Abstract
Covalent modifications of the histone N tails play important roles in eukaryotic gene expression. Histone acetylation, in particular, is required for the activation of a subset of eukaryotic genes through the targeted recruitment of histone acetyltransferases. We have reported that a histone C tail modification, ubiquitylation of H2B, is required for optimal expression of several inducible yeast genes, consistent with a role in transcriptional activation. H2B was shown to be ubiquitylated and then deubiquitylated at the GAL1 core promoter following galactose induction. We now show that the Rad6 protein, which catalyzes monoubiquitylation of H2B, is transiently associated with the GAL1 promoter upon gene activation, and that the period of its association temporally overlaps with the period of H2B ubiquitylation. Rad6 promoter association depends on the Gal4 activator and the Rad6-associated E3 ligase, Bre1, but is independent of the histone acetyltransferase, Gcn5. The SAGA complex, which contains a ubiquitin protease that targets H2B for deubiquitylation, is recruited to the GAL1 promoter in the absence of H2B ubiquitylation. The data suggest that Rad6 and SAGA function independently during galactose induction, and that the staged recruitment of these two factors to the GAL1 promoter regulates the ubiquitylation and deubiquitylation of H2B. We additionally show that both Rad6 and ubiquitylated H2B are absent from two regions of transcriptionally silent chromatin but present at genes that are actively transcribed. Thus, like histone H3 lysine 4 and lysine 79 methylation, two modifications that it regulates, Rad6-directed H2B ubiquitylation defines regions of active chromatin.
Keywords: Rad6, gene activation, H2B ubiquitylation
Chromatin remodeling plays a key role in the regulation of eukaryotic gene expression through the activity of both ATP-dependent nucleosome remodeling factors and histone modifying activities (Belotserkovskaya and Berger 1999). Histone modifications such as acetylation, phosphorylation, and methylation occur on the core histone N tails, and they are frequently targeted to nucleosomes in the promoter regions of active genes (Grunstein 1997; Spencer and Davie 1999). These modifications are regulated by a large group of enzymes, often subunits of multiprotein factors that are themselves recruited to specific genes through interactions with site-specific DNA-binding proteins (Kuo et al. 2000; Peterson and Workman 2000; Bhaumik and Green 2001; Hassan et al. 2001; Larschan and Winston 2001). Although the histone N tails carry the majority of the modifications, the histone H2A and H2B C tails are also posttranslationally modified. One such C tail modification, ubiquitylation, has also been linked to transcription (Bohm et al. 1980; West and Bonner 1980; Hensold et al. 1988; Davie et al. 1991; Bradbury 1992; Jason et al. 2002). Early work showed that nucleosomes in the 5′ regulatory regions of highly transcribed genes frequently contained elevated levels of ubiquitylated histones H2A and H2B, suggesting that the modified histones play a role in transcriptional activation (Levinger and Varshavsky 1982; Barsoum and Varshavsky 1985; Nickel et al. 1989). In addition, recent studies have implicated ubiquitylated H2B in heterochromatic gene silencing and gene repression in yeast (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002). However, the role of histone ubiquitylation in transcription is poorly understood. Because histones are primarily monoubiquitylated, which is not associated with protein turnover, the presence of ubiquitylated histones in chromatin has been postulated to confer a structural or signaling role (Briggs et al. 2002; Henry and Berger 2002; Jason et al. 2002; Sun and Allis 2002).
Histone H2B is the only histone species in Saccharomyces cerevisiae currently known to be ubiquitylated (Swerdlow et al. 1990; Robzyk et al. 2000). Between 1 and 10% of total H2B in this organism is modified by the attachment of a single ubiquitin moiety, although this number could be higher if ubiquitin conjugation is a highly dynamic process as has been suggested (Busch and Goldknopf 1981; Seale 1981; Wu et al. 1981; Mueller et al. 1985; Robzyk et al. 2000; Sun and Allis 2002; Henry et al. 2003). The evolutionarily conserved ubiquitin conjugating enzyme, Rad6/Ubc2, catalyzes monoubiquitylation of H2B in yeast (Jentsch et al. 1987; Sung et al. 1988; Robzyk et al. 2000). Rad6 is a multifunctional protein with roles in postreplication DNA damage repair, meiosis, transcriptional silencing, and protein turnover (Montelone et al. 1981; Borts et al. 1986; Dohmen et al. 1991; Huang et al. 1997; Hoege et al. 2002; Turner et al. 2002). Its role in meiosis and transcriptional silencing appears to be directly related to its role in ubiquitylation of H2B because mutation of RAD6 or loss of the H2B ubiquitylation site confer similar phenotypes in these two processes (Huang et al. 1997; Robzyk et al. 2000; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002). However, Rad6 targets a number of other substrates for modification besides H2B, and its substrate specificity appears to be determined by its association with different E3 ubiquitin ligases (Kornitzer et al. 1994; Kaplun et al. 2000; Pickart 2001; Hoege et al. 2002). Until recently, there was no known E3 that specified H2B ubiquitylation by Rad6. The Bre1 protein, a RING finger protein with structural similarities to other E3 ubiquitin ligases, has emerged as an E3 that directs Rad6 to H2B (Joazeiro and Weissman 2000; Hwang et al. 2003; Wood et al. 2003). Deletion of BRE1 globally reduces ubiquitylated H2B levels in yeast and confers several phenotypes that are similar to the loss of the H2B ubiquitylation site. Thus, Bre1 is predicted to play a role in the same cellular processes that are regulated by H2B ubiquitylation.
H2B ubiquitylation has been linked to transcription in yeast through its role in the trans-histone regulation of histone H3 lysine 4 and lysine 79 methylation (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002). Both of these H3 methylation marks have been associated with transcriptionally active euchromatin, and their presence is reduced in regions of silent chromatin (Strahl et al. 1999; Bernstein et al. 2002; Santos-Rosa et al. 2002; van Leeuwen et al. 2002; Ng et al. 2003a). Moreover, a genomewide analysis of H3 lysine 4 methylation showed that lysine 4 trimethylation is concentrated at the 5′ regions of transcriptionally active genes (Ng et al. 2003b). Given the role of H2B ubiquitylation in regulating H3 lysine 4 methylation, this suggested that the ubiquitin modification would also play a role in active transcription. In support of this view, we recently reported that monoubiquitylation of H2B is required for the optimal expression of two highly inducible yeast genes, GAL1 and SUC2 (Henry et al. 2003). Upon induction of the GAL1 gene, H2B was found to be sequentially ubiquitylated and then deubiquitylated at the GAL1 core promoter prior to the significant accumulation of transcripts. Deubiquitylation was shown to be controlled by the activity of the ubiquitin protease, Ubp8, a stoichiometric subunit of the SAGA coactivator, which is one of the earliest factors recruited to the GAL promoter following galactose induction (Sanders et al. 2002; Bryant and Ptashne 2003). However, the requirements for H2B ubiquitylation were not defined in this study. In this report, we investigated the role of Rad6 in the ubiquitylation of H2B at the GAL1 gene during galactose activation. We found that, like SAGA, Rad6 was also recruited to the GAL1 promoter, but, unlike SAGA, it remained there only transiently. Rad6 promoter association required the Gal4 activator and the Rad6-associated E3, Bre1, and thus in addition to its role in DNA damage repair, Rad6 can also be classified as a transcriptional coactivator. The period of Rad6 promoter association closely paralleled the period of H2B ubiquitylation and appeared to precede the recruitment of SAGA. This suggests that the staged recruitment of Rad6 and SAGA controls the ubiquitylation and deubiquitylation of H2B at the GAL promoter. We also show that H2B ubiquitylation, like the two H3 methylation marks that it controls, is generally excluded from silent chromatin but present in regions of transcriptionally active euchromatin. Thus, H2B ubiquitylation can also be considered as a mark of active chromatin.
Results
Rad6 is transiently associated with the GAL1 promoter
The GAL genes provide an excellent model for gene activation in eukaryotes. The genes are repressed in glucose-containing medium and induced when cells are shifted to the presence of galactose (Johnston and Carlson 1992; Trumbly 1992; Johnston et al. 1994). Upon gene activation, repression by Mig1/Tup1-Ssn6 is first relieved, and then a cascade of transcription factor recruitment to the GAL promoters is initiated by the enhanced binding of the Gal4 activator to UAS sites in each promoter. Studies performed with the GAL1-GAL10 genes have shown that one of the earliest factors to be recruited is the SAGA histone acetyltransferase complex, which requires Gal4 for its association with the GAL promoter (Bhaumik and Green 2001; Larschan and Winston 2001; Bryant and Ptashne 2003). Subsequently, Mediator, TBP, and RNA polymerase II become associated with the core promoter, and transcription initiates (Bryant and Ptashne 2003).
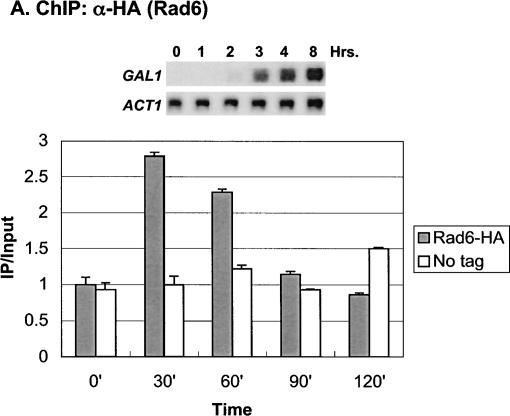
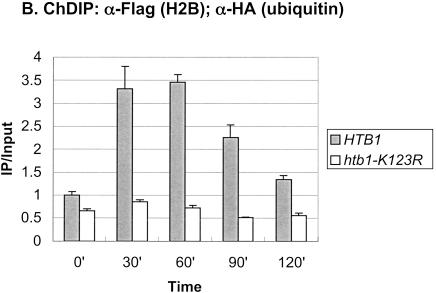
In addition to these factors, we recently showed that H2B ubiquitylation contributes to the maximal transcription of the GAL genes (Henry et al. 2003). In the absence of this histone modification, the levels of GAL1 transcript were reduced following galactose activation. Consistent with a role in the initiation of GAL1 transcription, H2B was found to be transiently ubiquitylated at the GAL1 core promoter at a time that preceded the accumulation of GAL1 mRNA. We therefore asked if Rad6, which directs monoubiquitylation of H2B, was also recruited to the GAL promoter upon gene induction using the method of chromatin immunoprecipitation (ChIP;Orlando and Paro 1993; Hecht et al. 1996; Kuo and Allis 1999). ChIP was carried out during growth in glucose medium and at 30-min intervals after a shift to galactose medium in a yeast strain that contained an HA epitope-tagged RAD6 gene (Fig. 1A). The data showed that Rad6 was dynamically associated with the GAL1 UAS. Maximal occupancy occurred at 30–60 min after the shift to galactose, and by 120 min Rad6 binding to the GAL1 UAS was substantially reduced. The peak of association significantly preceded the appearance of GAL1 transcripts as measured by Northern blot analysis (Fig. 1A, inset), suggesting that Rad6 recruitment to the UAS regulates a very early event in GAL1 transcription. This event is most likely the Rad6-mediated ubiquitylation of H2B, as the period of Rad6 association with the UAS and the period of H2B ubiquitylation at the core promoter almost exactly coincided (Fig. 1B).
Figure 1.
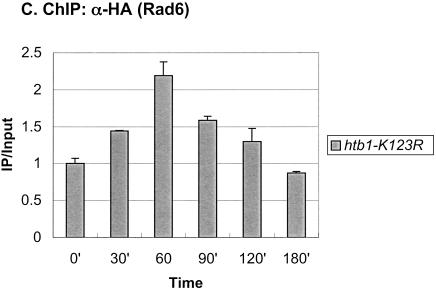
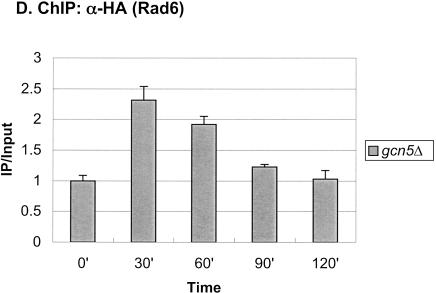
Rad6 is transiently associated with the GAL1 promoter. (A) Strains YKH010 (Rad6-HA HTB1) and JR5-2A (No tag control HTB1) were grown in YPD medium and shifted to YP + 2% galactose medium, and chromatin immunoprecipitation (ChIP) was performed with anti-HA antibodies at the indicated times. PCR analysis in real time was used to measure the abundance of GAL1 UAS sequences in immunoprecipitated (IP) DNA relative to input DNA. The Northern blot inset is from Figure 7A. (B) Strains YKH045 (Flag-HTB1; HA-ubiquitin) and YKH046 (Flag-htb1-K123; HA-ubiquitin) were grown as described in panel A. Chromatin double immunoprecipitation (ChDIP) was sequentially performed with anti-Flag and anti-HA antibodies, and PCR in real time was used to measure the abundance of GAL1 core promoter sequences in the IP DNA (α-HA) relative to input DNA (α-Flag). The data were normalized to the IP/Input ratios for INT-V, which is not regulated by galactose and thus served as a control. (C,D) Strains YKH017 (Rad6-HA htb1-K123R) and YCH001 (Rad6-HA HTB1 gcn5Δ) were grown as described in panel A, and ChIP was performed with anti-HA antibodies, followed by PCR analysis in real time using primers that detect the GAL1 UAS sequences.
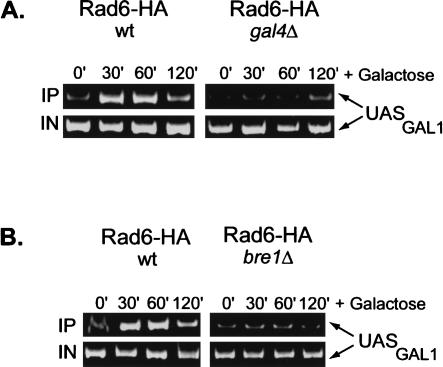
Next, we determined the genetic requirements for Rad6 association with the GAL1 promoter. We first asked if Rad6 recruitment occurred in an htb1-K123R mutant, where H2B is not ubiquitylated (Fig. 1C). In this mutant, Rad6 also transiently associated with the GAL promoter, although the peak of association was delayed by ∼30 min and the drop in promoter binding occurred somewhat more slowly. Thus, prior ubiquitylation of H2B is not a prerequisite for Rad6 promoter association, although it may control the kinetics of Rad6 recruitment and departure from the promoter. Because the Gal4 activator interacts in vivo and in vitro with a number of chromatin remodeling/modifying activities and is required to bring SAGA to the GAL1 promoter in vivo (Melcher and Johnston 1995; Bhaumik and Green 2001; Larschan and Winston 2001; Carrozza et al. 2002), we next asked if it was also required to bring Rad6 to GAL1. ChIP was performed in a gal4Δ strain, and the results showed that Rad6 did not occupy the GAL1 promoter when the activator was absent (Fig. 2A). Rad6 was recently reported to be in a complex with Bre1, an E3 ligase that directs Rad6 to monoubiquitylate H2B and is required for the presence of Rad6 at the constitutive PMA1 promoter (Hwang et al. 2003; Wood et al. 2003). The binding of Rad6 to the GAL1 UAS was also significantly reduced in a bre1Δ mutant, indicating that a Rad6–Bre1 complex is likely to be recruited to GAL1 (Fig. 2B). Finally, Rad6 continued to show transient occupancy of the GAL1 promoter in a gcn5Δ mutant, indicating that histone acetylation is also not required for its recruitment (Fig. 1D). Together, the results suggest that in response to galactose activation, the Gal4 activator recruits the Rad6–Bre1 complex to the GAL1 UAS, where it results in the ubiquitylation of H2B at the core promoter. BEcause we have shown that H2B ubiquitylation is important for the maximal transcription of GAL1 (Henry et al. 2003), the data indicate that Rad6, like SAGA, functions as a transcriptional coactivator at the GAL1 gene.
Figure 2.
Rad6 recruitment to the GAL1 promoter requires the Gal4 activator and the Bre1 E3 ligase. After a shift to YP + galactose medium, ChIP and conventional PCR analysis were carried out in a RAD6-HA HTB1 strain that contained wild-type genes and (A) gal4Δ (YCH001) or (B) bre1Δ (YCH002) deletions. The PCR reactions were electrophoresed on a 10% acrylamide gel, which was stained with ethidium bromide. All reactions were performed in the linear range of amplification.
SAGA and Rad6 show different patterns of GAL1 promoter association
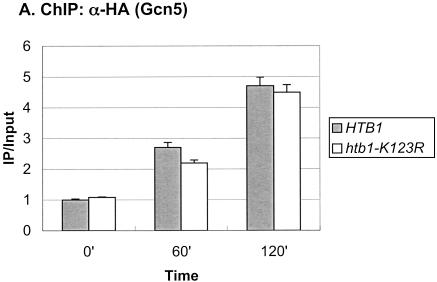
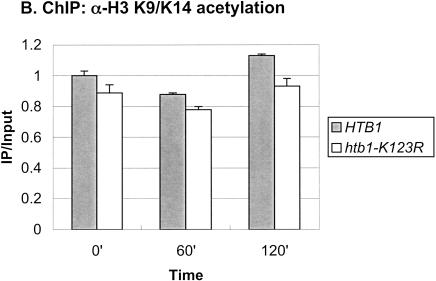
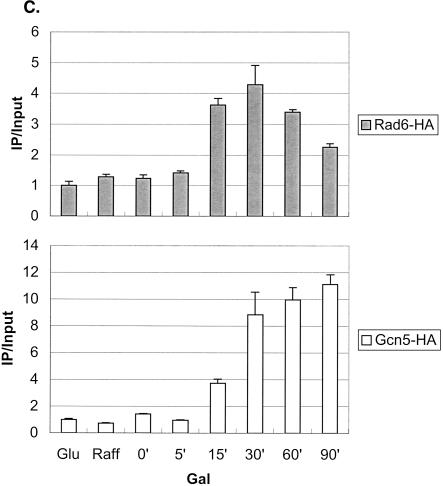
The SAGA complex is one of the earliest factors recruited to the GAL genes following galactose induction, and it has two known roles at the GAL1 core promoter (Bryant and Ptashne 2003). First, the Ubp8 subunit, a ubiquitin protease, targets H2B for deubiquitylation (Henry et al. 2003), and second, the Spt3 subunit is required for recruitment of the TATA-binding protein, TBP (Dudley et al. 1999; Bhaumik and Green 2001, 2002; Larschan and Winston 2001). Because deubiquitylation of H2B closely follows its ubiquitylation, we compared the recruitment of SAGA to that of Rad6 following a shift from glucose to galactose medium. As measured by ChIP in a strain containing a Gcn5-HA protein, SAGA showed increased association with the GAL1 UAS over the time course of the experiment (Fig. 3A), similar to the pattern we recently reported (Henry et al. 2003). At 120 min after the shift to galactose medium, a time when Rad6 was no longer associated with the promoter (Fig. 1A), SAGA was almost four times more concentrated at the GAL1 UAS than it was in glucose medium. Thus, the two coactivators show very different patterns of association with the GAL1 promoter: Rad6 binds transiently, whereas SAGA remains bound once it is recruited. To determine if Rad6 binds before SAGA, we examined the promoter association of the two factors at shorter time intervals following a shift from raffinose to galactose medium, which leads directly to GAL1 activation (Fig. 3C). The data show that Rad6 was again transiently associated with the GAL1 promoter. Moreover, Rad6 reached its maximal occupancy of the UAS earlier than SAGA (15–30 min vs. 30–90 min), supporting the view that Rad6 may be the first factor recruited to the GAL1 UAS upon galactose induction. As in the glucoseto-galactose shift, the period of Rad6 association preceded the maximal accumulation of GAL1 transcripts, consistent with a role for Rad6 recruitment in transcriptional activation (data not shown). Finally, we asked if Rad6-directed ubiquitylation of H2B were required for SAGA association with the GAL1 promoter by performing ChIP in an htb1-K123R mutant. SAGA continued to accumulate at the GAL1 UAS in the absence of both the H2B ubiquitylation site and the Bre1 E3 ligase (Fig. 3A; data not shown). As previously noted, Gcn5-associated H3 Lys 9/Lys14 acetylation levels did not change during GAL1 induction (Bhaumik and Green 2001; Larschan and Winston 2001), and here we show that the htb1-K123R mutation has no effect on this histone modification (Fig. 3B). Thus, we propose that Rad6 precedes SAGA at the GAL1 promoter, but that SAGA is recruited to the promoter independently of Rad6-mediated H2B ubiquitylation.
Figure 3.
Rad6 and SAGA show different patterns of association with the GAL1 promoter. (A) Strains carrying Gcn5-HA and either HTB1 (YKH012) or htb1-K123R (YKH019) were subjected to ChIP and PCR analysis in real time as described in the legend to Figure 1A before and after a shift to YP + galactose medium. (B) ChIP was performed with anti-H3 K9/K14 acetylation antibodies in strains Y131 (HTB1) and Y133 (htb1-K123R) before and after a shift to YP + galactose medium, and the DNA was analyzed by PCR in real time to determine the levels of H3 acetylation at the GAL1 core promoter. (C) ChIP was performed with anti-HA antibodies in strains YKH010 (Rad6-HA HTB1) and YKH0112 (Gcn5-HA HTB1) during growth in YPD (Glu), after 2 h in YP + 2% raffinose (Raff), and at the indicated times after galactose was added to a final concentration of 2%. PCR in real time was performed to measure the association of Rad6 and Gcn5 with the GAL1 UAS. The data in each panel were normalized to the IP/Input ratio of the INT-V control sequences.
Histone H2B is transiently ubiquitylated at the PHO5 promoter
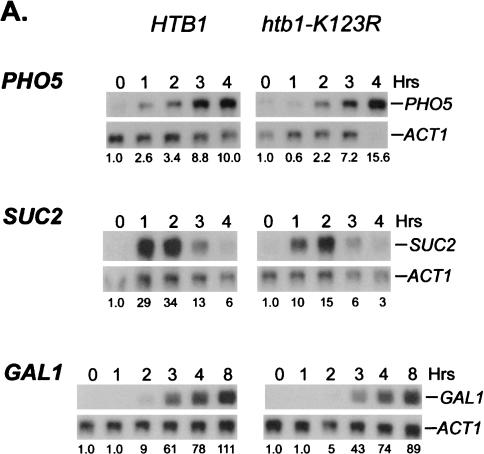
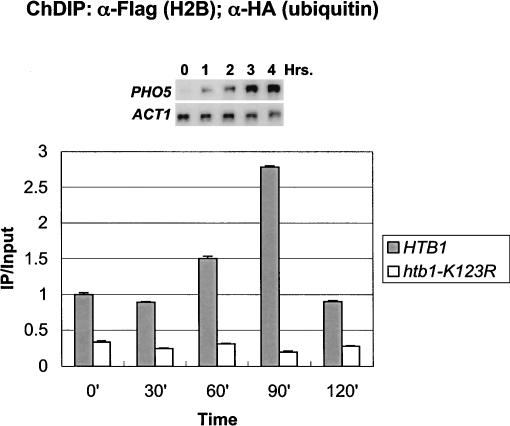
To determine if H2B ubiquitylation plays a role in the expression of other inducible genes, the levels of the modified histone were also examined during activation of the PHO5 gene. This gene is regulated by an entirely different mechanism from GAL1, but like GAL1, PHO5 transcription also shows a modest dependence on H2B ubiquitylation (see Fig. 7A, below;Vogel et al. 1989; Svaren and Horz 1997). Using the chromatin double immunoprecipitation assay, ubiquitylated H2B was found to transiently accumulate at the PHO5 core promoter after phosphate was depleted from the medium (Fig. 4). The level of ubiquitylated H2B peaked ∼2.3-fold above the repressed level at 90 min after the shift and declined to the uninduced level by 120 min, a time that preceded the maximal accumulation of PHO5 transcripts. This rise strictly depended on the presence of the H2B ubiquitylation site and was specific to the PHO5 gene, as the levels of ubiquitylated H2B remained unchanged at two intergenic regions under the same growth conditions (Fig. 4; data not shown). Thus, the data suggest that H2B ubiquitylation and deubiquitylation may play general roles in the initiation of transcription following gene activation.
Figure 7.
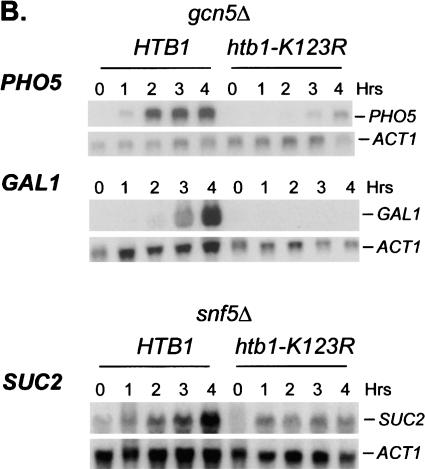
H2B ubiquitylation has overlapping functions with chromatin remodeling factors. (A) Cells from strain JR5-2A that contained either wild-type HTB1 or the htb1-K123R allele were shifted from YPD medium to: no phosphate synthetic medium (PHO5), YP medium + 2% galactose (GAL1), or YP medium + 0.05% glucose (SUC2) for the indicated times. Northern blot analysis was performed with the indicated probes. The numbers beneath each lane represent transcript levels normalized to ACT1 mRNA. (B) Northern blot analysis was performed in gcn5Δ (JR7-2B) or snf5Δ (JR16-6A) mutants that contained either wild-type HTB1 or the htb1-K123R allele after cells were shifted to the media described in panel A. The SUC2 blot was overexposed to show the effect of the htb1-K123R mutation on residual SUC2 mRNA accumulation in the snf5Δ mutant.
Figure 4.
H2B is transiently ubiquitylated at the PHO5 core promoter. ChDIP was carried out in strains YKH045 (Flag-HTB1) and YKH046 (Flag-htb1-K123R) during growth in YPD medium and at 30-min intervals after a shift to a medium lacking inorganic phosphate, and extracted DNA was analyzed by quantitative PCR in real time to determine the levels of H2B ubiquitylation at the PHO5 core promoter. The data were normalized to the INT-V IP/Input ratios. The Northern blot inset is from Figure 7A.
Distribution of H2B ubiquitylation
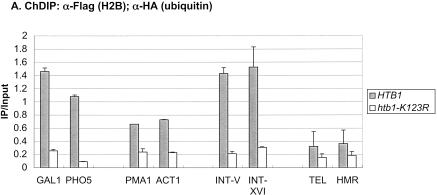
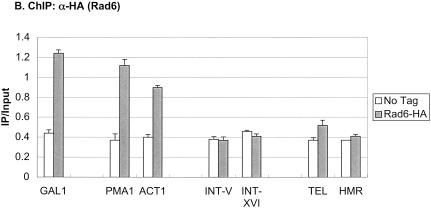
To determine if H2B ubiquitylation was restricted to transcribed genes, the levels of the modified histone were measured at different regions of the yeast genome by a chromatin double immunoprecipitation assay or ChDIP (Fig. 5A). Besides the induced GAL1 and PHO5 genes, two constitutively transcribed genes, ACT1 and PMA1, also contained ubiquitylated H2B at their promoters, albeit at lower relative amounts. In addition to these transcribed genes, two ORF-free intergenic regions on chromosomes V and XVI contained relatively high levels of the H2B modification, whereas two regions of silenced chromatin, TELVI-R and HMRa, had the lowest levels of ubiquitylated H2B. This distribution is remarkably similar to the pattern of H3 Lys 79 methylation at the same genomic regions, which is dependent on H2B ubiquitylation (data not shown). Together, the results suggest that ubiquitylated H2B, like H3 Lys 4 and Lys 79 methylation, may be broadly distributed in transcriptionally active euchromatin and excluded from heterochromatin-like regions (Bernstein et al. 2002; Ng et al. 2003a,b). Next, we measured Rad6 association with the same genomic regions by ChIP (Fig. 5B). Like ubiquitylated H2B, Rad6 was present at the promoters of each of the induced or constitutive genes, but absent from the two regions of silent chromatin. Surprisingly, Rad6 was also absent from the two ORF-free regions, even though these regions contain relatively high levels of H2B ubiquitylation. This suggests that the ubiquitylated species may be differentially stabilized depending on its genomic location. For example, if the ORF-free regions are not transcribed, then there may be no targeted recruitment of a ubiquitin protease, such as occurs at the GAL1 promoter upon SAGA association (Henry et al. 2003). Using similar reasoning, these data also imply that Rad6 may ubiquitylate H2B by both targeted and untargeted mechanisms.
Figure 5.
Distribution of ubiquitylated H2B. (A) ChDIP was sequentially performed with anti-Flag and anti-HA antibodies in strains YKH045 (Flag-HTB1) and YKH046 (Flag-htb1-K123R) after growth in YPD medium. PCR in real time was used to measure the level of H2B ubiquitylation at two transcriptionally silenced regions (TELVI-R and HMRa), two ORF-free intergenic regions (INT-V and INT-XVI), the core promoters of two highly transcribed constitutive genes (PMA1 and ACT1), and the core promoters of the GAL1 and PHO5 genes after induction with galactose or no phosphate, respectively. (B) ChIP was performed with anti-HA antibodies in strains YKH010 (Rad6-HA HTB1) and JR5-2A (No tag HTB1) grown in YPD or induced with galactose, and PCR in real time was used to measure the level of Rad6-HA at the genomic regions described in panel A.
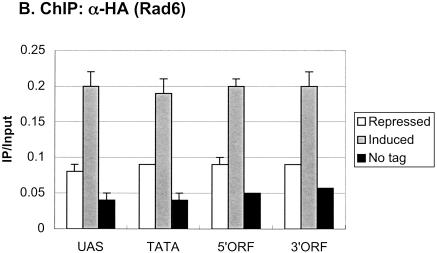
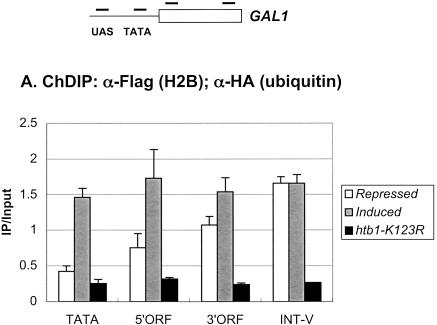
We then asked how H2B ubiquitylation was distributed at an individual transcribed gene. We predicted that it might be associated only with the core promoters of active genes, consistent with a role in transcription initiation. We addressed this question at the GAL1 gene by performing ChDIP in glucose-repressed and galactose-induced cells, using PCR primers that corresponded to the GAL1 core promoter, 5′ ORF, and 3′ ORF regions (Fig. 6A). Surprisingly, under repressed conditions, the 3′ ORF region was found to contain a significant level of ubiquitylated H2B, whereas the 5′ ORF region and core promoter had progressively lower levels of the modified histone. However, after 60 min of galactose induction, the level of ubiquitylated H2B rose disproportionately at the 5′ end of GAL1, resulting in an approximately equal distribution of the modified histone across the promoter and ORF. Thus, H2B ubiquitylation is, in fact, targeted to the 5′ end of GAL1 upon gene induction, consistent with a role in transcription initiation, but its presence throughout the gene suggests that it might also play a role in transcription elongation. When the distribution of Rad6 was examined at the same GAL1 regions by ChIP, little Rad6 was found to be associated with any region of the gene when cells were grown under repressing conditions (Fig. 6B). However, after galactose induction, ∼2.5-fold more Rad6 was present at the GAL1 UAS and core promoter as well as at the two ORF regions. This is consistent with Rad6 and ubiquitylated H2B having a role in both transcription initiation and transcription elongation.
Figure 6.
Ubiquitylated H2B and Rad6 are associated with the GAL1 promoter and ORF. (A) ChDIP was performed in the Flag-HTB1 strain YKH045 during growth in YPD medium (repressed, white bars) and after galactose induction (induced, gray bars). ChDIP was also performed in the Flag-htb1-K123R strain YKH046 after galactose induction (htb1-K123R, dark gray bars). DNA was analyzed at the GAL1 TATA, 5′ ORF, and 3′ ORF regions by PCR in real time. The INT-V sequences served as a control for media differences. (B) ChIP was performed in the Rad6-HA strain YKH010 during growth in YPD medium (repressed, white bars) and after galactose induction (induced, gray bars). ChIP was also performed in the untagged control strain JR5-2A after galactose induction (No tag, dark gray bar). DNA was analyzed by PCR in real time at the GAL1 UAS and the GAL1 regions described in panel A. The approximate location of primers used for PCR analysis is shown relative to GAL1 promoter regulatory elements and ORF.
H2B ubiquitylation plays an overlapping role with chromatin remodeling factors
Although our data indicate that H2B ubiquitylation might be fairly widespread in euchromatin, we have previously shown that the absence of the ubiquitylated species confers relatively modest effects on cell growth and gene expression (Henry et al. 2003). This suggests that the H2B modification might be functionally redundant with other histone modifying activities or nucleosome remodeling factors. In support of this view, we have noted two synthetic phenotypes in double mutants constructed between the htb1-K123R and gcn5Δ or snf5Δ alleles. First, htb1-K123R snf5Δ double mutants grow significantly more slowly than either single mutant, whereas htb1-K123R gcn5Δ double mutants show a Ts- growth defect (data not shown). Second, transcription of several inducible genes is severely compromised in the double mutants (Fig. 7). Whereas transcription of the GAL1, PHO5, and SUC2 genes was reduced less than twofold in htb1-K123R single mutants (Fig. 7A), transcription of the SAGA-dependent GAL1 and PHO5 genes and the Swi/Snf-dependent SUC2 gene was almost abolished in htb1-K123R gcn5Δ and htb1-K123R snf5Δ double mutants, respectively (Fig. 7B). This phenotype is unrelated to the growth defects of the double mutants because expression of the SAGA-dependent INO1 gene and the Swi/Snf- and SAGA-dependent HO gene was unaffected in the same double mutant backgrounds (data not shown). Thus, H2B ubiquitylation becomes very important for the expression of specific genes in the absence of either Gcn5-dependent histone acetylation or ATP-dependent nucleosome remodeling, indicating that the H2B modification plays a functionally overlapping role with these two chromatin remodeling factors.
Discussion
In this study, we show that upon galactose induction, Rad6 is dynamically associated with the GAL1 UAS, leading to a concomitant transient accumulation of ubiquitylated H2B at the core promoter. The temporal overlap between Rad6 promoter association and H2B ubiquitylation suggests that the H2B modification is a direct consequence of the recruitment of Rad6 to the GAL1 promoter. Both the Gal4 activator and the Rad6-associated E3 ligase, Bre1, are required for Rad6 to bind to the GAL1 promoter, suggesting that Gal4 might directly recruit the Rad6–Bre1 complex. Because we have previously shown that H2B ubiquitylation plays a role in the activation of GAL1 transcription (Henry et al. 2003), the data indicate that Rad6 functions as a transcriptional coactivator in addition to its role in mediating postreplication DNA repair. H2B ubiquitylation was also found to increase transiently at the PHO5 core promoter upon gene activation, suggesting that the dynamic regulation of this H2B modification could play a general role in the initiation of transcription. Consistent with the idea that H2B ubiquitylation is associated with transcriptionally active chromatin, the modified form of H2B was present at the promoters of both induced and constitutive genes but absent from regions of silent chromatin (Fig. 6A).
Rad6 and SAGA are sequentially recruited to the GAL1 promoter
Previous studies have shown that galactose induction is accompanied by the sequential appearance of proteins at the GAL1–GAL10 promoter, with the SAGA complex being the first factor to bind after the Gal4 activator (Bhaumik and Green 2001; Larschan and Winston 2001; Bryant and Ptashne 2003). In this study, we have identified Rad6 as a new protein that appears at the GAL1 promoter during galactose activation. Using ChIP, we found that the binding of Rad6 appears to temporally precede that of SAGA (Fig. 3C), suggesting that Rad6 may, in fact, be the earliest factor to associate with the GAL1 promoter. Moreover, the recruitment of SAGA also appears to be independent of Rad6's activity in H2B ubiquitylation because Gcn5-HA associates with the GAL1 promoter in both an htb1-K123R and bre1Δ mutant (Fig. 3B; data not shown). Thus, it is likely that Rad6 and SAGA are independently recruited to the GAL1 UAS, with Rad6 binding first. Both factors bind to the GAL promoter only if the activator Gal4 is present (Fig. 2A; Bhaumik and Green 2001; Larschan and Winston 2001; Bryant and Ptashne 2003). SAGA interacts with Gal4 in vitro (Bhaumik and Green 2001), but we do not know whether Rad6 contacts Gal4 directly, or whether Rad6 and SAGA might contact the same surface of the activator. Bre1 is also required for Rad6 association with the GAL1 UAS (Fig. 2B), and we therefore assume that a Rad6–Bre1 complex is recruited to the GAL1 promoter. Thus, Bre1 could also provide a binding surface for Gal4. Although Gal4 is required to bring Rad6 to the promoter, we do not know how Rad6 is displaced. In the absence of H2B ubiquitylation, Rad6 is still recruited to the GAL UAS, but its turnover is slightly delayed (Fig. 1C). Perhaps the ubiquitylated template itself plays a role in releasing Rad6 from the GAL promoter. Alternatively, the recruitment of SAGA might contribute to the displacement of Rad6.
Rad6 not only appears to precede SAGA at the GAL1 UAS, but it also binds transiently whereas SAGA binds stably. What is the significance of the different binding patterns between the two factors? We propose that the temporal difference in promoter association, which results in the staged recruitment of Rad6 and SAGA, provides a mechanism, first, to ubiquitylate H2B through Rad6-Bre1 and, subsequently, to deubiquitylate H2B through the Ubp8 subunit of SAGA (Henry et al. 2003). This would restrict both the period of H2B ubiquitylation as well as the level of the modified histone at the GAL1 promoter, two conditions that we have previously shown to be important for the optimal transcription of GAL1 (Henry et al. 2003). The transience of Rad6 promoter association might also contribute to the limited period and level of H2B ubiquitylation. SAGA, on the other hand, provides another function at the GAL1 core promoter besides deubiquitylating H2B, which might account for its more stable association with the UAS. The Spt3 subunit of SAGA is required for the recruitment of TBP, and the continued presence of SAGA at the UAS could help to “tether” TBP to the core promoter during successive rounds of transcription initiation (Larschan and Winston 2001; Bhaumik and Green 2002).
Rad6 functions either directly or indirectly as a transcriptional repressor in silent chromatin and at the repressible ARG1 gene, and this role is directly related to its activity in H2B ubiquitylation (Huang et al. 1997; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002; Turner et al. 2002). In contrast, Rad6 functions as a coactivator during galactose induction, also through its H2B ubiquitylation activity. Unlike SAGA, which is an essential coactivator at GAL1 (Bhaumik and Green 2001; Larschan and Winston 2001), Rad6 is dispensable for GAL1 transcription (data not shown). Gcn5 itself is also not required for GAL1 transcription, and gcn5Δ mutants appear phenotypically similar to htb1-K123R mutants with respect to their modest defect in GAL1 transcription (Fig. 7;Bhaumik and Green 2001; Larschan and Winston 2001). However, the presence of ubiquitylated H2B becomes very important for GAL1 expression when the Gcn5 subunit of SAGA is absent (Fig. 7B), suggesting that Rad6-directed ubiquitylation of H2B and Gcn5-mediated acetylation of H3 play overlapping roles during galactose induction. Consistent with this view, GAL1 transcript levels are significantly lower in an htb1-K123R hht1-K14A double mutant compared to each single mutant (C. Hillyer, unpubl. data). We do not know what step in the galactose induction pathway is sensitive to the combined loss of H2B ubiquitylation and Gcn5. The Gal4 activator binds robustly to the GAL UAS elements in an htb1-K123R gcn5Δ double mutant upon galactose induction;however, we have found that the Swi/Snf complex, which is required for GAL transcription in some strain backgrounds, is not recruited to the GAL1 promoter when both factors are absent (C. Hillyer, unpubl. data).
H2B ubiquitylation is a mark of active chromatin
H2B ubiquitylation is not restricted to the GAL1 promoter but is found at other actively transcribed genes as well, including the inducible PHO5 gene (Fig. 4). H2B is also transiently ubiquitylated at the PHO5 promoter during gene induction, and, as seen for GAL1, the period of H2B ubiquitylation precedes the maximal accumulation of PHO5 transcripts. This is consistent with the view that the H2B modification might play a general role during an early stage of gene activation. We assume, although we have not yet tested, that Rad6 ubiquitylates H2B at the PHO5 promoter when phosphate is depleted. SAGA is recruited to this promoter by the Pho4 activator, and thus the Ubp8 subunit of SAGA might also regulate the deubiquitylation of H2B during PHO5 activation (Barbaric et al. 2003). However, it was recently reported that SAGA-dependent hyperacetylation of nucleosomes at the PHO5 promoter leads to histone eviction, and this mechanism could also account for the loss of ubiquitylated H2B (Reinke and Horz 2003). Ubiquitylated H2B is also present at the promoters of the constitutively transcribed PMA1 and ACT1 genes, and Rad6 is associated with the same promoters (Fig. 6). If transient ubiquitylation of H2B plays a general role in the initiation of transcription, it is predicted that the ubiquitin moiety will be continuously turned over at constitutive as well as inducible genes. Although neither the PMA1 nor ACT1 gene is regulated by the SAGA complex, other Ubps also target H2B for deubiquitylation (C.F. Kao, unpubl. data), and these Ubps might play a role in removing the ubiquitin moiety at SAGA-independent genes.
Ubiquitylated H2B appears to be excluded from regions of silenced chromatin, just like the H3 Lys 4 and Lys 79 methylation marks that it controls (Bernstein et al. 2002; van Leeuwen et al. 2002; Ng et al. 2003a). Hypomethylation of the H3 N termini is a prerequisite for Sir protein binding to silenced chromatin, and Sir binding to hypomethylated H3 N tails is postulated to block the Set1 and Dot1 HMTases from accessing their substrates (van Leeuwen and Gottschling 2002; van Leeuwen et al. 2002; Ng et al. 2003a). Rad6 is also excluded from silent chromatin, but Sir proteins are not known to interact with the H2B C tail. Thus, it could be that the structure of silenced chromatin generally inhibits Rad6 access to these regions, thereby blocking H2B ubiquitylation. Regardless, the combined data support the view that H2B ubiquitylation defines domains of active chromatin, either directly or through its role in the trans-histone regulation of histone H3 Lys 4 and Lys 79 methylation.
Materials and methods
S. cerevisiae strains, plasmids, and growth conditions
Yeast strains are all derivatives of W303-1A and are listed in Table 1. The genomic copies of the RAD6 and GCN5 genes were tagged with the triple HA epitope at their C termini using his5 as a marker and sequences flanking the stop codons to direct integration at the appropriate chromosomal location (Longtine et al. 1998). Integrations were confirmed by PCR analysis and by Western blot analysis with anti-HA antibodies. Gene knockouts were performed by transformation with PCR products that contained the URA3 gene flanked by 50–70 bp of DNA surrounding the ATG and stop codon of the appropriate gene. All gene knockouts were confirmed by PCR analysis and phenotypic screens. Plasmids carrying Flag-tagged HTB1 or htb1-K123R genes have been previously described (Recht and Osley 1999; Robzyk et al. 2000). An HA-UBI4 gene driven by the constitutive GAPDH promoter was carried on plasmid pRG145, which was kindly provided by R. Gardner and D. Gottschling (Fred Hutchinson Cancer Research Center). This gene was integrated at the ura3-1 locus in strains carrying Flag-HTB1 or Flag-htb1-K123R after digestion with StuI, following standard genetic procedures. Expression was confirmed by Western blot analysis with anti-Flag and anti-HA antibodies.
Table 1.
S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| JR5-2A | MATa htb-1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 <pRS314-HTB1 or pRS314-htb1-K123R> | Robzyk et al. 2000 |
| JR7-2B | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 gen5Δ::TRP1 <pRS413-HTB1 or pRS413-htb1-K123R> | Recht and Osley 1999 |
| JR6-16A | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 snf5Δ <pRS314-HTB1 or pRS314-htb1-K123R> | Recht and Osley 1999 |
| YKH010 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 <pRS314-HTB1> | This study |
| YKH017 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 <pRS314-htb1-K123R> | This study |
| YKH012 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GCN5-HA::his5 <pRS314-HTB1> | Henry et al. 2003 |
| YKH019 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GCN5-HA::his5 <pRS314-htb1-K123R> | This study |
| YKH045 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GAPDH::HA-UB14::URA3 <pRS314-Flag-HTB1> | Henry et al. 2003 |
| YKH046 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GAPDH::HA-UB14::URA3 <pRS314-Flag-htb1-K123R> | Henry et al. 2003 |
| Y131 | MATa hta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can 1-100 <pRS426-HTA1-HTB1> | Robzyk et al. 2000 |
| Y133 | MATa hta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 <pRS426-HTA1-htb1-K123R> | Robzyk et al. 2000 |
| YCH001 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 gal4Δ::URA3 <pRS314-HTB1> | This study |
| YCH002 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 bre1Δ::URA3 <pRS314-HTB1> | This study |
| YCH003 | MATa htb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 gen5Δ::URA3 <pRS314-HTB1> | This study |
Cells were grown at 30°C in YPD medium or in SD medium supplemented with appropriate amino acids and bases. For gene induction experiments, cells were washed two times with sterile water and shifted into medium containing 2% galactose for GAL1 induction, into medium containing 0.05% glucose for SUC2 induction, and into no phosphate synthetic medium for PHO5 induction (Vogel et al. 1989). For analysis of Rad6-HA and Gcn5-HA GAL1 UAS association at short times after a shift to galactose-inducing conditions, cells were pregrown in YPD, washed, and resuspended in YP + 2% raffinose for 2 h, at which point galactose was added to a final concentration of 2%.
RNA analysis
Northern blot analysis was performed with 10–20 μg of total yeast RNA extracted from cells just before (0′) and at 1-h intervals after the shift to the appropriate medium for gene induction. Hybridization probes corresponding to the GAL1, SUC2, or PHO5 ORFs were labeled by the method of random priming. Hybridization intensities were quantitated with a Molecular Dynamics PhosphorImager.
Chromatin immunoprecipitation
Yeast strains were grown in YPD medium to an O.D.600 of 0.5–0.7 and then shifted to YP medium containing 2% galactose. Immediately before and at 30-min intervals up to 120 min after the shift, 50 mL of cells were removed and fixed with 1% formaldehyde at room temperature for 15 min for analysis of histone modifications and for 30 min for analysis of Rad6-HA and Gcn5-HA binding. Fixation was stopped by the addition of glycine to 125 mM for 5 min, the cells were collected by centrifugation and washed two times with ice-cold TBS (100 mM Tris at pH 7.5, 0.9% NaCl). After the cell pellets were quick-frozen in EtOH-dry ice, they were stored at -80°C. The cell pellets were thawed on ice, resuspended in 500 μL of FA lysis buffer (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1 sodium deoxycholate, 1 mM PMSF) supplemented with a fresh protease inhibitor cocktail (Sigma), and lysed by vortexing with glass beads for 15 min at 4° C. The cell lysates were sonicated six times for 10 sec each on a Branson Sonifier 250 set at 30% output and 100% duty cycle, with 30 sec cooling between each sonication cycle. The average size of the DNA fragments produced was between 300 and 500 bp. Following centrifugation at 15K for 30 min at 4° C in a microfuge, the solubilized chromatin was transferred to a fresh microfuge tube, and the volume was adjusted to 1 mL with FA lysis buffer. For analysis of input chromatin, 50–100 μL were removed.
For analysis of Rad6-HA and Gcn5-HA chromatin immunoprecipitations, 2–5 μl of anti-HA antibody (12CA5; Roche) were added to ∼20 O.D. equivalents of solubilized chromatin and incubation was continued overnight at 4°C. Immune precipitates were collected by a 2-h incubation at 4°C with 40 μL of protein G sepharose beads (Amersham) equilibrated with FA lysis buffer and protease inhibitors. The beads were washed sequentially with 1.4 mL FA-lysis buffer, 1.4 mL FA-lysis buffer + 0.5 M NaCl, 1.4 mL LiCl solution (10 mM Tris at pH 8, 250 mM LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA), and 1.4 mL TE (pH 8) for 5 min each at 4°C, and the immune complexes were eluted twice at room temperature with 250 μL of 1% SDS, 0.1 M NaCO3.
Chromatin double immunoprecipitation (ChDIP) to detect ubiquitylated H2B was carried out in strains YKH045 (Flag-HTB1; HA-UBI4) and YKH046 (Flag-htb1-K123R; HA-UBI4) by the method outlined in Henry et al. (2003), with the following modifications. One hundred microliters of M2 agarose beads (Sigma) equilibrated in FA lysis buffer were incubated with 40 O.D. equivalents of solubilized chromatin overnight at 4°C, the beads were collected by centrifugation and washed four times with FA lysis buffer, and the immune complexes were eluted with 500 μL FA lysis buffer containing 200 μg/mL 3× Flag peptide (Sigma) overnight at 4°C. One-tenth of the eluate was reserved for “input”, and anti-HA antibody (12C5A, Roche) was added to a final concentration of 15 μg/mL to the remaining eluate, followed by incubation at 4°C overnight. The immune complexes were collected by incubation with 80 μL protein G sepharose beads (Amersham) for 3–5 h at 4°C and the beads were washed as described above. The beads were resuspended in 100 μL TE (pH 8) with 20 μg RnaseA (Sigma) and incubated at 37°C for 30 min. Following a wash with 1 mL TE, the immune complexes were eluted from the beads by sequential incubation in 500 μL of 1% SDS/50 mM Tris (pH 8) for 5 min at 100°C and for 10 min at 65°C. After adjusting the NaCl concentration in each sample to 0.2 M, the cross-links were reversed by incubation at 65°C overnight. DNA was extracted according to published procedures (Hecht et al. 1996; Kuo and Allis 1999) and resuspended in 100 μL TE (“input”) or 25 μl TE (“IP”).
PCR analysis
Quantitative PCR in real time was performed using a SYBR Green master mix and an ABI 7000 Prism Sequence Detection system, both from Applied Biosystems. Triplicate samples containing 5 μL of 1:20–1:100 diluted IP DNA and 1:500–1:1000 diluted input DNA were analyzed, and all reactions were performed in the linear range of amplification. PCR primers were designed using the software program Primer Express (ABI). An intergenic sequence on chromosome V (INT-V) that is not regulated by carbon source or low phosphate served as an internal control. Conventional PCR was carried out with 5 μL of IP DNA and typically 5 μL of 1:50 diluted input DNA, using a MJ Research Thermocycler. PCR products were separated on a 10% polyacrylamide-SDS gel, which was stained with ethidium bromide and photographed on a Nucleotech gel documentation system.
Acknowledgments
We thank Sarah Gladden, Rebecca Hamel, Kim Johnson, and Jennifer Weissblum for their expert technical assistance, and Dana Underwood is acknowledged for her comments on the manuscript. Richard Gardner and Dan Gottschling are gratefully acknowledged for their generous gift of plasmid pRG145, and Zu-Wen Sun is thanked for his advice on the elution of Flag-H2B from fixed chromatin. Supported by NIH grants GM40118 (M.A.O.) and GM55360 (S.L.B.) and NSF grant MCB-0078940 (S.L.B.). K.W.H. was supported by NIH grant CA09171 (Training Program in Basic Cancer Research to The Wistar Institute).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1149604.
References
- Barbaric S., Reinke, H., and Horz, W. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23: 3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J. and Varshavsky, A. 1985. Preferential localization of varient nucleosomes near the 5′ end of the mouse dihydrofolate reductase gene. J. Biol. Chem. 260: 7688-7697. [PubMed] [Google Scholar]
- Belotserkovskaya R. and Berger, S.L. 1999. Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit. Rev. Eukaryot. Gene Expr. 9: 221-230. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Humphrey, E.L., Erlich, R.L., Schneider, R., Bouman, P., Liu, J.S., Kouzarides, T., and Schreiber, S.L. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. 99: 8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green, M.R. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 15: 1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22: 7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm L., Crane-Robinson, C., and Sautiere, P. 1980. Proteolytic digestion studies of chromatin core-histone structure. Identification of a limit peptide of histone H2A. Eur. J. Biochem. 106: 525-530. [DOI] [PubMed] [Google Scholar]
- Borts R.H., Lichten, M., and Haber, J.E. 1986. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics 113: 551-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E.M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays 14: 9-16. [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Xiao, T., Sun, Z.W., Caldwell, J.A., Shabanowitz, J., Hunt, D.F., Allis, C.D., and Strahl, B.D. 2002. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Bryant G.O. and Ptashne, M. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11: 1301-1309. [DOI] [PubMed] [Google Scholar]
- Busch H. and Goldknopf, I.L. 1981. Ubiquitin–protein conjugates. Mol. Cell. Biochem. 40: 173-187. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., John, S., Sil, A.K., Hopper, J.E., and Workman, J.L. 2002. Gal80 confers specificity on HAT complex interactions with activators. J. Biol. Chem. 277: 24648-24652. [DOI] [PubMed] [Google Scholar]
- Davie J.R., Lin, R., and Allis, C.D. 1991. Timing of the appearance of ubiquitinated histones in developing new macronuclei of Tetrahymena thermophila. Biochem. Cell Biol. 69: 66-71. [DOI] [PubMed] [Google Scholar]
- Dohmen R.J., Madura, K., Bartel, B., and Varshavsky, A. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitinconjugating enzyme. Proc. Natl. Acad. Sci. 88: 7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J., Schneider, J., Tawiah-Boateng, M.A., Wood, A., Dean, K., Johnston, M., and Shilatifard, A. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368-28371. [DOI] [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle, C., and Winston, F. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 13: 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389: 349-352. [DOI] [PubMed] [Google Scholar]
- Hassan A.H., Neely, K.E., Vignali, M., Reese, J.C., and Workman, J.L. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6: D1054-D1064. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger, S., and Grunstein, M. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92-96. [DOI] [PubMed] [Google Scholar]
- Henry K.W. and Berger, S.L. 2002. Trans-tail histone modifications: Wedge or bridge? Nat. Struct. Biol. 9: 565-566. [DOI] [PubMed] [Google Scholar]
- Henry K., Wyce, A., Lo, W.S., Duggan, L., Emre, N.C., Kao, C.F., Pillus, L., Shilatifard, A., Osley, M.A., and Berger, S.L. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 17: 2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensold J.O., Swerdlow, P.S., and Housman, D.E. 1988. A transient increase in histone H2A ubiquitination is coincident with the onset of erythroleukemic cell differentiation. Blood 71: 1153-1156. [PubMed] [Google Scholar]
- Hoege C., Pfander, B., Moldovan, G.L., Pyrowolakis, G., and Jentsch, S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135-141. [DOI] [PubMed] [Google Scholar]
- Huang H., Kahana, A., Gottschling, D.E., Prakash, L., and Liebman, S.W. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.W., Venkatasubrahmanyam, S., Ianculescu, A.G., Tong, A., Boone, C., and Madhani, H.D. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11: 261-266. [DOI] [PubMed] [Google Scholar]
- Jason L.J., Moore, S.C., Lewis, J.D., Lindsey, G., and Ausio, J. 2002. Histone ubiquitination: A tagging tail unfolds? Bioessays 24: 166-174. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath, J.P., and Varshavsky, A. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329: 131-134. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman, A.M. 2000. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549-552. [DOI] [PubMed] [Google Scholar]
- Johnston M. and Carlson, M. 1992. Regulation of carbon and phosphate utilization. In The molecular and cellular biology of the yeast Saccharomyces; Gene expression, pp. 193-281. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.1450661
- Johnston M., Flick, J.S., and Pexton, T. 1994. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 3834-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplun L., Ivantsiv, Y., Kornitzer, D., and Raveh, D. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. 97: 10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D., Raboy, B., Kulka, R.G., and Fink, G.R. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13: 6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H. and Allis, C.D. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods 19: 425-433. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., vom Baur, E., Struhl, K., and Allis, C.D. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6: 1309-1320. [DOI] [PubMed] [Google Scholar]
- Larschan E. and Winston, F. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 15: 1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L. and Varshavsky, A. 1982. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell 28: 375-385. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- Melcher K. and Johnston, S.A. 1995. GAL4 interacts with TATA-binding protein and coactivators. Mol. Cell. Biol. 15: 2839-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelone B.A., Prakash, S., and Prakash, L. 1981. Recombination and mutagenesis in rad6 mutants of Saccharomyces cerevisiae: Evidence for multiple functions of the RAD6 gene. Mol. Gen. Genet. 184: 410-415. [DOI] [PubMed] [Google Scholar]
- Mueller R.D., Yasuda, H., Hatch, C.L., Bonner, W.M., and Bradbury, E.M. 1985. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. Disappearance of these proteins at metaphase and reappearance at anaphase. J. Biol. Chem. 260: 5147-5153. [PubMed] [Google Scholar]
- Ng H.H., Xu, R.M., Zhang, Y., and Struhl, K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277: 34655-34657. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Ciccone, D.N., Morshead, K.B., Oettinger, M.A., and Struhl, K. 2003a. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. 100: 1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Robert, F., Young, R.A., and Struhl, K. 2003b. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709-719. [DOI] [PubMed] [Google Scholar]
- Nickel B.E., Allis, C.D., and Davie, J.R. 1989. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry 28: 958-963. [DOI] [PubMed] [Google Scholar]
- Orlando V. and Paro, R. 1993. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75: 1187-1198. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Workman, J.L. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10: 187-192. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503-533. [DOI] [PubMed] [Google Scholar]
- Recht J. and Osley, M.A. 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 18: 229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H. and Horz, W. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11: 1599-1607. [DOI] [PubMed] [Google Scholar]
- Robzyk K., Recht, J., and Osley, M.A. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501-504. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Jennings, J., Canutescu, A., Link, A.J., and Weil, P.A. 2002. Proteomics of the eukaryotic transcription machinery: Identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22: 4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C., Schreiber, S.L., Mellor, J., and Kouzarides, T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407-411. [DOI] [PubMed] [Google Scholar]
- Seale R.L. 1981. Rapid turnover of the histone-ubiquitin conjugate, protein A24. Nucleic Acids Res. 9: 3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer V.A. and Davie, J.R. 1999. Role of covalent modifications of histones in regulating gene expression. Gene 240: 1-12. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Ohba, R., Cook, R.G., and Allis, C.D. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. 96: 14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.W. and Allis, C.D. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104-108. [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash, S., and Prakash, L. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes & Dev. 2: 1476-1485. [DOI] [PubMed] [Google Scholar]
- Svaren J. and Horz, W. 1997. Transcription factors vs. nucleosomes: Regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22: 93-97. [DOI] [PubMed] [Google Scholar]
- Swerdlow P.S., Schuster, T., and Finley, D. 1990. A conserved sequence in histone H2A which is a ubiquitination site in higher eucaryotes is not required for growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 4905-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly R.J. 1992. Glucose repression in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 6: 15-21. [DOI] [PubMed] [Google Scholar]
- Turner S.D., Ricci, A.R., Petropoulos, H., Genereaux, J., Skerjanc, I.S., and Brandl, C.J. 2002. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol. Cell. Biol. 22: 4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F. and Gottschling, D.E. 2002. Genome-wide histone modifications: Gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 14: 756-762. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken, P.R., and Gottschling, D.E. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745-756. [DOI] [PubMed] [Google Scholar]
- Vogel K., Horz, W., and Hinnen, A. 1989. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 9: 2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M.H. and Bonner, W.M. 1980. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8: 4671-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A., Krogan, N.J., Dover, J., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J.F., et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11: 267-274. [DOI] [PubMed] [Google Scholar]
- Wu R.S., Kohn, K.W., and Bonner, W.M. 1981. Metabolism of ubiquitinated histones. J. Biol. Chem. 256: 5916-5920. [PubMed] [Google Scholar]