Abstract
Chromatin assembly is required for the duplication of chromosomes. ACF (ATP-utilizing chromatin assembly and remodeling factor) catalyzes the ATP-dependent assembly of periodic nucleosome arrays in vitro, and consists of Acf1 and the ISWI ATPase. Acf1 and ISWI are also subunits of CHRAC (chromatin accessibility complex), whose biochemical activities are similar to those of ACF. Here we investigate the in vivo function of the Acf1 subunit of ACF/CHRAC in Drosophila. Although most Acf1 null animals die during the larval-pupal transition, Acf1 is not absolutely required for viability. The loss of Acf1 results in a decrease in the periodicity of nucleosome arrays as well as a shorter nucleosomal repeat length in bulk chromatin in embryos. Biochemical experiments with Acf1-deficient embryo extracts further indicate that ACF/CHRAC is a major chromatin assembly factor in Drosophila. The phenotypes of flies lacking Acf1 suggest that ACF/CHRAC promotes the formation of repressive chromatin. The acf1 gene is involved in the establishment and/or maintenance of transcriptional silencing in pericentric heterochromatin and in the chromatin-dependent repression by Polycomb group genes. Moreover, cells in animals lacking Acf1 exhibit an acceleration of progression through S phase, which is consistent with a decrease in chromatin-mediated repression of DNA replication. In addition, acf1 genetically interacts with nap1, which encodes the NAP-1 nucleosome assembly protein. These findings collectively indicate that ACF/CHRAC functions in the assembly of periodic nucleosome arrays that contribute to the repression of genetic activity in the eukaryotic nucleus.
Keywords: ACF, ISWI, CHRAC, chromatin assembly, position-effect variegation, cell cycle
Eukaryotic DNA is packaged into a periodic nucleoprotein complex termed chromatin (van Holde 1989; Wolffe 1995). The nucleosome is the basic repeating unit of chromatin, and the nucleosomal core consists of 146 bp of DNA wrapped around an octamer of histones H2A, H2B, H3, and H4 (Luger et al. 1997). In addition to the core histones, chromatin contains other components such as linker histones and high mobility group proteins. Chromatin is involved in the regulation of transcription and other DNA-directed processes via posttranslational modifications of core histones (Grunstein 1997; Strahl and Allis 2000; Rice and Allis 2001; Vaquero et al. 2003), the reorganization of nucleosomes by chromatin remodeling factors (Kingston and Narlikar 1999; Kornberg and Lorch 1999), and the alteration of higher-order structures (Wallrath 1998; Henikoff 2000).
The assembly of chromatin is a fundamental biological process that occurs in proliferating cells during DNA replication and in quiescent cells during maintenance and repair of chromosomes (for recent reviews, see Verreault 2000; Mello and Almouzni 2001; Tyler 2002; Haushalter and Kadonaga 2003). During DNA replication, chromatin structure is transiently disrupted at the replication fork (Gruss et al. 1993), and the preexisting nucleosomes are segregated randomly between the daughter DNA strands (Sogo et al. 1986). Then, additional nucleosomes are formed with newly synthesized histones. In this process, it appears that histones H3 and H4 are deposited prior to the incorporation of histones H2A and H2B (Worcel et al. 1978; Smith et al. 1984). Chromatin assembly also occurs in nonreplicating DNA, and several examples of replication-independent assembly of chromatin have been described (Shelby et al. 2000; Ahmad and Henikoff 2002). These latter processes may occur during histone replacement, DNA repair, and transcription.
The basic chromatin assembly process is mediated by core histone chaperones and an ATP-utilizing motor protein. The histone chaperones include CAF-1 (chromatin assembly factor-1), NAP-1 (nucleosome assembly protein-1), Asf1 (anti-silencing function-1), nucleoplasmin, N1/N2, and Hir (histone regulatory) proteins. These proteins appear to deliver the histones from the cytoplasm to the sites of chromatin assembly in the nucleus. The ATP-utilizing assembly factor ACF (ATP-utilizing chromatin assembly and remodeling factor) can catalyze the transfer of histones from the chaperones to the DNA to yield periodic nucleosome arrays. The assembly reaction can also be catalyzed by purified RSF (remodeling and spacing factor), which appears to possess both chaperone and motor activities (Loyola et al. 2001).
In this work, we investigate the biological function of ACF. ACF was purified from Drosophila embryos as an activity that mediates the ATP-dependent assembly of regularly spaced nucleosome arrays in vitro (Ito et al. 1997). During the assembly process, ACF commits to and translocates along the DNA template (Fyodorov and Kadonaga 2002b). ACF consists of two subunits, Acf1 and ISWI (Ito et al. 1999), which cooperatively catalyze nucleosome assembly in conjunction with histone chaperone proteins NAP-1 or CAF-1 (Ito et al. 1997, 1999). Acf1 is the larger subunit of ACF, and it possesses WAC, DDT, WAKZ, PHD finger, and bromo-domain motifs (Ito et al. 1999; Fyodorov and Kadonaga 2002a). ISWI belongs to the SNF2-like family of DNA-dependent ATPases (Gorbalenya and Koonin 1993; Elfring et al. 1994; Eisen et al. 1995), and is a subunit of the ACF, CHRAC (chromatin accessibility complex), NURF, and TRF2 complexes. NURF and TRF2 complexes share only the ISWI subunit with ACF (Xiao et al. 2001; Hochheimer et al. 2002), whereas CHRAC is closely related to ACF. CHRAC was purified on the basis of its ability to increase the access of restriction enzymes to DNA in chromatin (Varga-Weisz et al. 1997), and it consists of Acf1, ISWI, and two small subunits, CHRAC-14 and CHRAC-16, which are detected only during early embryonic development (Corona et al. 2000). The biochemical activities of ACF and CHRAC are indistinguishable. We will refer to these Acf1-containing species as “ACF/CHRAC.” To study the function of ACF/CHRAC in vivo, we carried out a genetic analysis of the Drosophila acf1 gene. The results indicate that Acf1 programs ACF/CHRAC to perform functions that are distinct from those of the NURF complex, which shares a common ISWI ATPase subunit with ACF/CHRAC. In addition, the phenotypes of flies lacking Acf1 suggest that ACF/CHRAC does not disrupt chromatin, as might be expected for a nucleosome remodeling factor, but rather promotes the formation of chromatin, as would be expected for a chromatin assembly factor.
Results
Isolation and characterization of a null allele of acf1
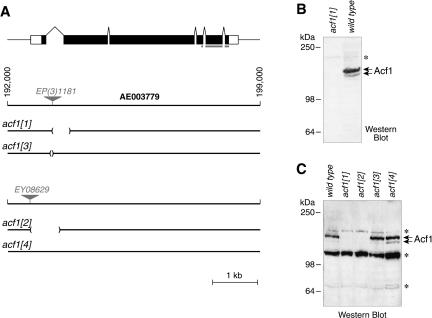
The gene that encodes Acf1 maps to the cytological interval 100E1–100E2 in the right arm of Drosophila chromosome 3. To investigate the function of ACF in vivo, we generated deletions in the acf1 gene by imprecise excision of the P-elements EP(3)1181 and EY08629 (Fig. 1A). We then analyzed the resulting alleles by PCR with primers that flank the sites of insertions. In this study, we used three alleles with small deletions in the acf1 gene. In Df(3R)acf11, there is a 499-bp deletion that encompasses part of the first intron, the splice acceptor site, and part of the second exon. In Df(3R)acf12, an 871-bp deletion eliminates the promoter region, the translation initiation codon, the first exon, and most of the first intron of acf1. In Df(3R)acf13, 83 bp of sequence entirely within the first intron of the gene are deleted. Finally, acf14 is a precise excision of the EY08629 insertion (Fig. 1A).
Figure 1.
Generation of null alleles of the acf1 gene. (A) The Drosophila acf1 gene. The structure of the acf1 gene and the location of the transposon insertions in EP(3)1181 and EY08629 are shown. The acf11 allele was generated by imprecise excision of acf1EP(3)1181, in which 499 bp of the first intron and second exon has been replaced by an 11-bp sequence from the P-element. The allele acf13 was generated by imprecise excision of acfEP(3)1181, in which 83 bp entirely within the first intron have been replaced by a 15-bp sequence from the P-element. The acf12 allele, in which 871 bp of the acf1 promoter and 5′ transcribed region is deleted, was generated by imprecise excision of acf1EY08629. The acf14 allele was generated by precise excision of acf1EY08629. AE003779 refers to the contig number of the Drosophila genome project (Adams et al. 2000). The noncoding and coding regions of the acf1 transcript are depicted as open and filled boxes, respectively. The gray bar indicates the Acf1 polypeptide that was used to prepare anti-Acf1 polyclonal antibodies. (B) Western blot analysis reveals that acf11 is a null allele. Nuclear extracts derived from 0- to 12-h embryos of homozygous acf11 flies or wild-type (Canton S) flies were subjected to Western blot analysis with anti-Acf1 antibodies. (C) Western analysis of Acf1 expression in embryos of acf1 alleles. Crude lysates of 0- to 12-h embryos (Ito et al. 1999) of homozygous wild-type, acf11, acf12, acf13, and acf14 flies were subjected to Western blot analysis with anti-Acf1 antibodies. Acf1 is not expressed in acf11 or acf12 embryos, but its expression is not significantly affected in acf13 or acf14 embryos. Asterisks indicate nonspecific bands.
Acf1 is strongly expressed in wild-type embryos between 0 and 12 h of development, but Acf1 polypeptide cannot be detected in homozygous embryos derived from homozygous acf11 parents (Fig. 1B) or acf12 parents (Fig. 1C). In contrast, the levels of Acf1 in acf13/acf13 or acf14/acf14 embryos appear to be the same as that in wild-type embryos (Fig. 1C). Therefore, acf11 and acf12 are null mutant alleles of acf1 gene, whereas acf13 and acf14 can be used as isogenic wild-type controls for acf11 and acf12, respectively. In this work, we have focused mainly on the characterization of the acf11 null allele in conjunction with the acf13 control allele.
acf1 homozygosity causes semilethality and delayed development, but adult “escapers” are fertile and pheno-typically normal. Only about 60% of the expected number of homozygous acf1 adults were produced after inter se matings of acf11/TM3 (n = 150, P < 0.001) or acf12/TM3 heterozygous parents (n = 100, P < 0.001). However, viable flies could have arisen due to maternal contribution of Acf1 from the TM3 balancer chromosome; for this reason, all subsequent experiments described here used progeny from homozygous acf1 mothers. The lethality of homozygous acf11 animals increased when crosses were performed with homozygous parents: 75% of acf11 larvae failed to pupariate and died in the third larval stage (L3). In addition, surviving acf11 animals committed to pupariation 1 to 2 d later than their wild-type (acf13) counterparts (data not shown). However, viable adult escapers were produced (25% of expected), and did not display significant defects in ovodeposition, larva hatching, or adult eclosion. We conclude that complete loss of Acf1 function results in significant lethality during the transition from larvae to pupae, as well as developmental delay, but that Acf1 is not absolutely essential for viability or fertility. It is likely that Acf1 null animals can proceed to L3, and even to adulthood, because of the presence of redundant factors (see following).
The homozygous mutation of ISWI, which encodes the ATPase subunit of ACF/CHRAC, was observed to result in lethality at late L3 as well as a gross alteration in the structure of the male X chromosome (Deuring et al. 2000). In contrast, flies lacking Acf1 are semilethal and fertile and do not exhibit any morphological abnormalities in larval polytene chromosomes or in mitotic chromosomes in syncytial embryos or larval neuroblasts (data not shown). It is important to note, however, that ISWI is also a subunit of multiprotein complexes other than ACF/CHRAC. For instance, the NURF remodeling factor is a complex of ISWI with NURF301, NURF55, and NURF38 (Xiao et al. 2001). Mutation of nurf301 results in phenotypes, such as the L3 lethality and abnormal male X chromosome, that are essentially identical to those seen on mutation of ISWI (Badenhorst et al. 2002). These results reveal that ACF/CHRAC performs different biological functions than NURF, even though ACF/CHRAC and NURF both share a common ISWI ATPase. Thus, the unique subunits of ACF/CHRAC and NURF can program the ISWI ATPase to perform distinct functions in vivo.
ACF/CHRAC is a major ATP-dependent chromatin assembly factor in Drosophila embryos
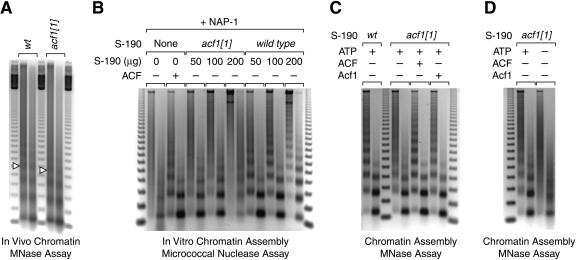
Because ACF/CHRAC is a chromatin assembly factor in vitro, we examined whether the mutation of the acf1 gene results in aberrant nucleosomal structure in vivo. To this end, we isolated nuclei from wild-type embryos and homozygous acf1 embryos derived from homozygous acf11 parents. The nuclear chromatin was digested in situ by micrococcal nuclease at two different concentrations, and the resulting partially digested DNA fragments were resolved by agarose gel electrophoresis (Fig. 2A). These experiments revealed a distinct decline in the periodicity of the nucleosome arrays in chromatin from the acf1 embryos relative to chromatin from wild-type embryos, as seven to eight bands can be seen in chromatin from wild-type embryos, whereas only four to five bands can be seen in the chromatin from the acf1 mutant embryos. In addition, the loss of Acf1 causes a significant decrease in the nucleosome repeat length (from 184 nt to 172 nt; compare locations of the open triangles that indicate tetranucleosome bands in Fig. 2A). Thus, in the absence of Acf1, chromatin in embryonic nuclei is less periodic and exhibits reduced average nucleosomal spacing. It is relevant to note that nucleosomal periodicity has been found to correlate with heterochromatin structure and transcriptional repression (see, for example, Vega-Palas et al. 1998; Weiss and Simpson 1998; Cryderman et al. 1999b). In addition, the nucleosome repeat length has been observed to be inversely correlated with genetic activity (see, for example, Compton et al. 1976; Lohr et al. 1977; Thomas and Thompson 1977). Hence, the properties of chromatin derived from the acf1 mutant embryos support a role for Acf1 in the assembly of repressive chromatin in vivo.
Figure 2.
Mutation of acf1 reduces chromatin assembly activity in embryos. (A) Mutation of acf1 results in reduced nucleosomal periodicity as well as a shorter nucleosome repeat length in Drosophila embryos. Nuclear chromatin from 0- to 12-h embryos was partially digested in situ with micrococcal nuclease by using two different concentrations of enzyme. The resulting partially digested DNA was analyzed by agarose gel electrophoresis and stained with ethidium bromide. The open triangles indicate DNA fragments derived from tetranucleosomes. (B) ACF/CHRAC is the major ATP-dependent chromatin assembly factor in Drosophila S-190 embryo extracts. S-190 extracts were prepared from wild-type and acf1 null embryos. Chromatin was assembled on relaxed plasmid DNA in the presence of the Drosophila core histone chaperone NAP-1. The source of ATP-dependent chromatin assembly activity was either recombinant ACF or the indicated amounts of the S-190 extracts. The reaction products were analyzed by using the micrococcal nuclease digestion assay, in which two different concentrations of the enzyme were used in the analysis of each assembly reaction. (C) ATP-dependent chromatin assembly activity in Acf1-deficient extracts can be restored by biochemical complementation with either purified recombinant ACF complex or purified recombinant Acf1 polypeptide. Chromatin was assembled in the presence of purified recombinant NAP-1 and either wild-type or Acf1-deficient S-190 extract (100 μg). Where indicated, reactions contained purified recombinant ACF or Acf1 (110 fmole). (D) The residual chromatin assembly activity in Acf1-deficient extracts is dependent on ATP. Chromatin was assembled with purified recombinant NAP-1 and Acf1-deficient S-190 extract (100 μg) in the presence or absence of 3 mM ATP (and ATP-regeneration system).
We also investigated whether the loss of Acf1 affects chromatin assembly activity in vitro. The ATP-dependent chromatin assembly activity of ACF was originally derived from a Drosophila embryo extract termed the S-190 (Kamakaka et al. 1993). We therefore prepared S-190 extracts from homozygous acf1 null embryos generated from acf11 homozygous parents, and compared the ability of Acf1-deficient and wild-type extracts to assemble nucleosome arrays in vitro (Fig. 2B). The micrococcal nuclease digestion assay was used to monitor the ATP-dependent assembly of periodic nucleosome arrays. In these experiments, we supplemented the reactions with purified recombinant Drosophila NAP-1 to ensure that core histone chaperone activity was not limiting. We observed that the Acf1-deficient extracts assemble nucleosomes much less efficiently than do wild-type extracts (Fig. 2B). In fact, extended nucleosome arrays could not be assembled with the Acf1-deficient extracts. Addition of purified recombinant ACF or Acf1 polypeptide to the assembly reactions restored full activity to the acf11 mutant embryo extracts (Fig. 2C). (The rescue of chromatin assembly activity by Acf1 alone is presumed to be due to the formation of ACF via the association of the exogenously added Acf1 with the endogenous ISWI in the mutant S-190 extracts.) Thus, the inability of the Acf1-deficient extracts to assemble extended nucleosome arrays is due to the absence of Acf1. We conclude that ACF/CHRAC is the major ATP-dependent chromatin assembly activity in Drosophila embryos.
The viability of flies lacking Acf1 suggests that there is at least one additional ACF-like factor that mediates chromatin assembly in the absence of Acf1. Consistent with this hypothesis, we found that the residual chromatin assembly activity in the Acf1-deficient extracts is dependent on ATP (Fig. 2D). One candidate for such an assembly factor is a potential Drosophila homolog of human RSF, which contains an ISWI-related ATPase subunit (hSNF2H) and has been shown to mediate the ATP-dependent assembly of nucleosome arrays (Loyola et al. 2001). Chromatin assembly might also be catalyzed by the putative Drosophila version of mammalian NoRC, which is a four-subunit complex that also contains the hSNF2H ATPase (Strohner et al. 2001; Santoro et al. 2002). In addition, it is possible that ATP-utilizing factor(s) that do not contain ISWI are responsible for the residual chromatin assembly activity in Acf1-deficient embryos.
acf1 genetically interacts with ISWI and nap1
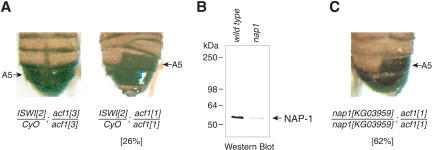
To investigate whether other ISWI-containing factors might assemble chromatin in a manner that is similar to that of ACF/CHRAC, we examined whether there are genetic interactions between acf1 and ISWI. To this end, we created fly lines that contained heterozygous ISWI1 or ISWI2 mutations (Deuring et al. 2000) along with homozygous acf11 or acf13 mutations, and monitored phenotypes associated with the simultaneous disruption of ISWI and Acf1 functions. Heterozygous ISWI/CyO flies, homozygous acf11/acf11 flies, double mutant ISWI1/CyO; acf13/acf13 flies, and double mutant ISWI2/CyO; acf13/acf13 flies exhibit normal segmentation of the abdomen (Fig. 3A, left panel and data not shown). In contrast, double mutant ISWI1/CyO; acf11/acf11 flies and double mutant ISWI2/CyO; acf11/acf11 flies exhibit a distinct homeotic transformation in the abdomen of males. This phenotype is manifested as ectopic reduced pigmentation of the abdominal A5 tergites, consistent with an A5 to A4 anterior transformation (Fig. 3A, right panel). The penetrance of this transformation is about 26% in the ISWI2/CyO; acf11/acf11 line. In addition, the double mutant male flies often display an aberrant, asymmetric segmentation of the lower abdomen (A3 through A5; Fig. 3A, right panel). Both ISWI alleles interacted with acf11 in the same manner, and the acf13 control flies did not exhibit interactions with ISWI.
Figure 3.
acf1 interacts with the ISWI and nap-1 genes. (A) acf1 genetically interacts with ISWI. Adult males of the double ISWI2/CyO; acf11/acf11 line exhibit homeotic transformation and aberrant segmentation of the abdomen. The phenotype penetrance is 26/100 scored individuals. This phenotype is not observed in single gene mutants or in ISWI2/CyO; acf13/acf13 flies. (CyO) Inversion balancer of the second chromosome carrying the Curly mutation. (B) nap1KG03959 is a hypomorphic allele of nap1. Total protein was prepared from L3 larvae with homozygous KG03959 (nap1KG03959) or unrelated KG06618 (control with wild-type nap1 gene) insertions, and the presence of NAP-1 was detected by Western blot analysis. (C) acf11 genetically interacts with nap1. Adult males of the double homozygous nap1KG03959/nap1KG03959; acf11/acf11 line exhibit homeotic transformation and aberrant segmentation of the abdomen. The phenotype penetrance is 62/100 scored individuals. This phenotype is not observed in single homozygous mutants or in nap1KG03959/nap1KG03959; acf13/acf13 flies.
These findings indicate that Acf1 and at least one other ISWI-containing factor affect fly segment identity. It is possible, for instance, that the homeotic transformation is due to the combined loss of assembly activity that results from both the mutation of acf1 and the dose reduction of ISWI (which appears to function in multiple ATP-utilizing assembly complexes). It should additionally be noted that the asymmetry of abdominal tergites that we see in the ISWI-acf1 double mutants is a somewhat commonly observed phenomenon (Bownes 1976; Madhavan and Madhavan 1980), and probably arises from cell death or proliferation defects in dorsal histoblast nests (Roseland and Schneiderman 1979).
If ACF/CHRAC functions in parallel with other ATP-dependent factors in the assembly of chromatin, it is possible that a similar phenotype would result from the reduction in assembly activity by the combined mutation of acf1 and a gene encoding a core histone chaperone that is involved in nucleosome assembly. In vitro, chromatin assembly by ACF/CHRAC requires the NAP-1 chaperone (Ito et al. 1997). We therefore examined the effect of mutation of both the acf1 and nap1 genes. The nap1KG03959 line contains a P-element inserted into the regulatory region of the nap1 gene. Individual homozygous nap1KG03959 adult flies are viable and phenotypically normal. Western blot analysis indicated that NAP-1 expression is ∼10-fold lower in nap1KG03959/nap1KG03959 L3 larvae compared with the corresponding wild-type larvae (Fig. 3B). Thus, nap1KG03959 is a hypomorphic allele of nap1. Double homozygous nap1KG03959/nap1KG03959; acf11/acf11 flies exhibit an adult male anterior transformation phenotype that is indistinguishable from that of ISWI2/CyO; acf11/acf11 flies (Fig. 3, cf. A, right panel, and C). The penetrance of the phenotype was found to be 62%. Homozygous nap1KG03959/nap1KG03959 flies, homozygous acf11/acf11 flies, and nap1KG03959/nap1KG03959; acf13/acf13 flies displayed normal abdominal segment identity (data not shown). These experiments thus reveal genetic interactions between acf1 and nap1. ACF (a motor protein) and NAP-1 (a histone chaperone involved in nucleosome assembly) have different biochemical functions. Unlike ACF/CHRAC, NAP-1 is not an ATP-dependent chromatin remodeling factor. Moreover, ACF/CHRAC and NAP-1 are known to cooperate in chromatin assembly in vitro (Ito et al. 1999). Therefore, the genetic interaction between acf1 and nap1 provides independent biological support for the role of ACF/CHRAC in chromatin assembly in conjunction with NAP-1 in vivo.
acf1 is a suppressor of position-effect variegation
To assess further whether ACF/CHRAC influences chromatin structure in vivo, we tested the effect of Acf1 on position-effect variegation (PEV), which is caused by chromatin-dependent transcriptional silencing. If ACF/CHRAC was involved in the disruption or destabilization of chromatin structure, then it would be expected that the loss of Acf1 would result in an enhancement of PEV. On the other hand, if ACF/CHRAC was involved in the formation of chromatin, as might be expected for a chromatin assembly factor, then it would be expected that the loss of Acf1 would result in a suppression of PEV.
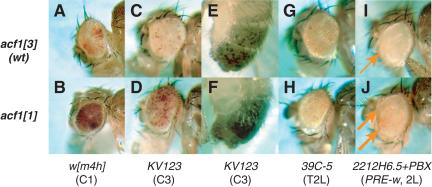
To test these possibilities, we analyzed the heterochromatic silencing displayed by In(1)wm4h. In wm4h, the white gene is partially repressed (variegated) because of an inversion in the X chromosome that places the gene in the proximity of pericentric heterochromatin (Fig. 4A; Tartof et al. 1989). In homozygous wm4h; acf11/acf11 flies, the white gene is strongly derepressed (Fig. 4B) compared with wild-type control flies (wm4h; acf13/acf13; Fig. 4A). PEV is not suppressed in crosses with heterozygous acf11/+ parents. Thus, acf1 is a recessive suppressor of PEV. This finding supports a role of ACF/CHRAC in the assembly of chromatin in vivo.
Figure 4.
acf1 contributes to Polycomb silencing and pericentric position-effect variegation (PEV). (A) Eye phenotypes of flies with In(1)wm4h in homozygous wild type control (acf13) genetic background. (B) Eye phenotypes of flies with In(1)wm4h in homozygous acf1 null (acf11) genetic backgrounds. Homozygous acf1 null flies exhibit strong suppression of PEV. (C,D) Eye phenotypes of flies with pericentric (C3) white transgene KV123 in heterozygous wild-type control (acf13) and acf1 null (acf11) genetic backgrounds. In E to H, flies were produced in a cross between y; KV123/KV123 males and yw/yw; acf1/acf1 females. The offspring of homozygous acf1 null mothers exhibit strong suppression of PEV. (E,F) Abdomen pigmentation phenotypes of flies with pericentric (C3) yellow transgene KV123 in heterozygous wild-type control (acf13) and acf1 null (acf11) genetic backgrounds. (G,H) Eye phenotypes of flies with a telomeric (T2L) hsp70-white transgene (39C-5) insertion (Cryderman et al. 1999a) in wild-type control (acf13) and acf1 null (acf11) genetic backgrounds. Telomeric PEV is not affected by loss of acf1. Similar results were obtained for insertions 39C-27 in T2R and 39C-72 in T4 (data not shown). (I,J) Eye phenotype of flies with homozygous insertion of 2212H6.5+PBX (PRE-white) transposon on the second chromosome (Chan et al. 1994) in homozygous wild-type control (acf13) and homozygous acf1 null (acf11) genetic backgrounds, respectively. Weak variegated expression of the white transgene is indicated by arrows. The PRE-white transgene is derepressed in acf1 null flies. In A, B, and G–J, representative phenotypes of 6- to 7-day-old females are shown. In C–F, representative phenotypes of 6- to 7-day-old males are shown. (Left column) Genetic backgrounds of the flies. (Bottom line) Names of stocks and location of aberrations/transposon inserts. (2L) Left arm of the second chromosome; (C1) centromere of the X chromosome; (C3) centromere of the third chromosome; (T2L) telomere of the left arm of the second chromosome; (T2R) telomere of the right arm of the second chromosome; (T4) telomere of the fourth chromosome; (PRE) Polycomb response element.
PEV can be affected by alterations in the timing of Drosophila development. For instance, mutations or environmental conditions (temperature, crowding, chemicals) that lead to a developmental delay have been observed to enhance variegation (Michailidis et al. 1988). Although the pupal development of homozygous acf1 flies is delayed relative to that of their wild-type counterparts, we observe suppression rather than enhancement of PEV on mutation of acf1. It thus appears that the suppression of PEV that results from the mutation of acf1 is probably not due to the developmental delay in flies lacking Acf1.
We further tested the effect of Acf1 on PEV associated with a different chromosome and reporter gene. In these experiments, we crossed males that carry a homozygous P-element insertion (Yan et al. 2002) in the pericentric region of chromosome 3 (P{SUPor-P}KV123) with yw/yw; acf11/acf11 or yw/yw; acf13/acf13 females. The P-element in the KV123 line contains variegating white and yellow genes (Fig. 4C,E; Yan et al. 2002). In the absence of maternal Acf1, the variegation of both white and yellow was suppressed (Fig. 4, cf. C,D and E,F). Thus, Acf1 contributes to pericentric PEV in different chromosomes with different reporter genes. We also investigated whether Acf1 influences telomeric PEV, and found that mutation of acf1 does not affect PEV with P-elements located in telomeric regions of chromosomes 2 and 4 (Fig. 4G,H; data not shown). Hence, Acf1 appears to suppress PEV in pericentric but not telomeric loci, as previously observed for almost all known suppressors and enhancers of PEV (Cryderman et al. 1999a; Donaldson et al. 2002). The involvement of acf1 in pericentric PEV suggests that ACF/CHRAC contributes to the assembly and/or maintenance of pericentric heterochromatin.
acf1 contributes to the establishment of Polycomb-dependent repressive chromatin
Polycomb regulation is another phenomenon that is caused by chromatin-dependent transcriptional silencing. The identity of body segments in Drosophila is specified by homeotic genes of the Antennapedia and bithorax complexes, which are in turn subject to regulation by Polycomb and trithorax group (PcG and trxG) genes (Kennison 1995; Pirrotta 1998). PcG genes encode protein complexes that can maintain chromatin-dependent transcriptional silencing via cis-acting DNA elements termed Polycomb response elements, or PREs (Jürgens 1985; Simon et al. 1993; Francis and Kingston 2001).
To determine the influence of Acf1 on Polycomb regulation, we tested whether the loss of Acf1 affects transcriptional repression by the Ubx PRE in a PRE-miniwhite reporter gene (Chan et al. 1994). In the wild-type control background(acf13/acf13), the expression of the PRE-miniwhite reporter gene was strongly repressed, with pigments limited to a small part of the adult fly eye (Fig. 4I). In the absence of Acf1 (acf11/acf11), we observed partial activation of the PRE-miniwhite reporter gene with pigments distributed over a larger area of the eye (Fig. 4J). This observed derepression in the homozygous acf11 background is comparable to derepression in a heterozygous Pc background(Poux et al. 2002; data not shown).
We also investigated whether acf1 interacts genetically with the segmentation function of Pc. The appearance of extra sex combs on distal portions of the second and third legs in F1 males was scored in the progeny from a cross between males with a heterozygous deficiency for Pc (Df(3L)Asc) and females homozygous for acf1 alleles. The mutation of acf1 significantly enhanced this Pc phenotype in a manner similar to that seen with other enhancers of the Pc gene (Landecker et al. 1994). Whereas only about 18% or 17% of the Df(3L)Asc/+; acf13/+ or Df(3L)Asc/+; acf14/+ males had extra sex combs on second and/or third pairs of legs (from the total number of male progeny scored, N = 250 for acf13, N = 100 for acf14), 61% or 58% of the Df(3L)Asc/+; acf11/+ or Df(3L)Asc/+; acf12/+ male flies had the extra sex comb phenotype (N = 150 for acf11, N = 100 for acf12, P < 0.001). In addition, >50% of males in the latter two crosses exhibited ectopic pigmentation of their A3 and A4 abdominal tergites (data not shown), which was never observed in crosses with acf13 or acf14 mothers. These results, combined with the derepression of PRE-mediated miniwhite silencing, demonstrate that acf1 is a Polycomb enhancer and suggest that ACF/CHRAC is involved in the assembly and/or maintenance of repressive chromatin in Polycomb-responsive loci.
The identity of Drosophila abdominal segments A5–A8 is determined by homeotic selector genes of the bithorax complex (for review, see Duncan 1987). For instance, in Pc/acf1 males, the posteriorly directed homeotic transformation may be caused by an increase in the expression of the bithorax complex gene Abd-B on loss of Acf1. On the other hand, the anterior transformation phenotype of ISWI/+; acf1/acf1 and nap1/nap1; acf1/acf1 animals (Fig. 3) is reminiscent of mutations in various trithorax group genes, which include the brm and kis genes that encode ATPase subunits of chromatin remodeling complexes (Tamkun et al. 1992; Daubresse et al. 1999). This anterior transformation is likely to result from a decrease in expression of Abd-B on loss of Acf1. These data suggest that Acf1 may be involved in repression or activation of Abd-B in different contexts. Transcriptional repression of Abd-B by Acf1 is consistent with its function in the assembly of repressive chromatin. In fact, genetic evidence in yeast (Goldmark et al. 2000) as well as polytene chromosome localization studies in Drosophila (Deuring et al. 2000) primarily implicate ISWI-containing complexes in transcriptional repression in vivo. Transcriptional activation of Abd-B by Acf1 could be due to its chromatin remodeling function, which could potentially facilitate transcription, or to an indirect effect, such as the repression of a transcriptional repressor of Abd-B.
Loss of Acf1 results in a shortening of S phase in Drosophila embryos
The packaging of DNA into chromatin appears to repress DNA replication (for reviews, see Spradling 1999; Fyodorov and Kadonaga 2001; Gerbi and Bielinsky 2002). We therefore sought to test the effect of Acf1 on chromatin-mediated repression of DNA replication. If ACF/CHRAC was involved in the disruption of chromatin structure, then the loss of Acf1 might be expected to cause an increase in chromatin-mediated repression of DNA replication. Alternatively, if ACF/CHRAC was involved in the assembly of chromatin, then the loss of Acf1 might be expected to result in a decrease in chromatin-mediated repression of DNA replication.
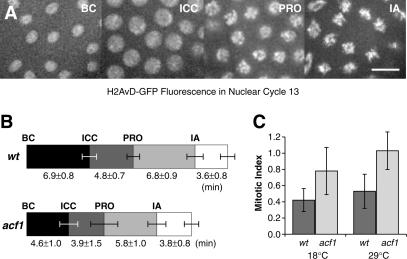
To test these hypotheses, we directly monitored the timing of nuclear divisions in wild-type versus Acf1-deficient Drosophila embryos. In these experiments, we visualized the nuclei in syncytial (cycle 13) embryos by the use of the fluorescent H2AvD-GFP histone (Clarkson and Saint 1999). At this stage of development, the nuclei undergo nearly synchronous divisions with only S and M phases. By using time-lapse microscopy, we measured the length of time for nuclei to transit from the beginning of cycle 13 (BC) to initiation of chromosome condensation (ICC), prophase (PRO), and initiation of anaphase (IA; Fig. 5A). These experiments revealed that acf11/acf11 embryos spend significantly less time in S phase than isogenic controls, as seen in the timing from the beginning of cycle 13 to the initiation of chromosome condensation (Fig. 5B). DNA replication appeared to proceed normally in the Acf1-deficient nuclei, because they exhibited no gross morphological defects in mitotic chromosome structure and passed through mitosis without any chromosome segregation defects. These results indicate that the loss of Acf1 results in a shortening of S phase in Drosophila embryos, which is consistent with a role of ACF/CHRAC in the assembly rather than disruption of chromatin in vivo.
Figure 5.
Loss of Acf1 results in faster progression through S phase in embryos and increased mitotic index in larval neuroblasts. (A) The effect of Acf1 on the cell cycle was determined by using time-lapse microscopy in syncytial embryos containing histone H2AvDGFP. Nuclear division cycle 13 progression was scored in embryos by morphological criteria as shown: beginning of cycle 13 (BC), initiation of chromosome condensation (ICC), prophase (PRO), and initiation of anaphase (IA). Scale bar, 5 μm. (B) Embryos lacking Acf1 have a shorter S phase than do wild-type embryos. Relative to wild-type embryos, Acf1-deficient embryos exhibit shorter times from the initiation of the cell cycle to the initiation of chromosome condensation (P = 0.002) as well as an overall shorter cell cycle (P = 0.003). The results represent an average of seven (acf11) and five (wild-type control) experiments. Mitotic progression was not significantly altered as judged by the length of time from anaphase onset to the end of the cell cycle (P = 0.1). (C) The loss of Acf1 results in an increase in the mitotic index of third instar larval neuroblasts. The mitotic indices of neuroblasts derived from acf11 null or wild-type control (acf13) larvae were determined at 18°C (P = 0.06) and 29°C (P = 0.015).
Mutation of acf1 leads to an increased mitotic index and accelerated S phase in replicating larval cells
We then investigated whether loss of Acf1 causes alterations in cell cycles that include G1 and G2 phases. To this end, we first examined the effect of mutation of acf1 on the mitotic index of larval neuroblasts. In these experiments, the mitotic index of acf11/acf11 mutant neuroblasts was approximately twice as high as that of isogenic (acf13/acf13) control neuroblasts (Fig. 5C). We did not detect mitotic abnormalities in the cells lacking Acf1 (data not shown). These findings are consistent with a shorter S phase in the acf1 neuroblasts, because a decrease in the time spent in S phase would result in an increase in the proportion of cells in mitosis. It is also possible, however, that the increase in the mitotic index is due to a delay in mitosis rather than a shortened S phase.
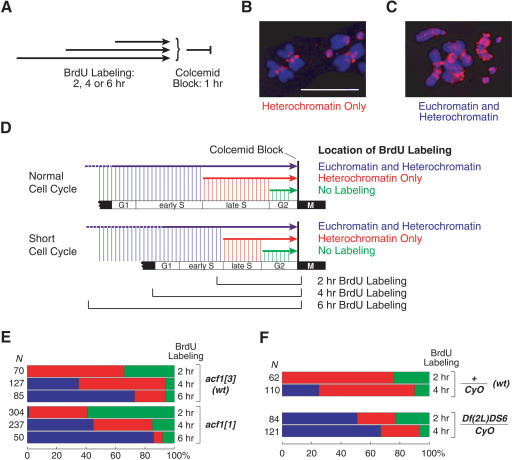
To determine the basis for the cell cycle defect in the Acf1-deficient neuroblasts, we monitored cell cycle progression in wild-type versus acf1 mutant neuroblasts by labeling the replicating DNA with BrdU and then blocking the cells in mitosis with Colcemid(Fig. 6A). The sites of BrdU incorporation were visualized by immunostaining of mitotic chromosomes. In Drosophila, heterochromatic regions mainly replicate late in S phase, whereas euchromatic regions generally replicate early in S phase (Sullivan and Karpen 2001; Schübeler et al. 2002). When the cells were in late S phase during the BrdU labeling period, we observed fluorescence mainly in centromeric heterochromatin (“Heterochromatin Only”; Fig. 6B). When the cells were in early S phase (or G1 phase) at the time of addition of BrdU, we observed fluorescence throughout the chromosomes because of the incorporation of BrdU in euchromatin in early S phase and in heterochromatin as the cells progressed through late S phase (“Euchromatin and Heterochromatin”; Fig. 6C). Also, when the cells were in G2 on addition of BrdU, we did not observe any fluorescence of the chromosomes (“No Labeling”; data not shown).
Figure 6.
The absence of Acf1 leads to a faster S phase in larval neuroblasts. (A) S phase progression was monitored by the incorporation of BrdU in replicating larval neuroblasts. Dissected neuroblasts were incubated with BrdU for 2, 4, or 6 h, and then blocked in mitosis by incubation with Colcemid for 1 h. The pattern of BrdU incorporation was scored as “No Labeling,” “Heterochromatin Only,” or “Euchromatin and Heterochromatin.” (B) Mitotic chromosomes with BrdU labeling of pericentric heterochromatin only. DNA, blue; BrdU, red; scale bar, 15 μm. (C) Mitotic chromosomes with BrdU labeling of euchromatin and heterochromatin. DNA, blue; BrdU, red. (D) Summary of the effects of the loss of Acf1 on the progression of larval neuroblasts through the cell cycle. (E) Neuroblasts lacking Acf1 exhibit a faster progression through S phase compared with isogenic controls (acf13/acf13), as judged by the earlier appearance of Euchromatin and Heterochromatin labeling. The colored bars represent the relative numbers of chromosomes with a particular type of BrdU staining: Euchromatin and Heterochromatin (blue), Heterochromatin Only (red), or No Labeling (green). The numbers at the left are the total numbers of chromosomes scored for BrdU incorporation in each experiment. (F) Heterozygous deficiency Df(2L)DS6 uncovering the entire histone gene cluster leads to a faster S phase relative to an isogenic control.
We carried out BrdU labeling of neuroblasts for 2, 4, or 6 h prior to blocking the cells in mitosis with Colcemid (depicted in Fig. 6D), and the results are summarized in Figure 6E. As expected, the proportion of cells with Euchromatin and Heterochromatin labeling increases with the duration of BrdU labeling (Fig. 6D,E). Notably, in each BrdU labeling period, there is a smaller proportion of cells in late S phase (“Heterochromatin Only”) in the acf1 mutant cells relative to the wild-type cells (Fig. 6E). In addition, a higher percentage of cells with euchromatic labeling (Euchromatin and Heterochromatin) was seen with Acf1-deficient larvae relative to wild-type larvae. For instance, in the 2-h labeling period, we observed labeling of euchromatin in the acf1 mutant cells but not in the wild-type cells (Fig. 6E). These results collectively indicate that progression through late S phase is faster in the Acf1-deficient neuroblasts than in the wild-type neuroblasts.
Thus, we have observed that embryonic nuclei and larval neuroblasts both exhibit a shortened S phase in the absence of Acf1. In a recent study (Collins et al. 2002), the depletion of Acf1 by RNAi was found to result in a decrease in the rate of BrdU incorporation during late S phase of cultured mammalian cells. The investigators concluded that the depletion of Acf1 causes a delay in the replication of heterochromatin, which is contrary to our conclusions. However, an alternate interpretation of their data is that siRNA-mediated depletion of Acf1 results in an acceleration of the rate of progression through S phase that causes a more rapid decline in the rate of BrdU incorporation in the later stages of S phase relative to that seen in control cells. Thus, the data of Collins et al. (2002) and the present study are not necessarily contradictory. It is also important to note that the behavior of siRNA-treated cultured mammalian cells, as in Collins et al. (2002), need not be identical to the behavior of Acf1-deficient cells in Drosophila embryos or larvae.
In our studies, the evidence for the shortened S phase is provided not only by the real-time measurement of the cell cycle progression in acf1 null mutant embryos (Fig. 5), but also by the analysis of chromosome BrdU labeling patterns in acf1 null mutant neuroblasts (Fig. 6). We therefore believe that the most straightforward interpretation of our data is that the loss of Acf1 results in an increase in the rate of progression through late S phase.
In studies of the CAF-1 histone chaperone, it was found that a dominant-negative mutant version of human CAF-1 inhibits DNA synthesis and causes S phase arrest (Ye et al. 2003). These results suggest that CAF-1, and perhaps chromatin assembly, is essential for S phase progression in mammalian cells. In contrast, we do not observe S phase arrest in cells that lack ACF/CHRAC in vivo. However, unlike ACF/CHRAC, CAF-1 is a core histone chaperone and interacts directly with PCNA, which is required for DNA replication. Thus, the effect of CAF-1 on the replication machinery is likely to be distinct from that of ACF/CHRAC, and the phenotypes observed on mutation of CAF-1 will not necessarily be identical to those seen on mutation of Acf1.
Reduction in the histone levels correlates with an accelerated S phase in replicating larval cells
We also sought to test further the notion that inefficient chromatin assembly results in derepression of DNA replication. To this end, we examined the cell cycle timing in the deficiency line Df(2L)DS6, which uncovers the entire histone gene cluster (Moore et al. 1983). Heterozygous deletion of the histone cluster slightly decreases the expression of the histone genes relative to that in wild-type flies (Bhadra et al. 1997). Thus, the Df(2L)DS6/CyO animals may have defects in chromatin structure that are similar to those in acf1 mutant flies. In fact, Df(2L)DS6/CyO animals exhibit suppression of pericentric PEV (Moore et al. 1983), which is also seen in acf11/acf11 flies (Fig. 4).
We monitored S phase progression in hemizygous Df(2L)DS6/CyO larval neuroblasts as well as in isogenic wild-type control cells (Fig. 6F). These experiments revealed that S phase is significantly accelerated in the Df(2L)DS6/CyO neuroblasts. We therefore conclude that the reduction of the histone levels in Df(2L)DS6/CyO flies is accompanied by an increase in the rate of S phase, and that these findings provide additional support for the hypothesis that defects in the integrity of chromatin result in the derepression of DNA replication.
Discussion
In this study, we generated and analyzed null mutant alleles of the Drosophila acf1 gene, which encodes the large Acf1 subunit of the ACF chromatin assembly factor. (CHRAC is identical to ACF except for the presence of two early embryonic 14- and 16-kD polypeptides.) Surprisingly, Acf1 is not absolutely required for viability. Chromatin from homozygous acf1 mutant embryos exhibits less nucleosomal periodicity as well as a shorter repeat length than chromatin from wild-type embryos. We further observed that extracts from Acf1-deficient embryos assemble nucleosomes in vitro much less efficiently than wild-type extracts, and also that the deficiency in chromatin assembly can be rescued on addition of purified recombinant ACF or Acf1. These findings indicate that ACF/CHRAC is a major chromatin assembly activity in Drosophila, but also that Acf1-deficient flies contain other ATP-utilizing chromatin assembly factor(s) that are able to sustain partial viability.
The analysis of the Acf1 null flies revealed that ACF/CHRAC performs different biological functions than NURF, even though ACF/CHRAC and NURF both share a common ISWI ATPase. Hence, the unique subunits of ACF/CHRAC and NURF can program the basic motor function of ISWI to perform specific biological tasks in vivo.
ATP-utilizing motor proteins could potentially assemble or disrupt chromatin structure. Through multiple lines of investigation, we further studied the function of ACF/CHRAC in vivo. First, we investigated whether there are genetic interactions between acf1 and nap1, because the ACF/CHRAC motor protein and the NAP-1 histone chaperone function together in chromatin assembly in vitro. Double mutant nap1/nap1; acf1/acf1 flies exhibit a homeotic transformation that is not seen in the corresponding single mutant flies. These results are consistent with the biochemical activities of ACF/CHRAC and NAP-1 in the chromatin assembly process.
Second, we tested the effect of Acf1 on heterochromatic transcriptional silencing. In these experiments, we observed suppression of pericentric position-effect variegation on loss of Acf1. We additionally found that Acf1-deficient flies exhibit reduced levels of Polycomb-mediated transcriptional silencing. These findings indicate that ACF/CHRAC is important for the establishment and/or maintenance of repressive chromatin states.
Third, we investigated whether Acf1 enhances or disrupts chromatin-mediated repression of DNA replication. We observed shortening of S phase in Acf1-deficient embryos and larval neuroblasts, which is consistent with a role of ACF/CHRAC in the assembly rather than disruption of chromatin in vivo. We further tested the effect of chromatin structure on the duration of S phase in larvae with a deficiency that uncovers the histone gene cluster. These animals contain reduced levels of histones and exhibit an acceleration of late S phase progression in larval neuroblasts relative to that in wild-type flies. Thus, the mutation of acf1 as well as the reduction in the level of histones each correlate with an increase in the rate of S phase progression. These data collectively support a role of Acf1 in the assembly of histones into chromatin.
In summary, several independent lines of experimentation implicate Acf1 in the formation of chromatin in vivo. These experiments provide evidence for the function of ACF/CHRAC (and other ATP-utilizing factors) in the assembly of chromatin in conjunction with the NAP-1 histone chaperone. They also include the unexpected finding of a role of ACF/CHRAC in Polycomb-mediated silencing as well as the discovery of mutations (acf1 and Df(2L)DS6) that result in an unusual increase in the rate of S phase. Lastly, the loss of Acf1 results in a decrease in the periodicity of nucleosome arrays as well as a shorter nucleosomal repeat length in bulk chromatin, which support a role of Acf1 in the assembly of repressive chromatin. Hence, the collective biochemical and genetic data indicate that ACF/CHRAC functions in the assembly of periodic nucleosome arrays that contribute to the repression of genetic activity in the eukaryotic nucleus.
Materials and methods
Fly strains and genetics
Flies were grown on standard corn meal, sugar, and yeast medium with Tegosept. Stocks and crosses were maintained at 23°C, except when otherwise indicated. The EP(3)1181 line (Rørth 1996) with a transposon in the first intron of the acf1 gene was obtained from Exelixis. ISWI1/SM5 and ISWI2/SM5 lines were a gift from John Tamkun. 2212H6.5+PBX transgenic line was a gift from Vincent Pirrotta. 39C-5, 39C-27, and 39C-72 variegating insertion lines (Cryderman et al. 1999a) were a gift from Lori Wallrath. Df(2L)DS6/CyO, Df(2L)DS8/CyO, Df(2L)DS9/CyO, and b pr cn/CyO were a gift from Tom Grigliatti and Adelaide Carpenter. Df(3L)Asc p/TM3 line (Gildea et al. 2000) was a gift from Allen Shearn. Df(3R)04661/TM3, Df(3R)faf-BP/TM6B, EY08629, P{SUPor-P}KG, and P{SUPor-P}KV insertion lines (Yan et al. 2002; Konev et al. 2003), balancer, and other common stocks were obtained from the Bloomington Stock Center.
The EP(3)1181 insertion line contains a P{EP} transposon within the first intron of acf1 and a homozygous lethal mutation that maps outside of the acf1 locus. This P-element insertion line was outcrossed into y1w1 background, F2 males were batch-crossed to w1; TM3/TM6B virgins, and individual acf1EP(3)1181/TM6B flies were crossed to generate homozygous insertion lines. The resulting flies are homozygous viable, and embryonic expression of Acf1 protein is not affected by the insertion (data not shown). Df(3R)acf11 and Df(3R)acf13 were generated by imprecise excision from acf1EP(3)1181. A total of 179 excision lines were analyzed by PCR across the acf1 locus followed by sequencing. Most of imprecise excision events resulted in small deletions entirely within the first intron. One deletion line, Df(3R)acf11, contained a 499-bp deletion (3′ of the site of insertion) that spanned the first intron and a part of the second exon of acf1 (Fig. 1A). Df(3R)acf13 contains a deletion of 83 bp entirely within the first intron of acf1 (Fig. 1A). Both acf11 and acf13 alleles are homozygous viable and can complement Df(3R)04661 and Df(3R)faf-BP, which uncover the acf1 gene.
The mRNA from embryos of acf11 and acf13 excision lines was analyzed by RT-PCR and sequencing. Because of the loss of the native splice acceptor site upstream of the second exon of Acf1 gene, the Df(3R)acf11 embryos express four acf11 messages with distinct patterns of splicing of the first intron. One of them contains the remainder of the first intron unspliced. In the other two, short introns (56 or 160 bp, respectively) are spliced out downstream of the first exon. Finally, in the fourth message, the first and second intron as well as the remainder of the second exon are spliced out. In all four cases, translation termination codons are placed in mRNA close to and downstream of the initiating ATG codon. Therefore, Df(3R)acf11 introduces a nonsense mutation in the acf1 gene. The acf13 mRNA is identical to that of the wild-type acf1 mRNA.
The EY08629 insertion line contains a P{EPgy2} transposon located 47 bp upstream of the acf1 putative transcription start site. Df(3R)acf12 and Df(3R)acf14 were generated by P-element excision from acf1EY08629. A total of 80 excision lines were analyzed by PCR across the acf1 locus followed by DNA sequencing. Df(3R)acf12 contains an 871-bp deletion (3′ of the site of insertion) that spans the putative promoter, first exon, and first intron of acf1 (Fig. 1A). acf14 is a precise excision allele (Fig. 1A). Both acf12 and acf14 alleles are homozygous viable and can complement Df(3R)04661 and Df(3R)faf-BP, which uncover the acf1 gene.
Male A5 to A4 transformation in ISWI2/CyO; acf1/acf1 and nap1/nap1; acf1/acf1 double mutants was defined as follows. Individual adult flies were examined under the dissecting microscope within 48 h of eclosion. The loss of at least 20% of A5 pigmentation in the acf11 background, compared with the A5 pigmentation in the acf13 background, was interpreted as anterior transformation. To study genetic interactions between acf1 and Pc, homozygous acf11/acf11, acf12/acf12, acf13/acf13, or acf14/acf14 females were crossed to males with hemizygous Df(3L)Asc, which uncovers Pc. At least 100 F1 males were examined in each cross. The appearance of sex comb teeth on the second and/or third leg was scored as transformation. Penetrance was presented as a percentage of the number of flies examined.
The flies with homozygous acf11 or acf13 alleles on the third and the wm4h allele on the X or PRE-white transgene insertion on the second chromosomes were produced by a series of crosses and maintained as homozygous stocks. Homozygous P{his2AvGFP} acf11 stock was produced by recombination of acf11 allele and H2AvD-GFP transposon in the third chromosome and crosses with balancers. In each case, acf1 genotype was confirmed by PCR.
Polyclonal antibodies and Western blotting
The fragment of acf1 coding sequence that encodes amino acid residues 1–5 and 1290–1479 of Acf1 polypeptide tagged with RGSHHHHHHDYKDDDDK at the C terminus (M23–6His-FLAG) was cloned into pET-28b vector (Novagen). The expression of the protein fragment was induced in BL21(DE3) cells with 0.42 mM IPTG at 37°C for 5 h. The bacteria were harvested and lysed by sonication in a phosphate buffer (pH 7) containing 500 mM NaCl and 8 M urea. The protein fragment was purified by nickel affinity chromatography in denaturing conditions as described by the manufacturer of the Ni-NTA resin (Qiagen). The protein sample containing urea was dialyzed into PBS, at which point the protein largely precipitated out of solution. The mixture of soluble protein and emulsion of insoluble protein was used for immunizing rabbits (Pocono Rabbit Farms). Antibodies were affinity purified by FPLC chromatography (low pH elution) on NHS-activated agarose (Amersham) cross-linked in urea to M23–6His-FLAG as described by Amersham. The antibody, which recognizes the C terminus of the Acf1 polypeptide, reacts strongly with the p185 isoform of the protein but only weakly with the p170 isoform (Fig. 1B). This result suggests that the p170 isoform of Acf1 is a C terminally truncated polypeptide that might arise by alternative splicing of Acf1 mRNA or by proteolytic clipping of p185. The NAP-1 polyclonal antiserum was described elsewhere (Ito et al. 1996).
Drosophila embryos (collected 0–12 h after egg deposition) or late L3 larvae were harvested and dechorionated with 50% bleach. Embryonic nuclear extracts were prepared as described (Soeller et al. 1988), and the protein concentration was determined by the Coomassie dye binding assay. For Acf1 Westerns (Fig. 1B), the nuclear extract (10 μg total protein per lane) was loaded on a 6% polyacrylamide-SDS gel. Alternatively (Fig. 1C), Acf1 was analyzed in crude embryo lysates as described previously (Ito et al. 1999). For NAP-1 Westerns, dechorionated larvae were dried by blotting, weighed, Dounce-homogenized, and lyzed by boiling in SDS-polyacrylamide gel electrophoresis loading buffer. The denatured protein (equivalent to 2 mg dechorionated L3 larvae per lane) was loaded on an 8% polyacrylamide-SDS gel. In all cases, after protein transfer to nitrocellulose, equal gel loading was verified by Ponceau S staining. In Acf1 Western blots, protein loading was additionally controlled by subsequent anti-NAP-1 Western analysis (data not shown). The membranes were blocked with 4% milk and probed with anti-Acf1 (1:2000 dilution of antiserum) or anti-NAP-1 (1:5000 dilution of antiserum) and HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories; 1:10,000 dilution). The HRP activity was detected with ECL-Plus reagents (Amersham) as described by the manufacturer.
In vivo chromatin analysis
For micrococcal nuclease digestion analysis, embryos were collected 0–12 h after egg deposition following inter se crosses of wild-type or homozygous acf11/acf11 parents. The nuclei were prepared as described(Cartwright et al. 1999). The amounts of nuclear material were adjusted based on A260 absorbance. Chromatin (0.25 A260 optical units) was digested for 10 min at room temperature with 0.005 or 0.02 units of micrococcal nuclease (Sigma) in 40-μL reactions that contained 15 mM Tris-HCl (pH 7.5), 150 mM sucrose, 60 mM KCl, 15 mM NaCl, 0.5 mM DTT, and 1 mM CaCl2. The reactions were stopped by the addition of EDTA to 10-mM final concentration. RNA and proteins were degraded by sequential digestion with RNase A and proteinase K. Micrococcal nuclease-digested DNA was extracted with phenol-chloroform and precipitated with ethanol. Approximately one-fourth of the DNA samples were analyzed by 1.2% agarose gel electrophoresis in 1× TBE, followed by ethidium bromide staining.
Chromatin assembly and analysis
Chromatin assembly (S-190) extracts were prepared from 10 to 50 g of acf11 or wild-type embryos (collected 0–6 h after egg deposition) as described previously (Fyodorov and Levenstein 2002). The protein concentration of each extract was determined by the Coomassie dye binding assay. Nucleosome arrays were assembled in reactions that contained 0.7 μg purified native Drosophila histones, 5.6 μg purified recombinant dNAP-1, and 0.7 μg pGIE-0 relaxed plasmid DNA in the presence of 50 mM KCl, 5 mM MgCl2, 3 mM ATP, ATP regeneration system, and an excess of recombinant Drosophila topoisomerase I (Fyodorov and Kadonaga 2003). Purified recombinant ACF (110 fmole; Ito et al. 1999) or 50- to 400-μg S-190 extracts were used in each reaction as the source of ATP-dependent chromatin assembly activity. In biochemical complementation experiments (Fig. 2C), reactions that contained 100 μg Acf1-deficient S-190 extracts additionally contained 110-fmole purified recombinant ACF or Acf1-FLAG. The assembled chromatin was analyzed by the micrococcal nuclease digestion assay with two different dilutions of the enzyme. The oligonucleosomal DNA was deproteinized and subjected to electrophoresis on 1.2% agarose-TBE gels and staining with ethidium bromide.
Analysis of cell cycle timing in early embryos and L3 neuroblasts
The homozygous P{his2AvGFP} acf11 or control embryos were collected and imaged as described (Yu et al. 2000; Blower and Karpen 2001). The cell cycle stage was scored according to the criteria described in Figure 5A.
To determine mitotic index, we dissected and squashed neuroblasts from L3 animals as described (Gatti et al. 1994). The mitotic index was scored from at least 350 optic fields (using a 100× objective) from at least five different brains for each genotype examined. Statistical analysis was performed using a Students t-test in Microsoft Excel.
The replication timing in Drosophila neuroblasts was determined as follows. Neuroblasts were dissected from wandering third instar larvae and incubated in 100-μM BrdU in Schneider's media (+10% FBS) for 2, 4, or 6 h. Neuroblasts were then transferred to Schneider's media (+10% FBS) containing 3 μg/mL Colcemid (Invitrogen) and incubated for 1 h. Brains were squashed, and BrdU was detected as described(Sullivan and Karpen 2001). For each mutant, at least two different brains were squashed for each time point.
Acknowledgments
We are grateful to Vincent Pirrotta (University of Geneva, Geneva, Switzerland), Lori Wallrath (University of Iowa, Iowa City, IA), John Tamkun (University of California, Santa Cruz, Santa Cruz, CA), Peter Harte (Case Western Reserve University, Cleveland, OH), Allen Shearn (Johns Hopkins University, Baltimore, MD), Tom Grigliatti (University of British Columbia, Vancouver, BC, Canada), and Adelaide Carpenter (University of Cambridge, Cambridge, UK) for fly stocks and antibodies. We thank Alexander Konev, Vecheslav Elagin, Jennifer Butler, Elizabeth Blackwood, and Konstantin Beirit for discussions and advice and Tanya Daigle for assistance with immunofluorescence analysis. We thank Alexander Konev, Jer-Yuan Hsu, Alexandra Lusser, Tamar Juven-Gershon, Buyung Santoso, Thomas Boulay, Karl Haushalter, Vassilios Alexiadis, Chin Yan Lim, and Darya Kopytova for critical reading of the manuscript. This work was supported by grants from the NIH to J.T.K. (GM58272) and to G.H.K (GM61169 and GM54549). D.V.F. was supported in part by the American Cancer Society Postdoctoral Fellowship # PF-99–329–01-CCG.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1139604.
References
- Adams M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Ahmad K. and Henikoff, S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191-1200. [DOI] [PubMed] [Google Scholar]
- Badenhorst P., Voas, M., Rebay, I., and Wu, C. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes & Dev. 16: 3186-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra U., Pal-Bhadra, M., and Birchler, J.A. 1997. A sex-influenced modifier in Drosophila that affects a broad spectrum of target loci including the histone repeats. Genetics 146: 903-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D. and Karpen, G.H. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M. 1976. Larval and adult abdominal defects resulting from microcautery of blastoderm staged Drosophila embryos. J. Exp. Zool. 195: 369-392. [DOI] [PubMed] [Google Scholar]
- Cartwright I.L., Cryderman, D.E., Gilmour, D.S., Pile, L.A., Wallrath, L.L., Weber, J.A., and Elgin, S.C.R. 1999. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 304: 462-496. [DOI] [PubMed] [Google Scholar]
- Chan C.-S., Rastelli, L., and Pirrotta, V. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13: 2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson M. and Saint, R. 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18: 457-462. [DOI] [PubMed] [Google Scholar]
- Collins N., Poot, R.A., Kukimoto, I., Garcia-Jimenez, C., Dellaire, G., and Varga-Weisz, P.D. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32: 627-632. [DOI] [PubMed] [Google Scholar]
- Compton J.L., Bellard, M., and Chambon, P. 1976. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc. Natl. Acad. Sci. 73: 4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona D.V.F., Eberharter, A., Budde, A., Deuring, R., Ferrari, S., Varga-Weisz, P., Wilm, M., Tamkun, J., and Becker, P.B. 2000. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC). EMBO J. 19: 3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman D.E., Morris, E.J., Biessmann, H., Elgin, S.C.R., and Wallrath, L.L. 1999a. Silencing at Drosophila telomeres: Nuclear organization and chromatin structure play critical roles. EMBO J. 18: 3724-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman D.E., Tang, H., Bell, C., Gilmour, D.S., and Wallrath, L.L. 1999b. Heterochromatic silencing of Drosophila heat shock genes act at the level of promoter potentiation. Nucleic Acids Res. 27: 3364-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubresse G., Deuring, R., Moore, L., Papoulas, O., Zakrajsek, I., Waldrip, W.R., Scott, M.P., Kennison, J.A., and Tamkun, J.W. 1999. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126: 1175-1187. [DOI] [PubMed] [Google Scholar]
- Deuring R., Fanti, L., Armstrong, J.A., Sarte, M., Papoulas, O., Prestel, M., Daubresse, G., Verardo, M., Moseley, S.L., Berloco, M., et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5: 355-365. [DOI] [PubMed] [Google Scholar]
- Donaldson K.M., Lui, A., and Karpen, G.H. 2002. Modifiers of terminal deficiency-associated position effect variegation in Drosophila. Genetics 160: 995-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan I. 1987. The bithorax complex. Annu. Rev. Genet. 21: 285-319. [DOI] [PubMed] [Google Scholar]
- Eisen J.A., Sweder, K.S., and Hanawalt, P.C. 1995. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 23: 2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfring L.K., Deuring, R., McCallum, C.M., Peterson, C.L., and Tamkun, J.W. 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 14: 2225-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N.J. and Kingston, R.E. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2: 409-421. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V. and Kadonaga, J.T. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106: 523-525. [DOI] [PubMed] [Google Scholar]
- ____. 2002a. Binding of Acf1 to DNA involves a WAC motif and is important for ACF-mediated chromatin assembly. Mol. Cell Biol. 22: 6344-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002b. Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418: 897-900. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol. 371: 501-518. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V. and Levenstein, M.E. 2002. Chromatin assembly in Drosophila systems. In Current Protocols in Molecular Biology, pp. 21.7.1-21..27. Wiley & Sons, New York. [DOI] [PubMed]
- Gatti M., Bonaccorsi, S., and Pimpinelli, S. 1994. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 44: 371-391. [DOI] [PubMed] [Google Scholar]
- Gerbi S.A. and Bielinsky, A.-K. 2002. DNA replication and chromatin. Curr. Opin. Genet. Dev. 12: 243-248. [DOI] [PubMed] [Google Scholar]
- Gildea J.J., Lopez, R., and Shearn, A. 2000. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homoloque of human retinoblastoma binding protein 2. Genetics 156: 645-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio, T.G., Estep, P.W., Church, G.M., and Tsukiyama, T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423-433. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.W. and Koonin, E.V. 1993. Helicases: Amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3: 419-429. [Google Scholar]
- Grunstein M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389: 349-352. [DOI] [PubMed] [Google Scholar]
- Gruss C., Wu, J., Koller, T., and Sogo, J.M. 1993. Disruption of the nucleosomes at the replication fork. EMBO J. 12: 4533-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haushalter K.A. and Kadonaga, J.T. 2003. Chromatin assembly by DNA-translocating motors. Nature Rev. Mol. Cell Biol. 4: 613-620. [DOI] [PubMed] [Google Scholar]
- Henikoff S. 2000. Heterochromatin function in complex genomes. Biochim. Biophys. Acta 1470: O1-O8. [DOI] [PubMed] [Google Scholar]
- Hochheimer A., Zhou, S., Zheng, S., Holmes, M.C., and Tjian, R. 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420: 439-445. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger, M., Kobayashi, R., and Kadonaga, J.T. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell Biol. 16: 3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Bulger, M., Pazin, M.J., Kobayashi, R., and Kadonaga, J.T. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145-155. [DOI] [PubMed] [Google Scholar]
- Ito T., Levenstein, M.E., Fyodorov, D.V., Kutach, A.K., Kobayashi, R., and Kadonaga, J.T. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes & Dev. 13: 1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153-155. [Google Scholar]
- Kamakaka R.T., Bulger, M., and Kadonaga, J.T. 1993. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes & Dev. 7: 1779-1795. [DOI] [PubMed] [Google Scholar]
- Kennison J.A. 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29: 289-303. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar, G.J. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 13: 2339-2352. [DOI] [PubMed] [Google Scholar]
- Konev A.Y., Yan, C.M., Acevedo, D., Kennedy, C., Ward, E., Lim, A., Tickoo, S., and Karpen, G.H. 2003. Genetics of P-element transposition into Drosophila melanogaster centric heterochromatin. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Kornberg R.D. and Lorch, Y. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9: 148-151. [DOI] [PubMed] [Google Scholar]
- Landecker H.L., Sinclair, D.A.R., and Brock, H.W. 1994. Screen for enhancers of Polycomb and Polycomblike in Drosophila melanogaster. Dev. Genet. 15: 425-434. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden, J., Tatchell, K., Kovacic, R.T., and Van Holde, K.E. 1977. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc. Natl. Acad. Sci. 74: 79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A., LeRoy, G., Wand, Y.-H., and Reinberg, D. 2001. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes & Dev. 15: 2837-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251-260. [DOI] [PubMed] [Google Scholar]
- Madhavan M.M. and Madhavan, K. 1980. Morphogenesis of the epidermis of adult abdomen of Drosophila. J. Embryol. Exp. Morphol. 60: 1-31. [PubMed] [Google Scholar]
- Mello J.A. and Almouzni, G. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11: 136-141. [DOI] [PubMed] [Google Scholar]
- Michailidis J., Murray, N.D., and Marshall Graves, J.A. 1988. A correlation between developmental time and variegated position effect in Drosophila melanogaster. Genet. Res. 52: 119-123. [Google Scholar]
- Moore G.D., Sinclair, D.A., and Grigliatti, T.A. 1983. Histone gene multiplicity and position effect variegation in Drosophila melanogaster. Genetics 105: 327-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. 1998. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93: 333-336. [DOI] [PubMed] [Google Scholar]
- Poux S., Horard, B., Christian, J.A., Sigrist, C.J.A., and Pirrotta, V. 2002. The Drosophila Trithorax protein is a coactivator required to prevent re-establishment of Polycomb silencing. Development 129: 2483-2493. [DOI] [PubMed] [Google Scholar]
- Rice J.C. and Allis, C.D. 2001. Code of silence. Nature 414: 258-261. [DOI] [PubMed] [Google Scholar]
- Rørth P. 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. 93: 12418-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseland C.R. and Schneiderman, H.A. 1979. Regulation and metamorphosis of the abdominal histoblasts of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 186: 235-265. [DOI] [PubMed] [Google Scholar]
- Santoro R., Li, J., and Grummt, I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32: 393-396. [DOI] [PubMed] [Google Scholar]
- Schübeler D., Scalzo, D., Kooperberg, C., van Steensel, B., Delrow, J., and Groudine, M. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat. Genet. 32: 438-442. [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Monier, K., and Sullivan, K.F. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151: 1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Chiang, A., Bender, W., Shimell, M.J., and O'Connor, M. 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158: 131-144. [DOI] [PubMed] [Google Scholar]
- Smith P.A., Jackson, V., and Chalkey, R. 1984. Two-stage maturation process for newly replicated chromatin. Biochemistry 23: 1576-1581. [DOI] [PubMed] [Google Scholar]
- Soeller W.C., Poole, S.J., and Kornberg, T. 1988. In vitro transcription of the Drosophila engrailed gene. Genes & Dev. 2: 68-81. [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Stahl, H., Koller, T., and Knippers, R. 1986. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J. Mol. Biol. 189: 189-204. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. 1999. ORC binding, gene amplification, and the nature of metazoan replication origins. Genes Dev. 13: 2619-2623. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature 403: 41-45. [DOI] [PubMed] [Google Scholar]
- Strohner R., Nemeth, A., Jansa, P., Hofmann-Roher, U., Santoro, R., Längst, G., and Grummt, I. 2001. NoRC—A novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20: 4892-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. and Karpen, G. 2001. Centromere identity in Drosophila is not determined in vivo by replication timing. J. Cell Biol. 154: 683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J.W., Deuring, R., Scott, M.P., Kissinger, M., Pattatucci, A.M., Kaufman, T.C., and Kennison, J.A. 1992. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561-572. [DOI] [PubMed] [Google Scholar]
- Tartof K.D., Bishop, C., Jones, M., Hobbes, C.A., and Locke, J. 1989. Towards an understanding of position effect variegation. Dev. Genet. 10: 162-176. [DOI] [PubMed] [Google Scholar]
- Thomas J.O. and Thompson, R.J. 1977. Variation in chromatin structure in two cell types from the same tissue: A short DNA repeat length in cerebral cortex neurons. Cell 10: 633-640. [DOI] [PubMed] [Google Scholar]
- Tyler J.K. 2002. Chromatin assembly. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur. J. Biochem. 269: 2268-2274. [DOI] [PubMed] [Google Scholar]
- van Holde K.E. 1989. Chromatin. Springer-Verlag, New York.
- Vaquero A., Loyola, A., and Reinberg, D. 2003. The constantly changing face of chromatin. Sci. Aging Knowl. Environ. 2003: RE4. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm, M., Bonte, E., Dumas, K., Mann, M., and Becker, P.B. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598-602. [DOI] [PubMed] [Google Scholar]
- Vega-Palas M.A., Venditti, S., and Di Mauro, E. 1998. Heterochromatin organization of a natural yeast telomere. Changes of nucleosome distribution driven by the absence of Sir3p. J. Biol. Chem. 273: 9388-9392. [DOI] [PubMed] [Google Scholar]
- Verreault A. 2000. De novo nucleosome assembly: New pieces in an old puzzle. Genes & Dev. 14: 1430-1438. [PubMed] [Google Scholar]
- Wallrath L.L. 1998. Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev. 8: 147-153. [DOI] [PubMed] [Google Scholar]
- Weiss K. and Simpson, R.T. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 18: 5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. 1995. Chromatin: Structure and function. Academic Press, San Diego.
- Worcel A., Han, S., and Wond, M.L. 1978. Assembly of newly replicated chromatin. Cell 15: 969-977. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos, R., Wang, H.-M., Hamiche, A., Ranallo, R., Lee, K.-M., Fu, D., and Wu, C. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8: 531-543. [DOI] [PubMed] [Google Scholar]
- Yan C.M., Dobie, K.W., Le, H.D., Konev, A.Y., and Karpen, G.H. 2002. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics 161: 217-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Franco, A.A., Santos, H., Nelson, D.M., Kaufman, P.D., and Adams, P.D. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11: 341-351. [DOI] [PubMed] [Google Scholar]
- Yu K.R., Saint, R.B., and Sullivan, W. 2000. The Grapes checkpoint coordinates nuclear envelope breakdown and chromosome condensation. Nat. Cell Biol. 2: 609-615. [DOI] [PubMed] [Google Scholar]