Abstract
In mammalian cells, spliced mRNAs yield greater quantities of protein per mRNA molecule than do otherwise identical mRNAs not made by splicing. This increased translational yield correlates with enhanced cytoplasmic polysome association of spliced mRNAs, and is attributable to deposition of exon junction complexes (EJCs). Translational stimulation can be replicated by tethering the EJC proteins Y14, Magoh, and RNPS1 or the nonsense-mediated decay (NMD) factors Upf1, Upf2, and Upf3b to an intronless reporter mRNA. Thus, in addition to its previously characterized role in NMD, the EJC also promotes mRNA polysome association. Furthermore, the ability to stimulate translation when bound inside an open reading frame appears to be a general feature of factors required for NMD.
Keywords: Gene expression, splicing, translation, polysome, exon junction complex
An essential step in eukaryotic gene expression is the removal of introns from nascent transcripts by pre-mRNA splicing. Although much has been learned about splicing by studying it in isolation, it is now clear that in cells numerous interconnections exist between splicing and other steps in gene expression (Maniatis and Reed 2002; Orphanides and Reinberg 2002; Le Hir et al. 2003). Some of these connections occur between cotemporaneous processes, such as splicing, transcription, and polyadenylation (Proudfoot et al. 2002). However, the act of pre-mRNA splicing can also affect downstream mRNA metabolic events that do not occur until well after splicing is complete. Examples of such downstream processes are mRNA export to the cytoplasm and mRNA turnover.
The effect of pre-mRNA splicing on mRNA export has been best documented in Xenopus oocytes, where some spliced RNAs are exported to the cytoplasm more rapidly than identical RNAs not produced by splicing (Luo and Reed 1999; Zhou et al. 2000; Le Hir et al. 2001). In a few cases, it has also been observed that an intron can alter the nucleocytoplasmic distribution of a particular mRNA in mammalian tissue culture cells (Ryu and Mertz 1989; Rafiq et al. 1997). However, more recent experiments indicate that splicing may not be crucial for the export of most mRNAs (Gatfield and Izaurralde 2002; Lu and Cullen 2003; Nott et al. 2003). Another effect of splicing on downstream mRNA metabolism is its role in nonsense-mediated decay (NMD). NMD is the process by which aberrant mRNAs containing premature termination codons are targeted for accelerated degradation. This process is thought to protect cells from the potentially deleterious effects of inappropriately truncated proteins. In mammalian cells, the mechanism by which authentic stop codons are distinguished from premature stop codons relies on their position relative to the last exon–exon junction. That is, if the first in-frame stop codon occurs more than 50–55 nt upstream of at least one exon–exon junction, the mRNA containing it is targeted for degradation (Maquat and Carmichael 2001; Wilusz et al. 2001; Wilkinson and Shyu 2002).
The means by which splicing affects downstream mRNA metabolism is by altering the complement of bound proteins that, together with the mRNA, comprise the mRNP. One such alteration is the exon junction complex (EJC), found exclusively on spliced mRNAs. The EJC consists of several proteins that, upon the completion of intron excision, are deposited on the mRNA product at a conserved position, 20–24 nt upstream of exon–exon junctions (Le Hir et al. 2000a). Some EJC components remain bound even after mRNA export to the cytoplasm, where their ultimate removal requires passage of ribosomes (Dostie and Dreyfuss 2002; Lejeune et al. 2002). Core components of the EJC include the Y14, Magoh, and REF/Aly proteins (Reichert et al. 2002), of which only Y14 and Magoh remain stably associated with mRNA after nuclear export (Le Hir et al. 2001; Dreyfuss et al. 2002). Other proteins that associate with the EJC include UAP56, TAP (NXF1), SRm160, RNPS1, and Upf3 in the nucleus and Upf2 in the cytoplasm (Gatfield et al. 2001; Kim et al. 2001; Le Hir et al. 2001; Luo et al. 2001).
UAP56, REF/Aly, and NXF1 (called Sub2, Yra1, and Mex67 in Saccharomyces cerevisiae) are all mRNA export factors, and their EJC association explains the enhanced nucleocytoplasmic export of spliced RNAs in Xenopus oocytes (Le Hir et al. 2000a; Zhou et al. 2000; Luo et al. 2001). Similarly, the EJC association of Upf2 and Upf3 can explain the role of splicing in NMD (Kim et al. 2001; Le Hir et al. 2001). These proteins, along with their binding partner Upf1, were originally defined as essential NMD factors in S. cerevisiae. In mammalian cells, the three Upf proteins and the EJC proteins RNPS1, Y14, and Magoh can all activate NMD when bound downstream from an in-frame stop codon (Lykke-Andersen et al. 2000, 2001; Fribourg et al. 2003; Gehring et al. 2003). Presumably, RNPS1, Y14, and Magoh function in NMD by recruiting one or more of the Upf proteins through direct or indirect interactions (Kim et al. 2001; Lykke-Andersen et al. 2001). Finally, Y14 and Magoh are both required for the proper localization of oskar mRNA during Drosophila oogenesis (Hachet and Ephrussi 2001; Mohr et al. 2001). Thus, splicing may also be important for proper localization of mRNAs in the cytoplasm.
In addition to the mRNA export and decay effects discussed above, several reports have indicated that spliced transcripts produce more molecules of protein per mRNA molecule than do otherwise identical transcripts not made by splicing (Braddock et al. 1994; Matsumoto et al. 1998; Lu and Cullen 2003; Nott et al. 2003; Wiegand et al. 2003). Recently we reported a quantitative analysis of the effects of introns on the expression of a T-cell receptor minigene and Renilla luciferase reporter in mammalian tissue culture cells (Nott et al. 2003). In both cases, introns internal to the open reading frame (ORF) were found to enhance mRNA translational yield. Here we show that spliced mRNAs are more efficiently incorporated into polysomes than otherwise identical mRNAs not produced by splicing. Experiments in Xenopus oocytes confirmed that this translational enhancement can be attributed to EJC deposition. In HeLa cells, both the enhanced translational yield and polysome association could be reproduced by tethering three different EJC proteins, RNPS1, Y14, and Magoh, to an intronless reporter. Remarkably, parallel tethering of hUpf1, hUpf2, and hUpf3b produced similar results. Thus, in addition to its previously characterized role in NMD, we show here that the EJC is capable of promoting mRNA translation. Furthermore, our data are consistent with the idea that proteins involved in NMD play broader roles in mRNA metabolism than simply the elimination of aberrant messages.
Results
Pre-mRNA splicing enhances polysome association of mRNA
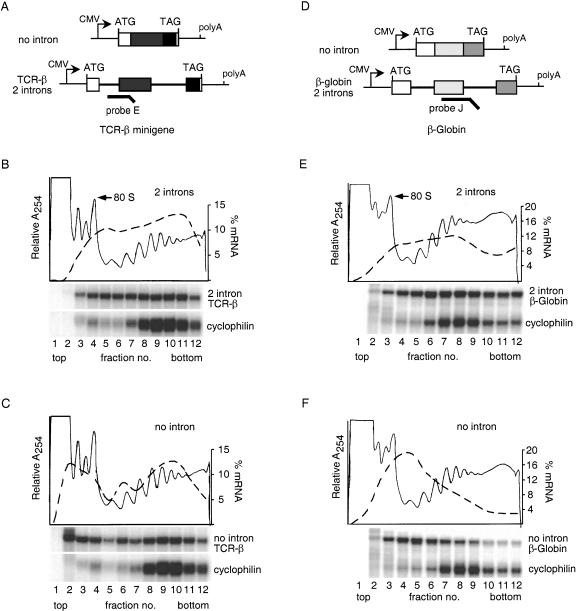
We previously reported that spliced mRNAs exhibit increased translational yield as compared with no-intron mRNAs in mammalian tissue culture cells (Nott et al. 2003). To determine if this effect was a consequence of increased translational efficiency, we analyzed the distribution of several spliced or no-intron mRNAs on cytoplasmic polyribosomes (polysomes; Fig. 1). Two constructs, TCR-β and β-globin, contained two introns each (Fig. 1A,D; Lykke-Andersen et al. 2000; Nott et al. 2003), whereas a third, Renilla luciferase, contained a single intron (Nott et al. 2003). Plasmids expressing these and their respective no-intron controls were transiently transfected into HeLa cells, and polysome analysis was performed 24 h later. Endogenous cyclophilin mRNA served as an internal control for relative polysome integrity. Treatment of the cells with puromycin, an antibiotic that causes nascent peptide release and polysome disruption, confirmed that sedimentation of the reporter mRNAs and cyclophilin in denser fractions did reflect polysome association (data not shown).
Figure 1.
Splicing enhances mRNA polysome association. (A,D) Schematic representation of TCR-β and β-globin constructs. Boxes represent exons, and lines connecting them denote introns. RPA probes are indicated by heavy lines. (B,C,E,F) Sucrose gradient fractionation of cytoplasmic extracts from cells expressing no-intron or intron-containing versions of TCR-β and β-globin. RNA extracted from each fraction (see Materials and Methods) was subject to RPA with probe E or J (A,D) and a probe specific to endogenous cyclophilin mRNA. Protected fragments were separated on a 10% denaturing polyacrylamide gel. Relative absorbance at 254 nm is depicted by thin lines, and the percent total mRNA in each fraction is shown by the dashed lines (scale on right).
Like the endogenous cyclophilin mRNA, the bulk of spliced TCR-β mRNA cofractionated with polysomes (Fig. 1B, fractions 5–12). In contrast, a large fraction of the no-intron TCR-β mRNA was found at the top of the gradient in the region containing individual ribosomal subunits and 80S monosomes (Fig. 1C). Similar results were observed for the β-globin (Fig. 1E,F) and the Renilla luciferase constructs (data not shown). Thus, for three different genes with different introns, splicing correlates with enhanced mRNA polysome association.
The exon junction complex plays a role in translation enhancement
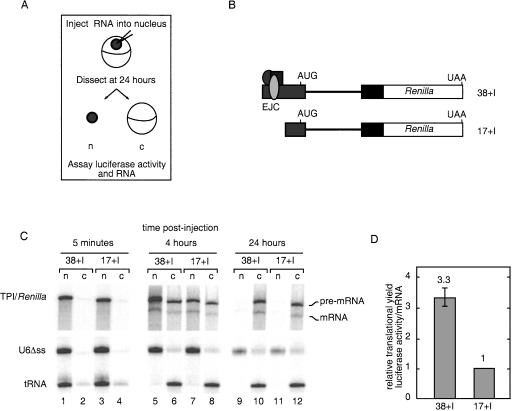
We next wanted to determine whether the enhanced translational yield of spliced mRNAs could be observed when splicing was uncoupled from other RNA processing events (e.g., transcription and polyadenylation) and to what extent it was attributable to EJC deposition. This was most easily addressable in the Xenopus oocyte system, in which we injected the Renilla luciferase pre-mRNAs schematized in Figure 2B. Pre-mRNAs were designed with a 38- or 17-nt 5′-exon plus the human triose phosphate isomerase intron 6 (TPI intron). Because the EJC is deposited at a fixed distance, 20–24 nt upstream of an exon–exon junction, the 38- and 17-nt exons are just long enough or too short, respectively, to accept an EJC (Le Hir et al. 2001). Uniformly radiolabeled and m7GpppG-capped transcripts were generated in vitro using DNA templates terminating 4 nt downstream from the first in-frame stop codon. Thus, none of the injected RNAs contained a poly(A) tail. The AUG start codon was located in the 5′-exon, and several in-frame stop codons in the intron ensured that functional luciferase could not be translated from the unspliced pre-mRNAs (38 + I and 17 + I). Coinjected controls included U6Δss snRNA (to control for nuclear injection and nuclear envelope integrity), human initiator methionyl-tRNA (which is rapidly exported via an export pathway distinct from mRNA; Jarmolowski et al. 1994), and unlabeled Firefly luciferase mRNA to control for translational differences between oocytes. By following both subcellular RNA distribution and luciferase activity as a function of time, we obtained information on both RNA export efficiency and its subsequent translation in the cytoplasm.
Figure 2.
EJC-dependent stimulation of translational yield in Xenopus oocytes. (A) Overview of experimental procedure. (B) Schematic representation of in vitro transcribed, radiolabeled RNAs injected into Xenopus oocytes. TPI intron 6 (thick line) and flanking TPI exons (gray and black boxes) and Renilla luciferase ORF (white box) are indicated. First exons were either 38 or 17 nt in length. (C) Denaturing polyacrylamide gel electrophoresis of RNA extracted from nucleus (n) and cytoplasm (c) at 5 min, 4 h, and 24 h post-injection. (Top panel) 6% gel for Renilla RNAs; (bottom panel) identical samples loaded on a 10% gel to separate the U6Δss RNA and tRNA. (D) Translational efficiencies at 24 h post-injection relative to 17 no I mRNA. Translational efficiencies from two independent experiments were calculated by first normalizing Renilla luciferase activity to Firefly luciferase activity and then dividing by cytoplasmic mRNA levels; error bars represent the range.
Immediately after injection (5-min time point), both precursor RNAs were found predominantly in the nuclear compartment (Fig. 2C). Examination of the 4- and 24-h time points revealed that both pre-mRNAs were spliced and the mRNAs exported with comparable efficiencies. As is often observed in oocyte experiments, a significant percentage of the injected pre-mRNAs was also exported (Fig. 2C; Le Hir et al. 2001). This is likely caused by an inability of the endogenous splicing apparatus to capture and retain all of the injected pre-mRNA before it can escape to the cytoplasm. Nonetheless, the presence of cytoplasmic pre-mRNA was of little consequence for our analysis because the pre-mRNAs were incapable of generating functional luciferase (see above), and no detectable luciferase activity was produced when the pre-mRNAs were injected directly into the cytoplasm (data not shown).
Protein expression was determined by monitoring cytoplasmic luciferase activities. Whereas neither construct produced detectable activity at 4 h (data not shown), light production was readily measurable in both cases by 24 h. A quantitative analysis of the relative translational yields was obtained by normalizing luciferase activities to cytoplasmic mRNA levels (Fig. 2D). This revealed that the 38 + I mRNA yielded 3.3 times more luciferase activity per mRNA molecule than did the 17 + I mRNA. This difference was not due to the 5′-UTR length as the respective no-intron control mRNAs were translated with equal efficiency in parallel experiments (data not shown). Thus, we conclude that the observed effect of splicing on translational yield is caused by EJC deposition and is independent of transcription and polyadenylation.
Individual EJC proteins enhance protein expression in a tethering assay
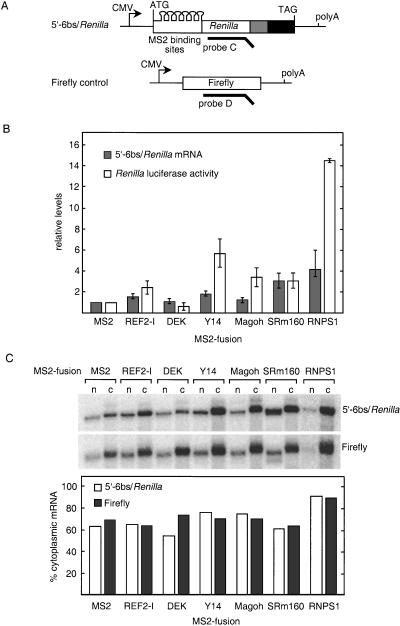
Having demonstrated that EJC deposition can mediate splicing-dependent enhancement of translational yield in Xenopus oocytes, we next wanted to determine whether this activity could be assigned to one or more specific protein(s). Thus, we tested whether the intron-dependent enhancement in translational yield could be replicated by tethering individual proteins within the ORF of Renilla luciferase. An intronless Renilla luciferase reporter carrying six MS2-binding sites at the 5′-end of the ORF (5′-6bs/Renilla; Fig. 3A) was cotransfected into HeLa cells along with constructs expressing individual EJC proteins as fusions with bacteriophage MS2 coat protein (see Materials and Methods). A plasmid encoding a no-intron Firefly luciferase gene was also cotransfected as a control for both luciferase activity measurements and mRNA comparisons. The MS2 coat protein alone served as the control for reporter expression in the absence of a fusion partner. Expression of all fusion proteins was confirmed by Western blotting (data not shown).
Figure 3.
Effects of tethering individual EJC proteins inside the Renilla luciferase ORF. (A) Schematic representation of 5′-6bs/Renilla reporter construct and Firefly control. Six MS2-binding sites were inserted 49-nt downstream of the start codon to be in-frame with the Renilla luciferase gene. (B) Levels of 5′-6bs/Renilla reporter mRNA and luciferase activity normalized to Firefly control and represented relative to MS2 alone. The data shown are the average of three to four independent experiments; error bars represent standard deviation. (C) Nuclear and cytoplasmic distribution of 5′-6bs/Renilla reporter and Firefly control mRNA in the presence of various MS2 fusion proteins. RNA was extracted from fractionated nuclear (n) and cytoplasmic (c) compartments and analyzed by RPA (top panel). The percent of total mRNA in the cytoplasm (lower panel).
The fusion proteins fell into four functional classes: (1) those that had little or no effect, (2) those that increased reporter mRNA abundance only, (3) those that increased mRNA translational yield only, and (4) one that increased both mRNA abundance and translational yield (Fig. 3B). Constituting the first class were the DEK and REF2-1 MS2-fusions. DEK had no effect on either mRNA abundance or luciferase activity, whereas tethering of REF2-1 only slightly increased both. In contrast, coexpression of MS2–SRm160 resulted in a threefold increase in Renilla luciferase activity. A concomitant threefold increase in 5′-6bs/Renilla mRNA abundance indicated that although tethered SRm160 can enhance mRNA levels, it had no detectable effect at the translational level. The third class consisted of Y14-MS2 and Magoh-MS2, both of which increased protein expression three- to fourfold without significantly affecting reporter mRNA abundance. Finally, coexpression of MS2–RNPS1 increased reporter mRNA abundance (∼fourfold) but enhanced luciferase expression to a much greater extent (∼14-fold). Thus, tethering RNPS1 to the reporter mRNA led to an ∼fourfold increase in mRNA levels and an ∼threefold increase in translational yield.
We next examined whether any of the fusion proteins altered the nucleocytoplasmic distribution of Renilla reporter mRNA. Comparison to the cotransfected Firefly control revealed that only MS2–DEK had any effect on reporter mRNA distribution, but this decrease in the proportion of mRNA that was cytoplasmic was minor at best (Fig. 3C). Because tethering of Y14, Magoh, or RNPS1 had no effect on the nucleocytoplasmic distribution of the Renilla reporter mRNA, the enhancements in translational yield produced by these proteins were not simply attributable to more efficient cytoplasmic localization of the message.
Taken together, the above results indicate that when tethered as MS2 fusions near the 5′-end of an ORF, individual EJC components can mediate significant enhancements of gene expression. The sole effect observed here of tethering SRm160 was to increase mRNA levels. Y14 and Magoh increased protein expression, mostly by increasing the amount of protein produced per mRNA. Only RNPS1 increased both mRNA levels and translational yield.
Tethered Upf proteins also enhance translational yield
Of all the EJC proteins tested here, the only ones that increased translational yield when tethered inside the Renilla luciferase ORF were RNPS1, Y14, and Magoh. Interestingly, all of these proteins were previously shown to trigger NMD when tethered downstream from a termination codon in β-globin mRNA (Lykke-Andersen et al. 2001; Fribourg et al. 2003; Gehring et al. 2003). In contrast, other EJC proteins that did not enhance translational yield in the experiments presented here also failed to trigger NMD (Lykke-Andersen et al. 2001). We next wanted to ask if the ability to trigger NMD generally correlates with the ability to enhance translational yield.
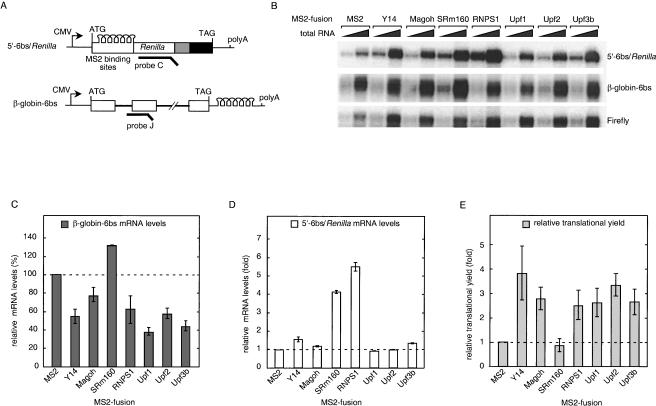
Three other proteins previously documented to trigger NMD when tethered downstream from the termination codon in β-globin mRNA are the NMD factors Upf1, Upf2, and Upf3b (Lykke-Andersen et al. 2000). To test the effects of tethering these proteins inside an ORF, plasmids encoding the appropriate MS2-fusion proteins were cotransfected into HeLa cells with the 5′-6bs/Renilla reporter and Firefly control as described above. A β-globin-6bs reporter carrying six MS2-binding sites downstream from the normal termination codon was also cotransfected to monitor NMD activation by the expressed proteins (Fig. 4A; Lykke-Andersen et al. 2000). The MS2-binding sites in the β-globin-6bs reporter were identical to the MS2-binding sites in the 5′-6bs/Renilla mRNA; they differed only in their position relative to the respective ORFs. The Y14, Magoh, and RNPS1 fusions were also tested in this set of experiments. MS2–SRm160 served as a negative control because it neither enhances translational yield (Fig. 3B) nor triggers NMD (Lykke-Andersen et al. 2001).
Figure 4.
Upf1, Upf2, and Upf3b also enhance the translational yield of 5′-6bs/Renilla mRNA. (A) Schematic representation of 5′-6bs/Renilla and β-globin-6bs constructs carrying identical MS2-binding sites. RPA probes are indicated with thick lines. (B) RPA of 2 and 8 μg of total mRNA extracted from transfected cells. (C) β-globin-6bs mRNA levels in the presence of MS2-fusion proteins relative to MS2 alone. The data reflect the average of two independent experiments; error bars represent the range. (D) Renilla reporter mRNA levels in the presence of MS2-fusion proteins relative to MS2 alone. The data reflect the average of two independent experiments; error bars represent the range. (E) Relative translational yields of Renilla luciferase. Luciferase activities were normalized to mRNA levels and are represented relative to MS2 alone. The data reflect the average of three independent experiments; errors were propagated using standard algorithms.
Consistent with previous reports (Lykke-Andersen et al. 2000, 2001; Fribourg et al. 2003; Gehring et al. 2003), tethering of Upf1, Upf2, Upf3b, Y14, Magoh, or RNPS1 downstream from the termination codon significantly reduced β-globin-6bs mRNA abundance (Fig. 4B,C). However, none of the Upf proteins had any significant effect on the coexpressed 5′-6bs/Renilla reporter (Fig. 4B,D). Thus, when tethered inside an ORF, the Upf proteins neither increase nor decrease mRNA levels. In contrast, both the RNPS1 and SRm160 MS2-fusions led to higher 5′-6bs/Renilla mRNA levels as was observed in Figure 3. MS2–SRm160 also yielded a slight increase in β-globin-6bs mRNA abundance, consistent with its ability to increase mRNA levels (Fig. 3), but not trigger NMD (Lykke-Andersen et al. 2001).
Whereas the Upf proteins failed to affect mRNA levels when tethered inside the Renilla ORF, they did enhance the amount of active luciferase produced per mRNA. When translational yields were calculated by normalizing the luciferase activity to mRNA levels, all three Upf proteins increased the translational output to extents indistinguishable from RNPS1, Y14, and Magoh (Fig. 4E). As was the case for the latter proteins, the increased translational yields mediated by the tethered Upf proteins were unlikely caused by enhanced mRNA export because the nucleocytoplasmic localization of the reporter mRNA was unchanged in the presence of the MS2–Upf proteins compared with MS2 alone (data not shown). Thus, all proteins tested to date that are capable of triggering NMD when tethered downstream of a termination codon also lead to increased translational yields when tethered inside an ORF.
Tethered EJC and Upf proteins promote mRNA polysome association
The data presented in Figure 1 demonstrated that the greater translational yields observed with spliced mRNAs correlated with their increased polysome association. To determine whether tethering of an EJC or Upf protein could replicate this effect, we examined the polysome distribution of the 5′-6bs/Renilla reporter mRNA in the absence or presence of coexpressed MS2 proteins. In the absence of any MS2 fusion (vector) or in the presence of MS2 alone (MS2), a large proportion of the 5′-6bs/Renilla mRNA was found in fractions that contained ribosomal subunits and monosomes, whereas only a small fraction of the reporter mRNA cosedimented with polysomes (Fig. 5A,D). Thus, expression of MS2 coat protein alone had no effect on polysome association of 5′-6bs/Renilla mRNA. In contrast, coexpression of the MS2–RNPS1 fusion significantly enhanced the fraction of 5′-6bs/Renilla mRNA cosedimenting with polysomes (Fig. 5A, lower panel; Fig. 5D). Distributions of endogenous cyclophilin mRNA and cotransfected Firefly control (data not shown) were unaffected in all experiments. A control experiment using puromycin to disrupt polysomes confirmed that the sedimentation of 5′-6bs/Renilla mRNA in denser fractions in the presence of MS2–RNPS1 was, indeed, due to polysome association (Fig. 5B).
Figure 5.
MS2-fusions of RNPS1, Y14. Magoh and Upf proteins enhance polysome association of Renilla reporter mRNA. (A) Sucrose gradient fractionation of cytoplasmic extracts from cells expressing 5′-6bs/Renilla reporter mRNA and individual MS2-fusion proteins (indicated on left). The absorbance profile is representative of all three gradients. RNA extracted from each fraction was subjected to RPA with probe C (Fig. 3A) and a probe specific to endogenous cyclophilin mRNA. Protected fragments were separated on a 10% denaturing polyacrylamide gel. (B) MS2–RNPS1 cotransfected cells were treated with Puromycin prior to lysis and fractionation. (C) Sucrose gradient fractionation of cytoplasmic extracts from cells expressing 5′-6bs/Renilla reporter mRNA plus MS2 alone, MS2–Y14, MS2–Magoh, MS2–Upf1, MS2–Upf2, or MS2–Upf3b. The absorbance profile is representative of all gradients. (D) Quantitative representation of 5′-6bs/Renilla mRNA distribution in polysome gradients in the presence of indicated MS2-fusion proteins. Relative mRNA levels in each fraction were calculated as a percent of the total, and amounts in fractions 1–4 and 5–12 were pooled to represent mRNAs cofractionating with monosomes and ribosomal subunits and polysomes, respectively. The data represent the averages of two independent experiments, and error bars reflect the range.
We also examined the polysome distribution of the 5′-6bs/Renilla reporter tethered to Y14, Magoh, Upf1, Upf2, or Upf3b (Fig. 5C). As observed with RNPS1 tethering of Y14, Magoh and all three Upf proteins shifted a greater percentage of reporter RNA into polysome-containing fractions (Fig. 5C), whereas distribution of endogenous cyclophilin mRNA was unaffected (data not shown). A quantitative measure of mRNA distributions upon tethering the respective MS2-fusion proteins was obtained by calculating the relative mRNA abundance in fractions 1–4 (monosomes and ribosomal subunits) and fractions 5–12 (polysomes; Fig. 5D). Thus, the translational enhancement observed upon tethering of the Upf proteins presumably occurs via the same mechanism as that mediated by splicing (Fig. 1) and EJC deposition.
Discussion
We have established that the increased translational yield observed with spliced mRNAs in mammalian cells correlates with their enhanced polysome association compared with no-intron transcripts. In Xenopus oocytes, this enhanced translation due to splicing can be uncoupled from transcription and polyadenylation and is attributable to deposition of an EJC. Splicing-dependent translational and polysome association enhancements in HeLa cells could be fully reproduced upon functional tethering of three different EJC proteins, Y14, Magoh, and RNPS1, to a reporter transcript. Thus, in addition to its previously documented role in NMD, the EJC also serves as a positive effector of eukaryotic mRNA translation. Notably, when tethered at the 5′-end of a reporter ORF, the NMD factors Upf1, Upf2, and Upf3b also enhanced mRNA translational yield and polysome association. Thus, all proteins presently known to trigger NMD when bound downstream of an ORF also enhance mRNA translational yield and polysome association when tethered inside an ORF. This suggests that factors involved in NMD likely play broader roles in translation than simply the elimination of aberrant transcripts.
Enhanced translation of spliced mRNAs
Previous studies have demonstrated that introns can enhance eukaryotic gene expression (Le Hir et al. 2003). Some of this stimulation is due to a splicing-dependent increase in mRNA levels. An additional effect is a splicing-dependent enhancement of mRNA translational yield (Braddock et al. 1994; Matsumoto et al. 1998; Lu and Cullen 2003; Nott et al. 2003). In the experiments presented here, we found that enhanced translational yield from spliced mRNAs correlates with a greater percentage of these mRNAs cofractionating with polysomes. These results are in agreement with a hypothesis that eukaryotic cells contain two pools of cytoplasmic mRNAs: translationally active (in polysomes) and translationally inactive (free mRNPs; Spirin 1996; Zong et al. 1999). Because initial ribosome recruitment is generally limiting for translation in vivo (Sonenberg 1996), the fraction of mRNA in either pool may well be a function of this recruitment efficiency. Consistent with this idea, we observed that the no-intron mRNAs that did cosediment with polysomes peaked in the same fractions as did their corresponding spliced mRNAs (Fig. 1; data not shown). This is most easily seen with the TCR-β constructs (Fig. 1B,C), where the polysomal peak for both spliced and no intron mRNAs occurs in fractions 8–11. This suggests that once an mRNA enters the translationally active pool, its ability to recruit additional ribosomes to form polysomes is independent of its synthetic history.
To enhance mRNA translational efficiency, pre-mRNA splicing could push a greater proportion of mRNA into the active pool either by preventing the association of proteins that inhibit or by depositing factors that stimulate the initial round of translation, or both. A previous study in Xenopus oocytes revealed that in vitro transcribed mRNA injected into the nucleus is translationally repressed upon export to the cytoplasm (Braddock et al. 1994). This repression could be overcome either by inclusion of a spliceable intron in the 3′-UTR or by coinjection of antibodies against FRGY2 proteins. The FRGY2 family of DNA/RNA-binding proteins are known translational repressors, as are CPEB and maskin (Bouvet and Wolffe 1994; Mendez and Richter 2001). Translational repression in mammalian cells may involve the known FRGY2 and CPEB homologs (Gebauer and Richter 1996; Herbert and Hecht 1999) and/or an additional abundant mRNP protein, p50 (Minich and Ovchinnikov 1992; Pisarev et al. 2002). Given the large number of RNA helicases known to be associated with the splicing machinery, it is possible that one or more of these proteins could serve as RNPases (Schwer 2001; Will and Luhrmann 2001) to prevent the association of such translational repressors. In the future it will be of interest to define the complete complement of proteins associated with spliced and unspliced mRNPs, and determine how the association of factors involved in translational repression might be altered by splicing or other nuclear events.
Our results from Xenopus oocytes indicate that much of the translational enhancement due to splicing can be attributed to EJC deposition. Using pre-mRNAs either too short or just long enough to accept an EJC, we found that a spliced mRNA capable of accepting an EJC produced >threefold more luciferase activity per cytoplasmic mRNA than a spliced mRNA incapable of harboring an EJC (Fig. 2). The magnitude of this effect was very similar to the two- to fourfold overall enhancement in translational yields observed for spliced mRNAs in HeLa cells (Nott et al. 2003). A similar short exon experiment carried out in transiently transfected HeLa cells (Wiegand et al. 2003) yielded findings consistent with our oocyte data. The oocyte experiments, however, allowed us to analyze the effects of EJC deposition on translational enhancement independent of any potentially confounding effects on transcription or polyadenylation (Le Hir et al. 2003).
Finally, in all cases we have so far examined, increased translational yield from spliced mRNAs is more consistent with their enhanced polysome association than with their enhanced nucleocytoplasmic export. We and others recently reported little or no alteration in the nucleocytoplasmic distribution of full-length mRNAs in mammalian tissue culture cells dependent on splicing (Lu and Cullen 2003; Nott et al. 2003). In the oocyte experiments reported here (Fig. 2), we saw no EJC-dependent difference in mRNA export at either 4 or 24 h. Importantly, at the 4-h time point, no luciferase activity was yet detectable, indicating that any EJC effect on protein expression in this experiment must have occurred subsequent to mRNA export.
Effects of individual EJC proteins on gene expression
Using an MS2 tethering strategy to recruit individual EJC proteins to an intronless reporter, we found that SRm160, Y14, Magoh, and RNPS1 all yielded significantly higher levels of gene expression than did MS2 alone (Fig. 3). Dependent on the bound protein, this enhancement was caused either by increased mRNA levels or by greater translational yield, or a combination of the two. Again, however, we observed no EJC protein-dependent effect on nucleocytoplasmic mRNA distribution. Thus, consistent with the above discussion, the main consequences of EJC factor recruitment for expression of the constructs examined here were to increase mRNA abundance and translational efficiency. From the tethering data, we can now assign these effects to individual EJC proteins.
Increased mRNA levels were observed upon tethering of either SRm160 or RNPS1 to a Renilla luciferase reporter. This could be caused by enhanced transcription, more efficient 3′-end formation, decreased degradation of nascent transcripts, or some combination of effects. The RNPS1-dependent increase in reporter mRNA levels could be caused by recruitment of factors involved in transcription. A general activator of pre-mRNA splicing in vitro, RNPS1 contains an RNA recognition motif (RRM) and a serine-rich domain (Badolato et al. 1995; Mayeda et al. 1999). The serine-rich domain is known to interact with the PITSLRE family of protein kinases (Loyer et al. 1998). Because of their presence in active transcription complexes and their interaction with transcriptional elongation factor ELL2, PITSLRE kinases were recently implicated in modulating pre-mRNA synthesis (Trembley et al. 2002). On the other hand, two independent groups have recently shown that both SRm160 and RNPS1 can promote 3′-end cleavage when tethered to reporter transcripts in vivo (McCracken et al. 2003; Wiegand et al. 2003). This is perhaps owing to more efficient recruitment of the components of the 3′-end cleavage machinery. SRm160 is known to interact with CPSF-160, a core component of the cleavage and polyadenylation machinery (McCracken et al. 2002), and more recently a genetic interaction has been reported between the Caenorhabditis elegans orthologs of SRm160 and the cleavage stimulation factor-I 50 kD subunit, CStf-50 (McCracken et al. 2003). Thus, MS2–SRm160, and MS2–RNPS1 by virtue of its association with SRm160 (McCracken et al. 2003), could have stimulated mRNA production by facilitating 3′-end formation of the tethered Renilla transcripts.
The increase in luciferase activity observed upon tethering SRm160 could be fully explained by the corresponding increase in reporter mRNA levels (∼threefold; Fig. 3B). Therefore, SRm160 has no apparent effect on translational yield (Fig. 4E). In contrast, tethering of RNPS1, Y14, or Magoh all yielded ∼threefold more luciferase activity per mRNA molecule than tethering of MS2 alone (Fig. 4E). This effect was most likely due to enhanced translational efficiency and not facilitated export, as none of the tethered proteins had any significant effect on nucleocytoplasmic distribution of the Renilla reporter mRNA (Fig. 4C). In an independent study, Wiegand et al. (2003) also demonstrated that Y14 and RNPS1 failed to enhance export of a tethered reporter mRNA. Rather, the increased polysome association of bound mRNAs in our experiments (Fig. 5) directly demonstrated that the tethered proteins enhanced translational efficiency. Consistent with an ability to affect cytoplasmic translation, RNPS1 and Y14 are both known shuttling proteins, and Magoh shows both nuclear and cytoplasmic localization (Kataoka et al. 2000, 2001; Lykke-Andersen et al. 2001).
A question yet to be addressed is how these EJC proteins are able to enhance translational yield when they are thought to be removed from spliced mRNAs upon the first passage of ribosomes (Dostie and Dreyfuss 2002; Lejeune et al. 2002). One possibility is that just as pre-mRNA splicing stably alters mRNP composition by EJC deposition (Le Hir et al. 2000b), the presence of an EJC may promote stable formation of a translationally active mRNP. This could be accomplished, for example, through EJC-dependent recruitment of translation initiation factors. Alternatively, before their removal, EJCs may target mRNPs to subcellular locations highly active in translation. In eukaryotic cells, most cytosolic ribosomes are known to be associated with the actin cytoskeleton, and active translation is highly dependent on the integrity of this structure (Bonneau et al. 1985; Negrutskii et al. 1994; Hovland et al. 1996; Stapulionis et al. 1997). The Y14 and Magoh requirement for proper localization of oskar mRNA in developing Drosophila embryos (Hachet and Ephrussi 2001; Mohr et al. 2001) could reflect a broader role for these proteins in targeting spliced mRNAs to particular locations in the cytoskeleton. In the future it will be of interest to examine the relationships between splicing, translation, and cytoskeletal mRNA localization, and the roles of individual EJC components in each of these processes.
Why would it be beneficial for spliced mRNAs to be better translation templates than no-intron mRNAs? In mammalian cells, where most mRNAs are spliced, the presence of an EJC may differentiate newly minted mRNAs from those that have already experienced translation. Enhanced translation initiation on EJC-carrying mRNAs would ensure that newly made mRNAs are assimilated into polysomes as rapidly as possible. Such coupling between mRNA synthesis and translation would therefore decrease the lag time between transcriptional activation and protein expression.
The relationship between translation and NMD
Previously, Lykke-Andersen et al. (2000, 2001) used the MS2 tethering strategy to determine which NMD and/or EJC proteins are responsible for communicating the positions of exon–exon junctions to the NMD machinery. The NMD factors Upf1, Upf2, and Upf3b, and the EJC proteins Y14, Magoh, and RNPS1 have now all been shown to trigger NMD when bound to the 3′-UTR of β-globin mRNA (Lykke-Andersen et al. 2000, 2001; Fribourg et al. 2003; Gehring et al. 2003). Here, we found that all of these same proteins stimulate translational yield to a similar extent when tethered inside the Renilla luciferase ORF. Thus, all proteins shown to date that target mRNAs to preferential decay when bound to the 3′-UTR also increase translational yield when tethered inside an ORF. Our experiments do not address whether these two activities are due to the tethered proteins themselves or some common binding partner. Nonetheless, the data do indicate a tight correlation between proteins capable of enhancing translation and proteins able to trigger NMD.
That proteins involved in NMD may play more general roles in translation is borne out by results from several other studies. For example, in S. cerevisiae, Upf1p, Upf2p, and Upf3p are required for both NMD and efficient translation termination (Czaplinski et al. 1998; Maderazo et al. 2000; Wang et al. 2001). There are several well-documented interactions between the Upf proteins and the release factors eRF1 and eRF3 (Czaplinski et al. 1998; Wang et al. 2001). Conversely, mutations in at least one general translation factor can also affect NMD. Certain mutations in two different subunits of the general translation initiation factor eIF3 are known to attenuate NMD. This role of eIF3 in NMD possibly reflects its activity in promoting ribosomal subunit dissociation upon translation termination (Cui et al. 1999; Welch and Jacobson 1999). Also documented are several structural similarities between NMD and translation initiation factors. Human Upf2 contains an eIF4G homology domain (Aravind and Koonin 2000; Ponting 2000) and, like eIF4G, has been shown to interact with both eIF4AI and eIF3 (Gingras et al. 1999; Mendell et al. 2000). Other proteins containing eIF4G homology domains include CBP80, a subunit of the nuclear cap-binding complex (McKendrick et al. 2001), and PAIP1 (Craig et al. 1998). It is possible that like eIF4G, CBP80, and PAIP1, human Upf2 has a function in modulating translation initiation. Finally, in S. cerevisiae, Upf1p, Upf2p, and Upf3p are known to regulate the decapping and degradation of both wild-type and nonsense-containing transcripts (He and Jacobson 2001). Thus, in all cases that have been so far examined, proteins required for NMD have additional functions either in translation initiation or termination or in general mRNA turnover. This is consistent with the hypothesis that NMD is a natural consequence of improper translation termination and not a specific process that evolved for the exclusive purpose of eliminating aberrant messages (Hilleren and Parker 1999; Schell et al. 2002). A question yet to be addressed is whether the effects of EJC proteins on translation and NMD are entirely separate functions, or are simply alternative outcomes of the same intermolecular interactions contingent on their position relative to the ORF.
Materials and methods
Plasmids, cell culture, and transfections
T-cell receptor-β minigene (TCR-β) and β-globin constructs with or without introns have been described previously (Lykke-Andersen et al. 2000; Nott et al. 2003). For the short exon experiments in Xenopus oocytes, TPI intron 6 with 5′-exons of different lengths were cloned in-frame with the Renilla luciferase gene. RNA was transcribed in vitro from linearized plasmid templates. The 5′-6bs/Renilla luciferase reporter was derived from a construct described previously (TPI/Renilla; Nott et al. 2003), by replacing the 5′ TPI cassette with six MS2 hairpin loops from β-Globin-6bs (Lykke-Andersen et al. 2000). N-terminal MS2 fusions of REF2-I, DEK, RNPS1, SRm160, Upf1, Upf2, and Upf3b have been described previously (Lykke-Andersen et al. 2000, 2001). C-terminal MS2 fusions of Y14 and Magoh were cloned and expressed from a pcDNA 3.1 vector. HeLa cells were cultured in DMEM supplemented with 10% fetal calf serum. All cells were transfected at 40%–50% confluency using Superfect (QIAGEN) according to the manufacturer's instructions. Luciferase assays and RNase protection analyses (RPAs) were performed as described previously (Nott et al. 2003).
Polysome analysis
Polysome profiles were performed essentially as described (Johannes and Sarnow 1998). For TCR-β, HeLa cells in 100-mm dishes were transfected with 16 μg of no-intron plasmid or 2 μg of intron-containing construct plus 14 μg of pcDNA 3.1. For β-globin experiments, 15μg of no-intron β-globin plasmid or 5 μg of two-intron β-globin plasmid and 10 μg of pcDNA3.1 were transfected. For tethering experiments using RNPS1, Upf1, Upf2, and Upf3b, 4 μg of MS2 fusion plasmid and 1.35μg of 5′-6bs/Renilla reporter were cotransfected along with 8.3 μg of pcDNA3.1 and 1.35μg of Firefly control plasmid. Also, 9 μg of MS2–Y14 and MS2–Magoh were cotransfected with 3 μg of 5′-6bs/Renilla reporter and 3 μg of Firefly control plasmid. Cultures were supplemented with 100 μg/mL cycloheximide 24–36 h post-transfection and incubated for 30 sec at room temperature. Cells were washed three times with ice-cold PBS containing 100 μg/mL cycloheximide and lysed directly on the plate at 4°C by addition of 500 μL of polysome extraction buffer (15 mM Tris-Cl at pH 7.4, 15 mM MgCl2, 60 mM NaCl, 0.5% Triton X-100, 100 μg/mL cycloheximide, 1 mg/mL heparin, 200 units RNasin [Promega], 5 mM PMSF) and gentle scraping. When indicated, puromycin (100 μg/mL) was added to the cultures 2 h prior to harvesting and cycloheximide was omitted from the gradient. For each construct, lysates from two 100-mm dishes were pooled into a microcentrifuge tube and incubated for 10 min on ice with occasional mixing. Nuclei and debris were removed by centrifugation at 12,000g for 5 min. Then 1 mL of each cytoplasmic lysate was layered onto an 11-mL 10%–50% sucrose gradient (Ruan et al. 1997) and centrifuged at 4°C in an SW40 rotor (36,000 rpm) for 2 h and 15 min. Fractions (1 mL) were collected from the top with concomitant measurement of absorbance at 254 nm, using an ISCO fraction collection system. Fractions were adjusted to 0.5% SDS and digested with Proteinase K (100 μg/mL final). RNA was extracted and treated with RQ1 RNase-free DNaseI (Promega) and analyzed by RPA.
Xenopus laevis oocyte microinjections
DNA templates for in vitro synthesis of U6Δss snRNA and human initiator methionyl-tRNA have been described (Jarmolowski et al. 1994). Synthesis of all transcripts was primed with the m7GpppG cap dinucleotide (NEB). Stage VI oocytes were injected as described by Jarmolowski et al. (1994), and ∼25 nL of RNA mixture along with Blue Dextran was injected per oocyte for all experiments. Five oocytes for each time point were dissected and verified for nuclear injection by visual assay for Blue Dextran, and a portion of the cytoplasm was used to measure Renilla and Firefly luciferase activity; RNA was extracted from the remainder. Extracted RNA equivalent to 1 oocyte nucleus or 1 oocyte cytoplasm was subject to denaturing polyacrylamide gel electrophoresis.
MS2 tethering experiments
For tethering experiments, HeLa cells in 60-mm dishes were transfected at 40% confluency and harvested 36 h post-transfection. For experiments shown in Figure 3, the following amounts of each plasmid were cotransfected: 1 μg of 5′-6bs/Renilla reporter, 1 μg of Firefly control, 3 μg of MS2 fusion construct, or 450 ng of MS2–Renilla reporter, 450 ng of Firefly control, 1.35μg of pNMS2–RNPS1, and 2.7 μg of empty vector. Although fusions other than MS2–RNPS1 gave similar results under both transfection conditions, MS2–RNPS1 gave consistent results only at lower DNA concentrations. Western blotting showed that RNPS1 is highly overexpressed when transfected at the higher amounts. For experiments shown in Figure 4, 450 ng of 5′-6bs/Renilla reporter, 450 ng of Firefly control, 200 ng of β-globin-6bs reporter, 1.35μg of pNMS2–RNPS1, and 2.5μg of empty vector were cotransfected. All other MS2-fusions were transfected at 3 μg plus 1 μg of empty vector. Cells were harvested by scraping in passive lysis buffer (Dual luciferase assay system; Promega). Part of the lysate was used to measure Renilla and Firefly luciferase activities using the dual luciferase assay system (Promega), and RNA was extracted from the remainder. Nuclear and cytoplasmic RNA were fractionated as described previously (Nott et al. 2003). Renilla luciferase activity and Renilla mRNA levels were normalized to Firefly luciferase activity and Firefly mRNA levels, respectively. β-Globin-6bs mRNA levels were also normalized to Firefly mRNA levels to control for transfection efficiencies.
Acknowledgments
We thank Michael Rosbash and Ken Dower for discussion and encouragement; Miles Wilkinson, Charles Query, and members of the Moore lab for critical comments on the manuscript; Bryan Cullen for sharing results prior to publication; and Jens-Lykke Andersen for providing us with constructs for MS2 fusions. M.J.M. is an HHMI associate investigator, and this work was supported, in part, by NIH grant GM53007.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1163204.
References
- Aravind L. and Koonin, E.V. 2000. Eukaryote-specific domains in translation initiation factors: Implications for translation regulation and evolution of the translation system. Genome Res. 10: 1172-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badolato J., Gardiner, E., Morrison, N., and Eisman, J. 1995. Identification and characterization of a novel human RNA-binding protein. Gene 166: 323-327. [DOI] [PubMed] [Google Scholar]
- Bonneau A.M., Darveau, A., and Sonenberg, N. 1985. Effect of viral infection on host protein synthesis and mRNA association with the cytoplasmic cytoskeletal structure. J. Cell Biol. 100: 1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P. and Wolffe, A.P. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77: 931-941. [DOI] [PubMed] [Google Scholar]
- Braddock M., Muckenthaler, M., White, M.R., Thorburn, A.M., Sommerville, J., Kingsman, A.J., and Kingsman, S.M. 1994. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 22: 5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.W., Haghighat, A., Yu, A.T., and Sonenberg, N. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392: 520-523. [DOI] [PubMed] [Google Scholar]
- Cui Y., Gonzalez, C.I., Kinzy, T.G., Dinman, J.D., and Peltz, S.W. 1999. Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA 5: 794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria, M.J., Paushkin, S.V., Han, X., Weng, Y., Perlick, H.A., Dietz, H.C., Ter-Avanesyan, M.D., and Peltz, S.W. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Dev. 12: 1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J. and Dreyfuss, G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12: 1060-1067. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195-205. [DOI] [PubMed] [Google Scholar]
- Fribourg S., Gatfield, D., Izaurralde, E., and Conti, E. 2003. A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat. Struct. Biol. 10: 433-439. [DOI] [PubMed] [Google Scholar]
- Gatfield D. and Izaurralde, E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159: 579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D., Le Hir, H., Schmitt, C., Braun, I.C., Kocher, T., Wilm, M., and Izaurralde, E. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11: 1716-1721. [DOI] [PubMed] [Google Scholar]
- Gebauer F. and Richter, J.D. 1996. Mouse cytoplasmic polyadenylylation element binding protein: An evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc. Natl. Acad. Sci. 93: 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., and Kulozik, A.E. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11: 939-949. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913-963. [DOI] [PubMed] [Google Scholar]
- Hachet O. and Ephrussi, A. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666-1674. [DOI] [PubMed] [Google Scholar]
- He F. and Jacobson, A. 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 21: 1515-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert T.P. and Hecht, N.B. 1999. The mouse Y-box protein, MSY2, is associated with a kinase on non-polysomal mouse testicular mRNAs. Nucleic Acids Res. 27: 1747-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker, R. 1999. mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5: 711-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland R., Hesketh, J.E., and Pryme, I.F. 1996. The compartmentalization of protein synthesis: importance of cytoskeleton and role in mRNA targeting. Int. J. Biochem. Cell Biol. 28: 1089-1105. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens, W.C., Izaurralde, E., and Mattaj, I.W. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124: 627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G. and Sarnow, P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673-682. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Diem, M.D., Kim, V.N., Yong, J., and Dreyfuss, G. 2001. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 20: 6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293: 1832-1836. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000a. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 19: 6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Moore, M.J., and Maquat, L.E. 2000b. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon–exon junctions. Genes & Dev. 14: 1098-1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Nott, A., and Moore, M.J. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28: 215-220. [DOI] [PubMed] [Google Scholar]
- Lejeune F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyer P., Trembley, J.H., Lahti, J.M., and Kidd, V.J. 1998. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J. Cell Sci. 111 (Pt 11): 1495-1506. [DOI] [PubMed] [Google Scholar]
- Lu S. and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.J. and Reed, R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. 96: 14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M., and Reed, R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644-647. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu, M.D., and Steitz, J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121-1131. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836-1839. [DOI] [PubMed] [Google Scholar]
- Maderazo A.B., He, F., Mangus, D.A., and Jacobson, A. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20: 4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499-506. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173-176. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Wassarman, K.M., and Wolffe, A.P. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17: 2107-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Badolato, J., Kobayashi, R., Zhang, M.Q., Gardiner, E.M., and Krainer, A.R. 1999. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 18: 4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Lambermon, M., and Blencowe, B.J. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22: 148-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Longman, D., Johnstone, I.L., Caceres, J.F., and Blencowe, B.J. 2003. An evolutionarily conserved role for SRm160 in 3′-end formation that functions independently of exon junction complex formation. J. Biol. Chem. 278: 44153-44160. [DOI] [PubMed] [Google Scholar]
- McKendrick L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m7 guanosine cap. Mol. Cell. Biol. 21: 3632-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi, S.M., Lake, R.G., Noensie, E.N., and Dietz, H.C. 2000. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20: 8944-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R. and Richter, J.D. 2001. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell. Biol. 2: 521-529. [DOI] [PubMed] [Google Scholar]
- Minich W.B. and Ovchinnikov, L.P. 1992. Role of cytoplasmic mRNP proteins in translation. Biochimie 74: 477-483. [DOI] [PubMed] [Google Scholar]
- Mohr S.E., Dillon, S.T., and Boswell, R.E. 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes & Dev. 15: 2886-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B.S., Stapulionis, R., and Deutscher, M.P. 1994. Supramolecular organization of the mammalian translation system. Proc. Natl. Acad. Sci. 91: 964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G. and Reinberg, D. 2002. A unified theory of gene expression. Cell 108: 439-451. [DOI] [PubMed] [Google Scholar]
- Pisarev A.V., Skabkin, M.A., Thomas, A.A., Merrick, W.C., Ovchinnikov, L.P., and Shatsky, I.N. 2002. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40 S ribosomal subunits to the initiation codon of β-globin mRNA. J. Biol. Chem. 277: 15445-15451. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. 2000. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25: 423-426. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Rafiq M., Suen, C.K., Choudhury, N., Joannou, C.L., White, K.N., and Evans, R.W. 1997. Expression of recombinant human ceruloplasmin—An absolute requirement for splicing signals in the expression cassette. FEBS Lett. 407: 132-136. [DOI] [PubMed] [Google Scholar]
- Reichert V.L., Le Hir, H., Jurica, M.S., and Moore, M.J. 2002. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev 16: 2778-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H., Brown, C.Y., and Morris, D.R. 1997. Analysis of ribosome loading onto mRNA species: Implications for translation control. In mRNA formation and function (ed. J.D. Richter), pp. 305-321. Academic Press, San Diego.
- Ryu W.S. and Mertz, J.E. 1989. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 63: 4386-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell T., Kulozik, A.E., and Hentze, M.W. 2002. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. 3: REVIEWS1006. [DOI] [PMC free article] [PubMed]
- Schwer B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8: 113-116. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. 1996. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In Translational control (eds. J.W.B. Hershey et al.), pp. 245-269. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Spirin A.S. 1996. Masked and translatable messenger ribonucleoproteins in higher eukaryotes. In Translational control (eds. J.W.B. Hershey et al.), pp. 319-334. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Stapulionis R., Kolli, S., and Deutscher, M.P. 1997. Efficient mammalian protein synthesis requires an intact F-actin system. J. Biol. Chem. 272: 24980-24986. [DOI] [PubMed] [Google Scholar]
- Trembley J.H., Hu, D., Hsu, L.C., Yeung, C.Y., Slaughter, C., Lahti, J.M., and Kidd, V.J. 2002. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem. 277: 2589-2596. [DOI] [PubMed] [Google Scholar]
- Wang W., Czaplinski, K., Rao, Y., and Peltz, S.W. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20: 880-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch E.M. and Jacobson, A. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18: 6134-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Lu, S., and Cullen, B.R. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. 100: 11327-11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.F. and Shyu, A.B. 2002. RNA surveillance by nuclear scanning? Nat. Cell Biol. 4: E144-E147. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann, R. 2001. Molecular biology. RNP remodeling with DExH/D boxes. Science 291: 1916-1917. [DOI] [PubMed] [Google Scholar]
- Wilusz C.J., Wang, W., and Peltz, S.W. 2001. Curbing the nonsense: The activation and regulation of mRNA surveillance. Genes & Dev. 15: 2781-2785. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401-405. [DOI] [PubMed] [Google Scholar]
- Zong Q., Schummer, M., Hood, L., and Morris, D.R. 1999. Messenger RNA translation state: The second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. 96: 10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]