10,11). Brackets I and II highlight the ATP-dependent changes in DNaseI digestion pattern increased by NUMAC and decreased by SWI/SNF; brackets III and IV highlight the opposite effect generated by SWI/SNF or NUMAC. (B) Stimulation of SWI/SNF remodeling activity by rCARM1 at limiting concentrations of SWI/SNF. The ATP-dependent remodeling of mononucleosomes by mock (0.1 μg of BSA, lanes 2,3), rCARM1 (0.1 μg, lanes 8,9), SWI/SNF (0.12 μg, lanes 12,13), NUMAC (0.12 μg, lanes 10,11), and rCARM1 and SWI/SNF mix [6 ng of rCARM1 pre-incubated with 0.12 μg of SWI/SNF, lanes 4,5, or 30 ng of rCARM1 preincubated with 0.12 μg of SWI/SNF, lanes 6,7, which gave 1:1 (lanes 4,5) or 5:1 (lanes 6,7) molar ratio for rCARM1/SWI/SNF]. Stars denote weak ATP-dependent changes by SWI/SNF and NUMAC.
To compare the ATPase activity of SWI/SNF and NUMAC, we normalized the amount of complex by Western blotting using BRG1 as a standard (Fig. 4A, left). The other components used in the in vitro ATPase assay are shown by SDS-PAGE analysis (Fig. 4A, right). Remarkably, by measuring the cumulative activity in the first 60 min of the reaction (Fig. 4B, inset), we found that NUMAC was at least twice as active as SWI/SNF in ATP hydrolysis when normalized by equimolar amounts of BRG1 (Fig. 4B). Because CARM1 is devoid of ATPase activity itself (Fig. 4B, inset), these results suggest that it functions as a modifier or competence factor within the large complex. Similar to previous reports for SWI/SNF (Phelan et al. 1999), the ATPase activity of NUMAC was stimulated by DNA and increased by about 10-fold in the presence of nucleosomes (Fig. 4B), although DNA-dependent stimulation of SWI/SNF ATPase activity was modest in our assay. We then examined whether addition of rCARM1 was sufficient to stimulate SWI/SNF ATP hydrolysis in either the presence or absence of nucleosomes, and whether the enzymatic activity of CARM1 was required. As observed above, addition of wild-type CARM1 or an HMT-deficient variant (CARM1 mutant 189VLD191→AAA) led to similar levels of activation (2.2-fold) of SWI/SNF-dependent ATP hydrolysis (Fig. 4C,D), indicating that the methyltransferase activity is not a component of the enhancement. This is mechanistically similar to NuRD, whose histone deacetylase activity is not required to stimulate chromatin remodeling (Xue et al. 1998).
Figure 4.
CARM1 stimulates ATPase activity of NUMAC and BRG1. (A) Western normalization of BRG1 amount in SWI/SNF and NUMAC complex. Approximately 100 ng of SWI/SNF and NUMAC were loaded on 8% SDS-PAGE, transferred to nitrocellulose membrane, and blotted with the mixture of BRG1/BAF155/CARM1 antibodies (left). A total of 0.5 μg of SWI/SNF, 0.1 μg of rCARM1, 2 μg of core histones, and 3 μg of nucleosomes were resolved on SDS-PAGE and Coomassie stained (right). (B) Stimulation of SWI/SNF (0.3 μg, white) and NUMAC (0.3 μg, black) ATP hydrolysis by 0.1 μg of BSA (lanes 3,7), 1 μg of 1-kb plasmid DNA (lanes 4,8), 3 μg of core histones (lanes 5,9), or 3 μg of nucleosomes (lanes 6,10). The hydrolysis of 1.7 pmole [γ-32P]ATP by SWI/SNF and NUMAC in the presence of BSA (mock), naked DNA, histones, or nucleosomes was analyzed by TLC, and the autoradiography (inset) is shown. The hydrolysis of [γ-32P]ATP in the presence of 0.1 μg of BSA (lane 1) or 0.1 μg of rCARM1 (lane 2) were shown as controls. SWI/SNF and NUMAC are normalized to equimolar amounts of BRG1. The molar amount of ATP hydrolyzed by per mole of BRG1 in each complex is presented on the y-axis. (C) CARM1 stimulates ATPase activity of SWI/SNF. Time course of [γ-32P]ATP hydrolysis is determined using 0.5 μg of SWI/SNF in the absence (lanes 1-5) or presence (lanes 6-10) of nucleosomes (3 μg) and wild-type or mutant rCARM1 (0.1 μg). (D) Quantitation of SWI/SNF ATPase activity in C. The SWI/SNF activity in the presence (▪) or absence of nucleosomes (♦) is depicted.
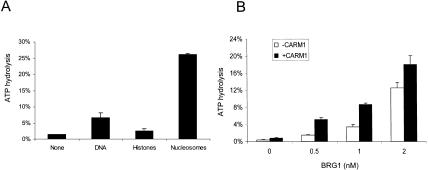
It has been reported that nucleosome remodeling and ATPase activity of other complexes are affected by non-ATPase subunits, that is, the stimulation of BRG1 by Ini-1/BAF155/BAF170 (Phelan et al. 1999) and the stimulation of ISWI by ACF1 (Eberharter et al. 2001). In view of this, we examined whether CARM1 could directly stimulate ATP hydrolysis by BRG1. The Flag-tagged BRG1 protein was baculovirally expressed and purified from Sf9 cells using Flag-M2 affinity resin as described (Phelan et al. 1999). First, as controls, we show that the BRG1 ATPase is activated by both nucleosomes and naked DNA (Fig. 5A) as described previously (Phelan et al. 1999). Next, as shown in Figure 5B, at concentrations of BRG1 (0.5-2 nM) in which its ATPase activity is at submaximal levels, we observe an approximate 2.5-fold stimulation by CARM1 (Fig. 5B). Because this effect is similar to that seen with the entire SWI/SNF complex, our results suggest that CARM1 may directly regulate BRG1 ATPase activity independent of other BAFs. We also explored whether CARM1 could promote ATP hydrolysis by ACF/ISWI, a different ATPase subclass (Langst and Becker 2001). Although we confirmed that ISWI-mediated ATP hydrolysis is stimulated by nucleosomes, the addition of CARM1 failed to show any effect (data not shown), suggesting that its action on BRG1 is specific.
Figure 5.
The effect of CARM1 on nucleosome-stimulated BRG1. (A) The [γ-32P]ATP hydrolysis is determined for 5-nM of BRG1 in the presence of 0.1 μg BSA (mock), 1 μg of 1-kb linearized plasmid DNA, 3 μg of core histones, or 3 μg of nucleosomes at 30°C for 30 min, and the percent of hydrolyzed [γ-32P]ATP by BRG1 is plotted. (B) BRG1 ATPase activity was determined in the presence (▪) or absence (□) of 2 nM CARM1. The ATPase activity is presented as the percent of hydrolyzed [γ-32P]ATP. Results are expressed as the mean value ± SD of duplicates.
CARM1 binds BRG1 and NUMAC enhances ER-dependent transcription
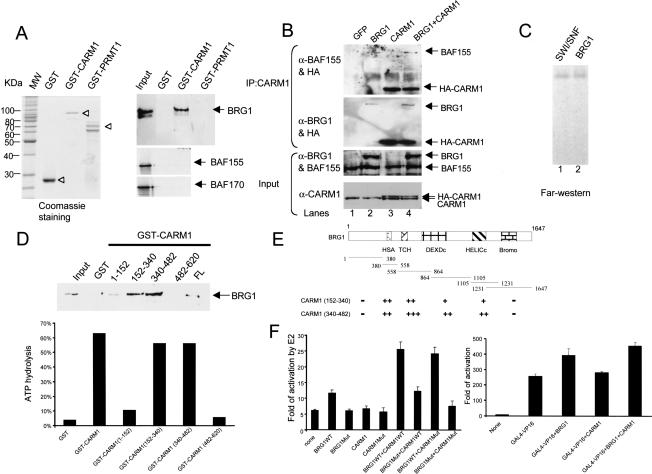
Because rCARM1 stimulates BRG1 ATPase activity, we determined whether these two proteins interact physically. In the GST pull-down assay using purified proteins (Fig. 6A, left, open arrow), direct association was observed between CARM1 and BRG1, but not between PRMT1 and BRG1 (Fig. 6A, right). In addition, neither BAF 155 nor BAF 170 displayed CARM1 binding (Fig. 6A, right). To illustrate that the formation of NUMAC is BRG1 dependent, we transfected expression vectors for GFP, CARM1, and BRG1, individually, or their combination into BRG1/hBRM-deficient SW13 cells, which express other functional BAFs and CARM1. Coimmunoprecipitation of endogenous BAF 155 with endogenous CARM1 was observed when BRG1 was transfected (Fig. 6B, cf. lanes 2 and 1), and increased amounts of BRG1 and BAF155 coimmunoprecipitated by CARM1 antibodies when cells were transfected with DNA constructs expressing HA-CARM1 (Fig. 6B, cf. lanes 4 and 2). This indicates that BRG1 can serve as the bridging factor between CARM1 and other BAFs. Likewise, of all proteins in the SWI/SNF complex (Fig. 6C, lane 1), only BRG1 was recognized by 35S-Met-labeled CARM1 in a Far-Western experiment (Fig. 6C). Taken together, these experiments suggest that BRG1 is the primary, and possibly the only subunit in the complex targeted by CARM1. We further mapped BRG1-interacting domains in CARM1 using a GST pull-down assay. GST-CARM1 (FL) and four GST-fusion CARM1 fragments (1-152, 152-340, 340-482, and 482-620) were expressed in Escherichia coli BL21 DE3 (pLysS) cells and purified by GST Sepharose beads. Purified BRG1 binds two central domains in CARM1 (amino acids 152-340 and 340-482; Fig. 6D, top), which correspond to substrate-binding regions in the related PRMTs, that is, the s-adenosyl methionine-binding domain and the domain responsible for binding to protein substrates (McBride and Silver 2001). Addition of CARM1 fragments spanning these domains was sufficient to elicit a robust stimulation of BRG1 ATP hydrolysis (Fig. 6D, bottom). Reciprocal mapping of the CARM1-binding region in BRG1 identified several helicase-like domains as targets with the amino terminus and carboxy-terminal bromo domain as negative controls (Fig. 6E, pulse represents the strength of interaction). The interaction of BRG1 with CARM1 through the central helicase regions presumably provides the physical basis for the modulation of its ATPase activity.
Figure 6.
Direct association between CARM1 and BRG1. (A) CARM1 binds to purified BRG1 in GST pull-down assays, whereas no interaction was observed between CARM1 and 35S-Met-labeled BAF155 or BAF 170 (right). Coomassie staining of GST, GST-CARM1, and GST-PRMT1 (left, arrowhead). BRG1 was visualized by Western blotting and 10% of fraction applied for binding was loaded as input. (B) Coimmunoprecipitation of CARM1 with BRG1 and BAF155 in SW13 cells. Nuclear extracts of SW13 cells transfected with indicated plasmids were immunoprecipitated with CARM1 antibody. The CARM1 immunoprecipitates were analyzed by Western blotting using HA/BRG1 or HA/BRG155 antibodies. The 1/10 of nuclear extracts from transfected cells were loaded as Input and probed with α-BRG1/BAF155 or α-CARM1 antibodies. (C) In the SWI/SNF complex, only BRG1 is recognized by CARM1 in a Far-Western assay. A total of 1.0 μg of SWI/SNF (lane 1) and 0.1 μg of BRG1 (lane 2) was loaded on 8% SDS-PAGE, transferred to PVDF membrane, and incubated with 35S-Met-labeled CARM1. The membrane was washed three times with buffer (20 mM Tris at pH 7.6, 10% glycerol, 100 mM NaCl, 1 mM EDTA, 0.1% Tween 20) and exposed to X-ray film. (D) Stimulation of ATPase activity by two BRG1-interacting fragments of CARM1. GST-CARM1 fragments were expressed in E. Coli and purified as described in the Materials and Methods. Recombinant BRG1 is purified from Flag-BRG1 expressing baculovirus-infected Sf9 cells and 1/10 of the total rBRG1 applied to the GST-CARM1 affinity column is shown as input. BRG1 is detected by Western blotting. (Bottom) A total of 0.3 μg of GST-CARM1 or fragments was applied in BRG1 ATPase assay (final concentration of BRG1 is 5 nM). (E) Mapping of CARM1-interacting regions in BRG1. Comparable amounts of 35S-labeled BRG1 fragments and purified GST-CARM1 proteins were used in each binding reaction as described (DiRenzo et al. 2000). When compared with the total input, strong (≥20%, +++), moderate (10%, ++), weak (5%, +), and no (-) binding are indicated. (HSA) Helicase domain with SANT association; (TCH) conserved in transcription and CHROMO domain helicases; (DEXDc) homology to DEAD-like RNA helicases superfamily; (HELICc) helicase superfamily carboxy-terminal domain; (BROMO) bromo domain. (F) Transactivation of CARM1 in ER signaling requires BRG1. BRG1 and BRM-deficient adrenal carcinoma SW13 cells were plated in 24-well dishes and transfected with indicated plasmids using FuGene (Roche). All experiments were normalized to internal β-gal controls and carried out in triplicates. The date represented the fold of induction by estrodial (1 μM). Similar conditions were used in GAL4-VP16 transfection assays.
We further investigated how CARM1 and BRG1 might cooperate to potentiate NR activation. BRG1 has been shown to stimulate ER activation, although its recruitment seemed to be indirect and mediated by additional factors, distinct from p160 proteins (Ichinose et al. 1997; DiRenzo et al. 2000). We therefore speculated that CARM1 might facilitate BRG1 recruitment through its binding to p160 proteins. First, we confirmed that both BRG1 and CARM1 are recruited to ER-target genes in a ligand-dependent and concurrent manner using a time-dependent chromatin immunoprecipitation (ChIP) assay on the cathepsin D promoter in MCF7 cells (data not shown; Shang et al. 2000; Burakov et al. 2002). To determine whether the coactivation of estrogen signaling by CARM1 is BRG1 dependent, we overexpressed CARM1 in SW13 cells. Overexpression of CARM1 alone failed to activate estrogen signaling in SW13 cells. However, cotransfection of BRG1 and CARM1 effectively restored ER activation. This restoration was lost with the BRG1 ATPase mutant (K785R; Fig. 6F). A similar effect was observed with GR (data not shown). The corequirement of CARM1 and BRG1 for activation of steroid hormone signaling suggests a functional cooperativity between the HMT CARM1 protein and ATP-remodeling activities. Such a synergistic effect between BRG1 and CARM1 was not observed with GAL4-VP16 (Fig. 6F) and GAL4-Sp1 (data not shown), suggesting that transactivation by GAL4-VP16 may not require NUMAC, but only require SWI/SNF (Neely et al. 1999). These results collectively suggest that the physical association of CARM1 and BRG1 mutually enhances their ATPase and methylase functions to promote ER signaling.
Discussion
Histone methylation on either lysines or arginines has been correlated with gene activation or repression. Methyl groups on histones seem to serve as critical marks through which transcription factors direct specific chromatin-related events (Bannister et al. 2002). For example, the ER-regulated gene pS2 is activated rapidly by estrogen, and this is accompanied by methylation of histone H3 at R17 within the promoter. During purification of native activities that generate R17 methylation on nucleosomal histone H3, we discovered that CARM1 is able to methylate nucleosomal histones, but only within the context of a native multisubunit complex. This phenomenon of redirecting enzymatic specificity from free histones to chromatin resembles that described previously for the GCN5-HAT complexes. In that case, purified GCN5 efficiently acetylates free histones but not nucleosomal histones, whereas association with Ada and Spt bestows on GCN5 its characteristic ability to modify nucleosomes (Grant et al. 1997; Fig. 7B). Here, we show that SWI/SNF alters CARM1 substrate specificity from free core histones to nucleosomes, suggesting that SWI/SNF (and specifically BRG1) enables CARM1 activity to be selectively activated and coordinated with the remodeling event (Bauer et al. 2002; Fig. 7B). Three possibilities might account for the SWI/SNF-modulated nucleosomal activity of CARM1. First, NUMAC might recognize certain conformations of histone tails that exist in nucleosomes,but not free histones. Second, ATP-dependent remodeling of chromatin by SWI/SNF might facilitate concurrent histone methylation. However, nucleosomal histone methylation activity is not completely inhibited by ATPγS (Fig. 2E) or ATP removal (Fig. 2F), even though the ATPase activity of NUMAC is almost completely abrogated. This leads to the third possibility, that SWI/SNF subunits might redirect CARM1 binding to nucleosomes. This later model is supported by the observation that nucleosomes avidly associate with SWI/SNF and stimulate its ATPase activity (Phelan et al. 1999). In spite of the existence of a large number of chromatin modifiers, they are generally grouped into two distinct classes, ATP-dependent remodeling complex and histone-modification enzymes. Perhaps NUMAC and NURD represent another distinct class of chromatin modifiers, in which the ATP-remodeling complexes are incorporated with histone enzymatic activities to influence the efficiency of histone modification.
A number of biochemically distinct SWI/SNF complexes have been described previously that display tissue specificity or confer a functional selectivity through association with transcription factors (Armstrong et al. 1998; Cho et al. 1998; Bochar et al. 2000; Underhill et al. 2000). In particular, PBAF has been shown to be necessary for ligand-dependent transactivation by several nuclear hormone receptors such as VDR and PPARs (Lemon et al. 2001), suggesting a functional specificity for certain signaling pathways. Our finding that HMT and SWI/SNF cooperatively regulate ER-reporter gene expression further extends this idea by establishing a new level of communication between chromatin remodeling activities and the histone methylation enzymes. Whether NUMAC activates many or a select subset of nuclear receptors is not yet known. We speculate that the NUMAC cascade might function in other pathways, for example, in muscle differentiation, as SWI/SNF has been shown to cooperate with MyoD to regulate muscle differentiation (de la Serna et al. 2001), and CARM1 is also implicated in this process (Chen et al. 2002).
It is known that the central ATPase subunit of SWI/SNF alone has chromatin-remodeling activity, whereas the remaining subunits can modulate reaction efficiency, substrate specificity, and promote targeting. In the best-studied ISWI-containing complexes, the ATPase ISWI subunit alone moves histone octamers to the ends of DNA fragments, whereas the octamer is moved to the center of the DNA in the presence of Acf1 (Langst and Becker 2001). In the SWI/SNF complex, conserved subunits INI1/BAF155/BAF170 stimulate the ATPase activity of BRG1 by three- to sevenfold under submaximal enzyme and substrate conditions (Phelan et al. 1999). The ability of rCARM1 to stimulate the ATPase/remodeling activity of SWI/SNF, and the observation that NUMAC exhibits a different DNase I digestion pattern from that of SWI/SNF in the mononucleosome disruption and restriction digestion assays (Fig. 3A,B; data not shown) suggests that CARM1 might affect both the efficiency and the outcome of remodeling.
Distinct classes of chromatin-modifying activities, consisting of ATP-driven remodeling complexes, histone acetyltransferases (HATs), and histone methyltransferases (HMTs), have evolved to collectively overcome the repressive structure of chromatin in NR-targeted gene activation. One fundamental question remaining is the precise order of coactivator recruitment. In thyroid hormone receptor and retinoic acid receptor signaling, p300 seems to be recruited before SWI/SNF (Dilworth et al. 2000; Huang et al. 2003). Previously, we have observed that either ATP remodeling or HAT activities assist in nucleosomal histone methylation by CARM1 in vitro (Xu et al. 2001), which suggests that histone methylation might preferentially take place on a loose chromatin template that emerges at the later stage(s) of transcriptional activator complex formation. Whether this is true for an endogenous NR-targeted promoter is unclear. Another important question concerns the mechanism by which SWI/SNF complexes are recruited to specific NR-targeted promoters. A widely accepted model is that SWI/SNF is targeted to specific promoters via direct interactions with sequence-specific transcription factors and their coactivators (Peterson and Workman 2000; Hassan et al. 2001). The central ATPase BRG1 and subunits BAF57, BAF60a, and BAF250 have been implicated in mediating the interaction with steroid receptors such as ER and GR (DiRenzo et al. 2000; Nie et al. 2000; Belandia et al. 2002; Hsiao et al. 2003). We suggest that the recruitment of NUMAC to NR-target genes is most likely mediated by the p160 proteins, which associate with CARM1 and NRs through distinct domains. These events appear to be ligand dependent both in vitro and in vivo. This hypothesis is supported by recent observations that blocking the recruitment of p160 family coactivators abrogates chromatin remodeling induced by liganded thyroid receptor (Huang et al. 2003). Thus, direct interaction between CARM1 and BRG1 identifies at least one route for targeting SWI/SNF to specific genes, albeit other BAFs might also participate in this process. Taken together, NR-cofactor, cofactor-cofactor, and cofactor-histone interaction might function in a combinatorial manner to stabilize coactivator complexes assembled on NR-activated promoters.
Materials and methods
Antibodies,cell lines and CARM1 complex purification, and glycerol gradient
Antibodies against BAF170, BAF155, hSNF2h, and Ini-1/hSNF5 were purchased from Santa Cruz-Biotechnology and used for Western blotting. Apyrase was purchased from Sigma-Aldrich. Antibodies against BRG1, BAF57, BAF250, and BAF180 were generous gifts from Weidong Wang (NIH). CARM1 with amino-terminal Flag-tag and carboxy-terminal HA-tag was subcloned into the pLNCX (Clontech) vector, and a stably transfected MCF7 cell line was established as described (Xu et al. 2001). The complex was purified from nuclear extracts with anti-Flag M2 affinity resin (Sigma), washed extensively with 20 mM HEPES (pH 7.9), 150 mM KCl, 0.2 mM EDTA, 0.2 mM EGTA, 0.1% NP-40, 10% glycerol, 1 mM DTT, and protease inhibitors (Roche), and then three times with the same buffer containing 300 mM KCl, and eluted with 0.2 mg/mL Flag peptides in 20 mM Tris (pH 8), 150 mM KCl, 1 mM DTT, and 5% glycerol. Approximately 5 μg of affinity-purified complexes were applied to a 5-mL 5%-30% glycerol gradient (glycerol in 20 mM HEPES at pH 7.9, 0.1 mM EDTA, 50 mM KCl with PMSF) and centrifuged in SW55 rotor at 36,000 rpm for 16 h at 4°C. The 100-μL fractions were collected, either directly analyzed by Western blotting or TCA precipitated, followed by silver-staining.
Cell culture and transient transfections
MCF7 (ATCC HTB-22) and SW13 (ATCC CCL-105) cells were cultured in DMEM supplemented with 10% FBS and L-glutamine at 37°C and 5% CO2. SW13 cells were plated into 24-well dishes in phenol red-free DMEM supplemented with 10% charcoal-stripped FBS 1 d prior to transfection and approached 60% confluence at the time of transfection. Cells were transfected with 10 ng of ERE-TK-luc and the indicated combinations of 50 ng of pCMX-hERα, 200 ng of pBJ5-hBRG1, 200 ng of pBJ5-hBRG1 (K785R), 200 ng of CMX-CARM1, or 200 ng of CMXCARM1(189VLD191→AAA) mutant (Xu et al. 2001) and 100 ng of β-galactosidase constructs. In GAL4-VP16 transient transfection assays, 30 ng of GAL4-VP16, 50 ng of MH2004-TKluc and 100 ng of indicated activators were used. Transfections were performed with Fugene (Roche) according to manufacturer's protocol. The medium was replaced with fresh medium containing 100 nM estradiol or ethanol a day before luciferase levels were measured and normalized with β-gal activities.
GST pull-down and Far-Western assay
Flag-BRG1 (Phelan et al. 1999) and Flag-CARM1 were expressed in a baculovirus expression system and purified using Flag-M2 affinity resin. GST-CARM1, GST-PRMT1, and GST-CARM1 (1-152, 152-340, 340-482, 482-620) are E. Coli-expressed proteins. GST pull-down assay was performed as described (Chen et al. 1999b). BRG1 fragments were PCR amplified with the amino-terminal primer containing T7 and ribosomal binding sites ATTAATACgACTCACTATAggAAACAgACACCATG, followed by the corresponding specific sequence. The PCR products were purified and used directly in TNT-coupled in vitro transcription/translation system (Promega; Inoue et al. 2002). Far-Western assay was performed as described (DiRenzo et al. 2000), except that we used 35S-Met-labeled CARM1 prepared by TNT reaction (50 μL) and free 35S-Met was removed with Bio-spin 30 columns (Bio-Rad).
HMT assay
Protein samples (indicated amount of rCARM1 or 10 μL glycerol gradient fractions) are incubated with 3 μg of free core histones or nucleosomes at 30°C for 60 min in reaction buffer containing 50 mM HEPES (pH 7.9), 5 mM MgCl2, 1 mM DTT, 1 μL3H-S-Adenosyl-L-methionine [3H]SAM, 85 Ci/mmole, and 1.0 mCi/mL; Amersham Pharmacia Biotech). The reaction is stopped by the addition of SDS sample buffer, and the proteins are then separated on a 15% SDS-polyacrylamide gel. The gel is Coomassie stained, destained, and immersed into Amplify solution (Amersham) for 15 min before drying. Usually, the dried gel is exposed to Kodak XAR film overnight, or the histone H3 band is cut out and counted in the presence of scintillation solvent.
ATPase assays
The 15-μL reactions contained 50 mM KCl, 8 mM MgCl2, 10 μM ATP, 3 μg nucleosomes or 2 μg histones or 1 μg of 1-kb DNA fragment and 5% glycerol. The reaction was initiated by adding 1 μL of 1 mCi/mL [γ-32P]ATP (3000 Ci/mmole; Amersham) and 0.3 μg of SWI/SNF or 0.3 μg of NUMAC to bring the reaction volume to 15 μL, and incubated at 30 °C for indicated times. Reactions were stopped at different time points by addition of 5 μL of 100 mM EDTA and 50 mM Tris (pH 7.5). A total of 0.3 μL of reaction was spotted on PEI-cellulose TLC plates (EM science) and run in 0.5 M LiCl and 1 M formic acid. The fraction of free 32P-Pi at each time point was quantitated on a PhosphorImager (Molecular Dynamics).
Mononucleosome disruption assays
Sequence-positioned mononucleosomes (5S) were reconstituted by salt dialysis and purified by sucrose gradient sedimentation as described (Rhodes and Laskey 1989) using purified HeLa core histones at a 1:1 histone:DNA ratio. DNase I footprinting analysis was carried out essentially as described (Kadam and Emerson 2003). Following salt dialysis and sucrose gradient purification, mononucleosomes were incubated with indicated complexes in either the presence (+) or absence (-) of 1 mM ATP for 20 min at 30°C and digested with DNaseI for 1 min. A total of 0.005 U and 0.15 U of DNaseI were used for naked DNA and mononucleosome digestion in 20 μL of reaction, respectively. When rCARM1 was premixed with SWI/SNF, they were incubated at 27°C for 15 min before the addition of 0.1 pmole (5 nM) mononucleosomes.
Quantitation of ATP and protein purification
The quantitative detection of ATP using bioluminescence was performed according to the instructions from the manufacturer (Roche, Cat. #1699695). Baculovirus-expressed Flag-tagged CARM1 was purified as described (Xu et al. 2001). Core histones and nucleosomes were purified from HeLa nuclear pellets by sequential steps, including micrococcal nuclease digestion, sucrose gradient purification, and hydroxyapatite column binding as described (Ausio and van Holde 1986; Laybourn and Kadonaga 1991). The human SWI/SNF complex was purified from HeLa cells stably expressing Flag-INI-1 subunit (Phelan et al. 1999). All protein concentrations were determined by Bradford assay (Bio-Rad) and normalized by Western blotting when indicated. Serial dilutions of BRG1 and CARM1 were normalized by Western blotting to obtain equal amounts of BRG1 in SWI/SNF versus NUMAC or CARM1 in NUMAC versus free form.
Acknowledgments
We thank Dr. Robert Kingston (Massachusetts General Hospital) for p2085S-G5E4 plasmid and his permission to use Flag-INI1 stable cell line and National Cell Culture Center for services. We thank Dr. Weidong Wang (National Institutes of Health) for pBJ5-BRG1 plasmids and BRG1 and BAF antibodies, and Drs. F. Dmitry and J. Kadonaga (University of California, San Diego) for providing recombinant viruses for expression of ACF/ISWI. We thank Dr. N. Tanese (New York University School of Medicine) for instruction in designing PCR products for in vitro translation. We thank H. Juguilon and G. Pfeifer for technical help, L. Ong and E. Stevens for administrative assistance, and C. Park and F. Wolfgang for the peptide analysis. We thank H. Chen, R. Yu, S. Hong, and T. Sternsdorf for critical reading of the manuscript. R.M.E. is an investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1141704.
References
- Aranda A. and Pascual, A. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81: 1269-1304. [DOI] [PubMed] [Google Scholar]
- Armstrong J.A., Bieker, J.J., and Emerson, B.M. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93-104. [DOI] [PubMed] [Google Scholar]
- Ausio J. and van Holde, K.E. 1986. Histone hyperacetylation: Its effects on nucleosome conformation and stability. Biochemistry 25: 1421-1428. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Schneider, R., and Kouzarides, T. 2002. Histone methylation: Dynamic or static? Cell 109: 801-806. [DOI] [PubMed] [Google Scholar]
- Bauer U.M., Daujat, S., Nielsen, S.J., Nightingale, K., and Kouzarides, T. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3: 39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P.B. 2002. Nucleosome sliding: Facts and fiction. EMBO J. 21: 4749-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B., Orford, R.L., Hurst, H.C., and Parker, M.G. 2002. Targeting of SWI/SNF chromatin remodeling complexes to estrogen-responsive genes. EMBO J. 21: 4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar D.A., Wang, L., Beniya, H., Kinev, A., Xue, Y., Lane, W.S., Wang, W., Kashanchi, F., and Shiekhattar, R. 2000. BRCA1 is associated with a human SWI/SNF-related complex: Linking chromatin remodeling to breast cancer. Cell 102: 257-265. [DOI] [PubMed] [Google Scholar]
- Burakov D., Crofts, L.A., Chang, C.P., and Freedman, L.P. 2002. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J. Biol. Chem. 277: 14359-14362. [DOI] [PubMed] [Google Scholar]
- Chen D., Ma, H., Hong, H., Koh, S.S., Huang, S.M., Schurter, B.T., Aswad, D.W., and Stallcup, M.R. 1999a. Regulation of transcription by a protein methyltransferase. Science 284: 2174-2177. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin, R.J., Xie, W., Wilpitz, D., and Evans, R.M. 1999b. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98: 675-686. [DOI] [PubMed] [Google Scholar]
- Chen S.L., Loffler, K.A., Chen, D., Stallcup, M.R., and Muscat, G.E. 2002. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 277: 4324-4333. [DOI] [PubMed] [Google Scholar]
- Cho H., Orphanides, G., Sun, X., Yang, X.J., Ogryzko, V., Lees, E., Nakatani, Y., and Reinberg, D. 1998. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol. 18: 5355-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S., Bauer, U.M., Shah, V., Turner, B., Berger, S., and Kouzarides, T. 2002. Crosstalk between CARM1 Methylation and CBP Acetylation on Histone H3. Curr. Biol. 12: 2090-2097. [DOI] [PubMed] [Google Scholar]
- de la Serna I.L., Carlson, K.A., and Imbalzano, A.N. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27: 187-190. [DOI] [PubMed] [Google Scholar]
- Dilworth F.J. and Chambon, P. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20: 3047-3054. [DOI] [PubMed] [Google Scholar]
- Dilworth F.J., Fromental-Ramain, C., Yamamoto, K., and Chambon, P. 2000. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol. Cell 6: 1049-1058. [DOI] [PubMed] [Google Scholar]
- DiRenzo J., Shang, Y., Phelan, M., Sif, S., Myers, M., Kingston, R., and Brown, M. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20: 7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A., Ferrari, S., Langst, G., Straub, T., Imhof, A., Varga-Weisz, P., Wilm, M., and Becker, P.B. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodeling. EMBO J. 20: 3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C.J. and Archer, T.K. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393: 88-91. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan, L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 11: 1640-1650. [DOI] [PubMed] [Google Scholar]
- Hassan A.H., Neely, K.E., Vignali, M., Reese, J.C., and Workman, J.L. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6: D1054-D1064. [DOI] [PubMed] [Google Scholar]
- Hsiao P.W., Fryer, C.J., Trotter, K.W., Wang, W., and Archer, T.K. 2003. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 23: 6210-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.Q., Li, J., Sachs, L.M., Cole, P.A., and Wong, J. 2003. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. EMBO J. 22: 2146-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H., Garnier, J.M., Chambon, P., and Losson, R. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188: 95-100. [DOI] [PubMed] [Google Scholar]
- Inoue H., Furukawa, T., Giannakopoulos, S., Zhou, S., King, D.S., and Tanese, N. 2002. Largest subunits of the human SWI/SNF chromatin remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 277: 41674-41685. [DOI] [PubMed] [Google Scholar]
- Kadam S. and Emerson, B.M. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11: 377-389. [DOI] [PubMed] [Google Scholar]
- Koh S.S., Chen, D., Lee, Y.H., and Stallcup, M.R. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276: 1089-1098. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12: 198-209. [DOI] [PubMed] [Google Scholar]
- Langst G. and Becker, P.B. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell. Sci. 114: 2561-2568. [DOI] [PubMed] [Google Scholar]
- Laybourn P.J. and Kadonaga, J.T. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254: 238-245. [DOI] [PubMed] [Google Scholar]
- Lemon B., Inouye, C., King, D.S., and Tjian, R. 2001. Selectivity of chromatin-remodeling cofactors for ligand-activated transcription. Nature 414: 924-928. [DOI] [PubMed] [Google Scholar]
- Logie C. and Peterson, C.L. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 16: 6772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Baumann, C.T., Li, H., Strahl, B.D., Rice, R., Jelinek, M.A., Aswad, D.W., Allis, C.D., Hager, G.L., and Stallcup, M.R. 2001. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 11: 1981-1985. [DOI] [PubMed] [Google Scholar]
- McBride A.E. and Silver, P.A. 2001. State of the arg: Protein methylation at arginine comes of age. Cell 106: 5-8. [DOI] [PubMed] [Google Scholar]
- McKenna N.J. and O'Malley, B.W. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108: 465-474. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan, H.Y., and Kingston, R.E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475-487. [DOI] [PubMed] [Google Scholar]
- Neely K.E., Hassan, A.H., Wallberg, A.E., Steger, D.J., Cairns, B.R., Wright, A.P., and Workman, J.L. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4: 649-655. [DOI] [PubMed] [Google Scholar]
- Nie Z., Xue, Y., Yang, D., Zhou, S., Deroo, B.J., Archer, T.K., and Wang, W. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20: 8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K., Rice, J.C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., Chuikov, S., Valenzuela, P., Tempst, P., Steward, R., et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9: 1201-1213. [DOI] [PubMed] [Google Scholar]
- Olave I.A., Reck-Peterson, S.L., and Crabtree, G.R. 2002. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71: 755-781. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. 2002. Chromatin remodeling enzymes: Taming the machines. Third in review series on chromatin dynamics. EMBO Rep. 3: 319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L. and Workman, J.L. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. De. 10: 187-192. [DOI] [PubMed] [Google Scholar]
- Phelan M.L., Sif, S., Narlikar, G.J., and Kingston, R.E. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3: 247-253. [DOI] [PubMed] [Google Scholar]
- Rhodes D. and Laskey, R.A. 1989. Assembly of nucleosomes and chromatin in vitro. Methods Enzymol. 170: 575-585. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu, X., DiRenzo, J., Lazar, M.A., and Brown, M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843-852. [DOI] [PubMed] [Google Scholar]
- Sif S., Saurin, A.J., Imbalzano, A.N., and Kingston, R.E. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes & Dev. 15: 603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup M.R. 2001. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20: 3014-3020. [DOI] [PubMed] [Google Scholar]
- Travers A. 1999. An engine for nucleosome remodeling. Cell 96: 311-314. [DOI] [PubMed] [Google Scholar]
- Underhill C., Qutob, M.S., Yee, S.P., and Torchia, J. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275: 40463-40470. [DOI] [PubMed] [Google Scholar]
- Washburn M.P., Wolters, D., and Yates III, J.R. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19: 242-247. [DOI] [PubMed] [Google Scholar]
- Westin S., Rosenfeld, M.G., and Glass, C.K. 2000. Nuclear receptor coactivators. Adv. Pharmacol. 47: 89-112. [DOI] [PubMed] [Google Scholar]
- Xu W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B.M., Montminy, M., and Evans, R.M. 2001. A transcriptional switch mediated by cofactor methylation. Science 294: 2507-2511. [DOI] [PubMed] [Google Scholar]
- Xue Y., Wong, J., Moreno, G.T., Young, M.K., Cote, J., and Wang, W. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2: 851-861. [DOI] [PubMed] [Google Scholar]