ABSTRACT
The human fungal pathogen Candida albicans causes lethal systemic infections because of its ability to grow and disseminate in a host. The C. albicans plasma membrane is essential for virulence by acting as a protective barrier and through its key roles in interfacing with the environment, secretion of virulence factors, morphogenesis, and cell wall synthesis. Difficulties in studying hydrophobic membranes have limited the understanding of how plasma membrane organization contributes to its function and to the actions of antifungal drugs. Therefore, the role of the recently discovered plasma membrane subdomains termed the membrane compartment containing Can1 (MCC) was analyzed by assessing the virulence of a sur7Δ mutant. Sur7 is an integral membrane protein component of the MCC that is needed for proper localization of actin, morphogenesis, cell wall synthesis, and responding to cell wall stress. MCC domains are stable 300-nm-sized punctate patches that associate with a complex of cytoplasmic proteins known as an eisosome. Analysis of virulence-related properties of a sur7Δ mutant revealed defects in intraphagosomal growth in macrophages that correlate with increased sensitivity to oxidation and copper. The sur7Δ mutant was also strongly defective in pathogenesis in a mouse model of systemic candidiasis. The mutant cells showed a decreased ability to initiate an infection and greatly diminished invasive growth into kidney tissues. These studies on Sur7 demonstrate that the plasma membrane MCC domains are critical for virulence and represent an important new target for the development of novel therapeutic strategies.
IMPORTANCE
Candida albicans, the most common human fungal pathogen, causes lethal systemic infections by growing and disseminating in a host. The plasma membrane plays key roles in enabling C. albicans to grow in vivo, and it is also the target of the most commonly used antifungal drugs. However, plasma membrane organization is poorly understood because of the experimental difficulties in studying hydrophobic components. Interestingly, recent studies have identified a novel type of plasma membrane subdomain in fungi known as the membrane compartment containing Can1 (MCC). Cells lacking the MCC-localized protein Sur7 display broad defects in cellular organization and response to stress in vitro. Consistent with this, C. albicans cells lacking the SUR7 gene were more susceptible to attack by macrophages than cells with the gene and showed greatly reduced virulence in a mouse model of systemic infection. Thus, Sur7 and other MCC components represent novel targets for antifungal therapy.
Introduction
The human fungal pathogen Candida albicans commonly exists as a harmless commensal organism on the skin and gastrointestinal tract of humans. Medical interventions or immunosuppression permits C. albicans to enter the bloodstream and invade tissues, which can lead to organ failure and death (1, 2). Changes in medical care and the lack of more efficient antifungal drugs are leading to increased candidiasis (3). Thus, a better understanding of the mechanisms that permit survival in the host is needed to develop novel therapeutic approaches. In particular, knowledge of the plasma membrane is limited, but it plays a multifaceted role in C. albicans pathogenesis by mediating environmental sensing, nutrient uptake, virulence factor secretion, cellular morphogenesis, and cell wall biogenesis (4, 5). The significance of studies on the plasma membrane is underscored by the fact that the most effective antifungal drugs currently used affect this essential barrier or its resident proteins (6).
Plasma membrane organization is poorly understood because of the difficulties in studying hydrophobic membrane components. For example, the structure and function of lipid raft domains in the plasma membrane remain controversial (7, 8). However, recent studies indicate that the fungal plasma membrane consists of at least three distinct protein-organized subdomains. One type of domain consists of a series of 300-nm-sized patches that were named the membrane compartment containing Can1 (MCC) because it contains the Can1 arginine permease (9–11). The MCC patches are immobile and are thus distinct from the mobile cortical actin patches detected at sites of endocytosis (10, 12). The MCC patches are also distinct in that they are associated with membrane invaginations that appear as 50-nm-deep furrows (8). Another domain, termed the membrane compartment occupied by Pma1 (MCP), contains proteins that readily diffuse, such as the plasma membrane ATPase Pma1, and are present throughout the plasma membrane but are excluded from the MCC (11). A third domain consists of punctate patches containing the TORC2 complex, which regulates cell polarity and ceramide synthesis (13).
Analysis of the MCC in Saccharomyces cerevisiae has identified other integral membrane protein constituents, including several nutrient symporters and two different families of proteins that are predicted to contain four membrane-spanning domains (14, 15). One family is represented by Nce102, which in S. cerevisiae is implicated in sphingolipid signaling and regulation of MCC formation (14). The other family of tetraspanners is represented by Sur7. Mutation of SUR7 in S. cerevisiae alters sphingolipid composition and causes defects in sporulation and osmotic stress (12, 16). Other Sur7 family members (Fmp45, Ynl194c, and Pun1/Ylr414c) are implicated in nitrogen stress, cell wall integrity, and survival in stationary phase (17–19). The MCC proteins also colocalize with a complex of cytoplasmic proteins that reside on the inner surface of the plasma membrane; this complex is known as an eisosome (20). Eisosome proteins include Pil1 and Lsp1, paralogs that contain BAR domains and are thought to promote membrane curvature at sites of the MCC and eisosomes (21, 22). Pil1 and Lsp1 are also needed for efficient endocytosis, cell wall structure, and MCC/eisosome formation (12, 20). Other important proteins present in eisosomes include the Pkh1/2 protein kinases, which regulate endocytosis, cell wall integrity, actin localization, and response to heat stress (23–25). Pkh1/2 also regulate the formation of eisosomes by phosphorylating Pil1 and Lsp1 (24, 25).
The MCC/eisosome proteins are widely conserved in fungi, but their functional roles have diverged, possibly because of differences in genetic redundancy (26–30). C. albicans encodes only two obvious members of the Sur7 family: Sur7 and Fmp45. Deletion of C. albicans SUR7 caused broad defects in cellular organization, including severe mislocalization of actin and septins (26, 27, 31). A striking phenotype of the sur7Δ mutant is that it forms abnormal extensions of cell wall growth into the cytoplasm. Cell wall function is also abnormal, as indicated by increased sensitivity to cell wall stress (32). Consistent with this, sur7Δ cell walls contain less of the β-1,3-glucan that is needed to confer cell wall strength and rigidity. The in vitro studies also revealed that sur7Δ mutants have defects in virulence-related functions, including hyphal morphogenesis, invasive growth into agar, and biofilm formation (27, 31, 32). In addition, sur7Δ cells are also more sensitive to the antifungal drug fluconazole, which perturbs ergosterol synthesis, and to agents that cause cell wall stress (e.g., the Pkc1 inhibitor cercosporamide) (27, 32). Therefore, the virulence properties of the C. albicans sur7Δ mutant were analyzed in this study. The sur7Δ mutant was found to be more sensitive to attack by macrophages than the wild type, which correlates with increased sensitivity to oxidative stress and copper. The sur7Δ mutant cells were also strongly defective in invasive growth and virulence in a mouse model of systemic candidiasis. These results indicate that Sur7 and the MCC/eisosome domains represent novel targets for therapeutic intervention of plasma membrane function.
RESULTS
sur7Δ C. albicans cells are phagocytosed less efficiently by macrophages.
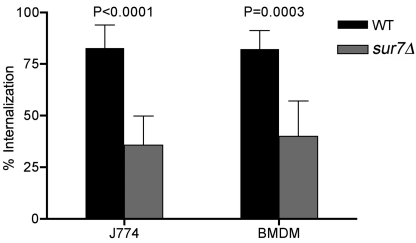
The altered cell wall phenotypes of the C. albicans sur7Δ mutant, including decreased β-glucan and increased mannose, may affect recognition by cells of the innate immune system (32). To examine this, sur7Δ cells were assayed for their ability to be phagocytosed by the mouse macrophage-like cell line J774, using a competition assay that compares the uptake of one cell type expressing green fluorescent protein (GFP) with another cell type expressing red fluorescent protein (RFP) (33). This assay can sensitively determine whether a given cell type is phagocytosed faster (e.g., S. cerevisiae) or slower (e.g., mannan mutants) than wild-type C. albicans (33). Interestingly, the competition assays showed that sur7Δ cells were phagocytosed at a lower efficiency than the wild-type control strain (Fig. 1). Similar results were obtained with mouse bone marrow-derived macrophages.
FIG 1 .
Competition assay for phagocytosis of sur7Δ cells by macrophages. The wild-type (WT) control strain expressing GFP (SKY43) and a sur7Δ strain expressing RFP (SKY66) were incubated with the J774 macrophage cell line or mouse bone marrow-derived macrophages (BMDM). The percentage of each cell type that had been phagocytosed was then determined by microscopic examination. The results are the averages of results from at least two independent assays.
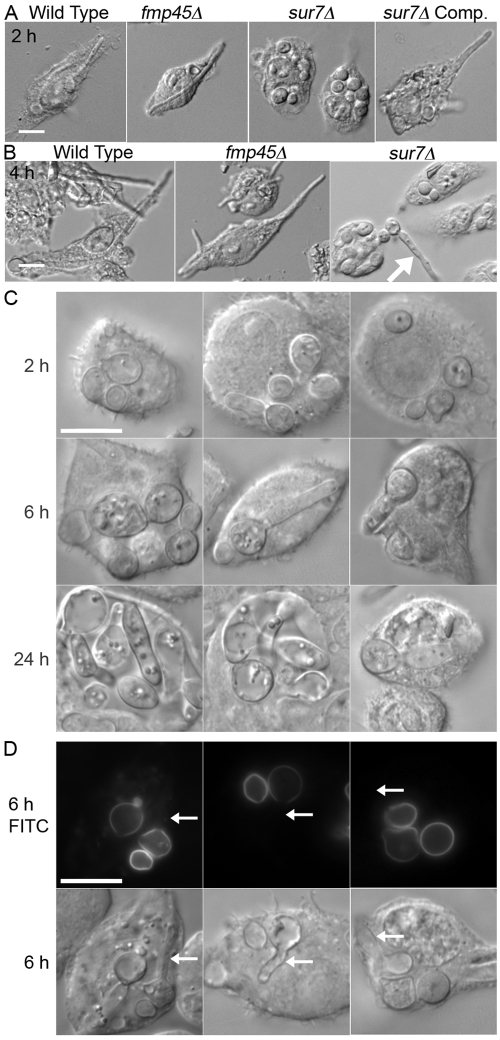
The ability of sur7Δ cells that have been phagocytosed to form hyphae and break free of a macrophage was analyzed in time course studies. As expected (34), the wild-type control strain formed elongated hyphae by 2 h postinfection (Fig. 2A) and by 4 h had typically lysed the J774 cells (Fig. 2B). Similar results were observed for an fmp45Δ mutant lacking the SUR7 paralog FMP45 and for a control strain in which the sur7Δ mutation was complemented by reintroduction of a wild-type copy of SUR7. In contrast, at 4 h postinfection, the majority of sur7Δ cells either formed smaller hypha-like outgrowths or failed to undergo detectable growth within the macrophage. Even after 6 h, the sur7Δ cells showed primarily reduced hyphal outgrowths that failed to lyse the macrophages (Fig. 2C). After 24 h, it was still possible to find sur7Δ mutant cells that did not escape from the J774 macrophage cells (Fig. 2C), whereas there were no macrophages left adhering to the substratum in cultures infected with wild-type C. albicans. These results are consistent with a previous report that sur7Δ cells were defective in killing macrophages when assayed at 24 h after infection, although the status of the sur7Δ cells was not reported in that study (31). The variable growth of sur7Δ cells within J774 cells may relate to the fact that the population of sur7Δ cells is heterogeneous, as the mutant cells become progressively more abnormal with age (32).
FIG 2 .
Intracellular growth of C. albicans after phagocytosis. The C. albicans strains indicated by their genotypes were added to the macrophage cell line J774, and then cell morphology was determined by microscopic examination after 2 h (A) or 4 h (B). The arrow indicates the greater extent of growth of a cell that is outside and had not been phagocytosed. Comp., complemented. (C) Images of the sur7Δ mutant captured after the indicated times of incubation with J774 cells. (D) sur7Δ cells were cross-linked to the fluorescent dye fluorescein, added to J774 cells, and then examined after 6 h by fluorescence microscopy. Regions of older cell growth cross-linked to fluorescein fluoresce, whereas the regions of newer cell growth do not. Arrows point to examples of new growth. The wild-type control strain was DIC185, the sur7Δ strain was YJA11, the complemented sur7Δ strain carrying a copy of the wild-type SUR7 gene was YJA12, and the fmp45Δ strain was YHXW3 (Table 1). FITC, fluorescein isothiocyanate. Bars, 10 μm.
To confirm that the sur7Δ cells initiated new growth within macrophages, the sur7Δ cells were chemically cross-linked to the fluorescent dye fluorescein and then imaged at 6 h postinfection. This method distinguishes between the input cells, which are fluorescent, and the new cell growth, which is not fluorescent (32). The results revealed that all of the small rounded cells were fluorescent, indicating that they were the original input cells that had not grown further. In contrast, the cells that showed various degrees of polarized growth to form elongated cell structures were fluorescent only on one end, revealing the extent of new growth that occurred in the macrophage (Fig. 2D). Thus, sur7Δ cells are better than wild-type cells at avoiding phagocytosis but grow poorly following phagocytosis and are defective at killing macrophages.
Increased sensitivity of sur7Δ cells to a subset of phagolysosomal conditions.
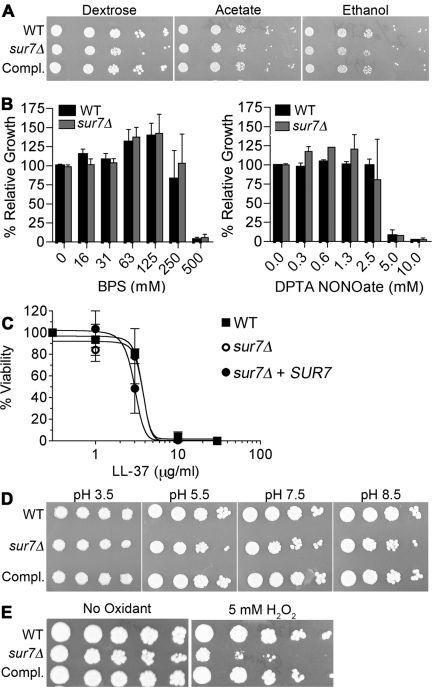
C. albicans cells that have been phagocytosed must switch to using two carbon molecules for nutrition, rather than the glucose that is present in the culture medium (35). However, the sur7Δ mutant cells grew similarly to the wild-type cells when spotted onto synthetic minimal medium containing ethanol or acetate (Fig. 3A). This indicates that the poor intraphagosomal growth is not due to a defect in growth on nonfermentable carbon sources.
FIG 3 .
Growth of sur7Δ under phagosome-like conditions. (A) Dilutions of the indicated cell types were spotted onto solid agar medium containing 2% dextrose, 2% acetate, or 2% ethanol as a carbon and energy source. (B) Effects on growth of the indicated concentration of the iron chelator BPS or the NO-generating agent DPTA NONOate. Cells were incubated in 96-well trays with different concentrations of BPS or DPTA NONOate for 2 days at 37°C, and then the extent of growth was determined using a spectrophotometer. (C) Susceptibility to killing by an antimicrobial peptide was assayed by incubating the indicated cells with the indicated concentrations of LL-37 for 90 min, after which cells were plated to quantify their viability. (D) Effects of pH on growth were determined by spotting dilutions of the indicated cells onto YPD agar plates buffered to the indicated pH. (E) Sensitivity to oxidation was determined by spotting dilutions of cells onto YPD agar plates with or without 5 mM H2O2. Plates were incubated for 2 days at 37°C and then photographed. Strains used included the wild-type control (DIC185), the sur7Δ mutant (YJA11), and the complemented sur7Δ strain carrying a copy of the wild-type SUR7 gene (YJA12).
Pathogens trigger macrophages to undergo changes in the phagosomal compartment that are designed to destroy the invading cells (34, 36). Therefore, sur7Δ cells were examined for sensitivity to conditions in vitro that mimic different aspects of the phagolysosomal environment. Growth assays showed that there were no changes in the sensitivity of sur7Δ cells to the iron chelator bathophenanthrolinedisulfonic acid (BPS), the nitric oxide generator of diethylenetriaminepentaacetic acid (DPTA) NONOate (Fig. 3B), or the antimicrobial host defense peptide LL-37 (Fig. 3C). The growth of the sur7Δ cells was also not affected by lowering the pH to 5.5, the typical pH of a phagosome, and the mutant cells even grew well on plates buffered to pH 3.5 (Fig. 3D). The oxidizing agent H2O2 caused the most apparent effect on the growth of the sur7Δ mutant. Dilutions of cells spotted onto a yeast extract-peptose-dextrose (YPD) medium plate containing 5 mM H2O2 showed an obvious defect in the growth of sur7Δ cells compared to the wild-type control strain or the sur7Δ strain complemented with SUR7 (Fig. 3E). Thus, sur7Δ cells are more sensitive to oxidation but not to all phagolysosomal conditions.
sur7Δ cells are more sensitive than the wild type to a variety of different oxidizing conditions.
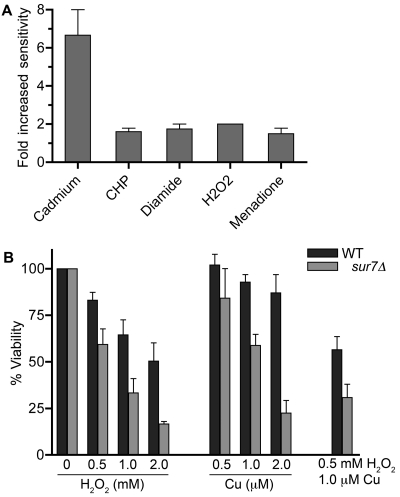
sur7Δ cells were next examined for sensitivity to chemicals that preferentially cause distinct types of oxidative damage. Cells were examined for sensitivity to, in addition to H2O2, menadione (superoxide-generating agent), cumene hydroperoxide (aromatic hydroperoxide), diamide (thiol oxidant), and cadmium chloride (increases reactive oxygen species) (37). These compounds were tested in broth dilution assays, and the effects on growth were measured after 48 h. The sur7Δ mutant was about 2-fold more sensitive than the wild type to H2O2, menadione, cumene hydroperoxide, and diamide (Fig. 4A). The sur7Δ cells showed a higher (6.6-fold-increased) sensitivity to cadmium, perhaps because cadmium also has additional effects on cells (38). For example, the Pkc1 cell wall integrity pathway is important for resistance to cadmium (39), and sur7Δ cells are also more sensitive to the Pkc1 inhibitor cercosporamide (32). Thus, sur7Δ cells are more sensitive to a broad range of chemicals that increase oxidative damage in cells.
FIG 4 .
Sensitivity to different oxidizing conditions. (A) Relative sensitivities of sur7Δ cells (YJA11) and the wild-type control (DIC185) when grown in the presence of the oxidants CdCl2, cumene hydroperoxide (CHP), diamide, H2O2, and menadione. Cells were grown in the presence of different dilutions of each oxidant, and then the extent of growth was determined after 2 days using a spectrophotometer. (B) Cells were incubated for 25 min in the presence of the indicated concentrations of H2O2, CuSO4, or both, and then the cells were washed and plated on YPD agar plates to determine the percentages of surviving cells.
The assays described above monitored the cumulative effects after 2 days of growth and could therefore be influenced by the ability of cells to induce genes that counteract oxidative stress and permit adaptation to higher levels of oxidant. To more closely mimic the oxidative burst that occurs in a phagosome, C. albicans cells were incubated for 25 min in various concentrations of H2O2 and then plated to determine their viability. The sur7Δ mutant was more sensitive than the wild type to H2O2, as the concentration required to cause 50% cell death was about 3-fold lower (Fig. 4B). Sensitivity to copper was also tested, since it has been reported that copper is imported into the phagosome, where it acts synergistically to enhance the effects of the oxidative burst (40). Copper was prepared for these assays by first reducing it with ascorbic acid, which potentiates its ability react with H2O2 to form hydroxyl radicals that are more toxic (40, 41). The results showed a significant increase in the sensitivity of sur7Δ cells to a 25-min exposure to copper. The combination of H2O2 and copper caused a synergistic effect on killing (Fig. 4B), demonstrating that copper can potentiate the effects of oxidation on the killing of C. albicans.
Increased copper sensitivity for sur7Δ mutant correlates with poor growth in macrophages.
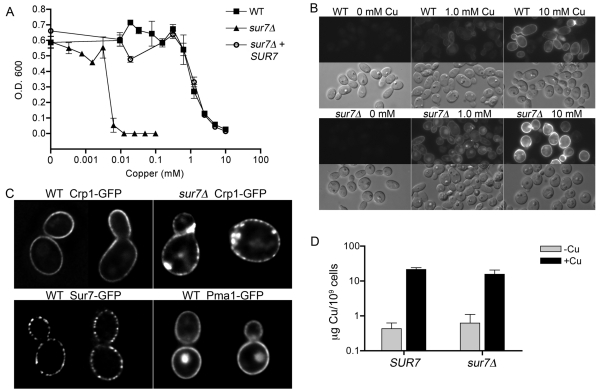
The effects of copper on sur7Δ cells were examined further by assaying the ability of cells to grow in the presence of different dilutions of copper. Interestingly, the sur7Δ cells were ~2,000-fold more sensitive to copper than the wild type (Fig. 5A), indicating that elevated copper levels may also contribute to the slow growth of sur7Δ cells in the phagosome. Increased sensitivity to copper was previously reported for mutation of C. albicans CRP1, which encodes a plasma membrane protein that exports copper out of the cell (42, 43). Analysis of strains carrying a CRP1-GFP fusion gene showed that Crp1-GFP was localized primarily to the plasma membrane in both sur7Δ cells and the wild type, indicating that the copper sensitivity of sur7Δ cells was not due to a defect in the membrane trafficking of Crp1-GFP (Fig. 5B). In some images, Crp1-GFP appeared to have a slightly punctate distribution in the plasma membrane that was usually more obvious in sur7Δ cells. This slightly punctate pattern was also seen for the MCP protein Pma1-GFP (11) and was not as distinct as the pattern seen for Sur7, suggesting that Crp1-GFP is present in the MCP domain (Fig. 5C). sur7Δ cells also displayed unusual patches of apparently intracellular Crp1-GFP that correlate with the expected formation of the ectopic intracellular growth of the plasma membrane and cell wall in this mutant.
FIG 5 .
Effects of copper on the growth of sur7Δ cells. (A) Cells were grown in media containing the indicated concentrations of CuSO4 for 2 days at 37°C, and then the relative levels of growth were determined using a spectrophotometer. The wild-type control strain was DIC185, the sur7Δ strain was YJA11, and the complemented sur7Δ strain carrying a copy of the wild-type SUR7 gene was YJA12. (B) Wild-type (YLD111-9) and sur7Δ (YLD105-8) cells expressing CRP1-GFP were grown in medium containing the indicated concentrations of CuSO4 for 2 h and then examined by fluorescence microscopy. (C) Cells expressing Sur7-GFP (YHXW4), Pma1-GFP (YHXW11), and Crp1-GFP (YLD111-9 and YLD105-8) were analyzed by deconvolution microscopy. A midsection of each cell type is shown, demonstrating that Crp1-GFP is not restricted to MCC domains, unlike Sur7-GFP. Crp1-GFP was induced by growing cells for 2 h in 10 mM CuSO4. Two representative cells from independent image deconvolution steps are shown for each cell type. (D) Copper contents in wild-type (DIC185) and sur7Δ mutant (YJA11) cells were quantified by inductively coupled plasma mass spectrometry (ICP-MS) for cells grown in the absence of added copper or in the presence of 100 µM CuSO4 for 4 h. The results represent the averages of results from two independent experiments in which the analysis was carried out in triplicate. Error bars, standard deviations (SD).
Dose-response assays showed that Crp1-GFP began to be detectably induced at 0.1 mM copper in both the wild type and the sur7Δ mutant and was induced to higher levels with increasing copper in the medium (Fig. 5B). Comparison of the signal intensity in digital images indicated that the dose-response curve for induction of Crp1-GFP was shifted for sur7Δ cells so that they were about 3-fold more sensitive than the wild-type control. The observation that the sur7Δ mutant is 2,000-fold more sensitive than the wild type to copper but is only 3-fold more sensitive for the induction of Crp1-GFP indicates that sur7Δ cells do not behave as though they are accumulating high levels of copper. Consistent with this, copper levels were similar in wild-type and sur7Δ cells grown in the absence of added copper, and both cell types showed similar increases in copper content when grown in the presence of 100 µM CuSO4 (Fig. 5D). These results suggest that another type of copper detoxification mechanism is defective in sur7Δ cells. An interesting possibility is that copper sensitivity may relate to the endosomal trafficking defect of sur7Δ cells, since endosomes have been implicated in copper sequestration (see Discussion).
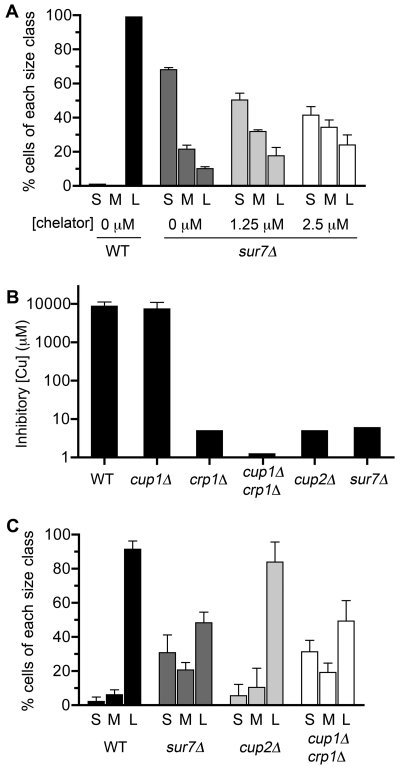
The significance of the copper-sensitive phenotype of sur7Δ cells with regard to their poor growth in macrophages was examined by infecting J774 macrophages with C. albicans cells in the presence of the cell-permeable copper chelator ammonium tetrathiomolybdate. To compare the extents of growth in macrophages, C. albicans cells were scored as small if they appeared as budding cells or had a germ tube smaller than the size of the mother cell, medium if they had a germ tube with a diameter less than that of the macrophage, and large if they had distorted the macrophage or lysed it. The sur7Δ cells incubated with 1.25 µM and 2.5 µM chelator showed increasing fractions of medium and large cells (Fig. 6A), consistent with copper contributing to their poor growth. Wild-type C. albicans was not affected by the chelator, as essentially all of the cells still formed large germ tubes. Higher concentrations of chelator (not shown) started to cause an inhibitory effect on C. albicans growth.
FIG 6 .
Effects of copper chelator and mutations that increase sensitivity to copper on the growth of C. albicans in J774 macrophage cells. (A) J774 macrophages were incubated with the indicated concentrations of the cell-permeable copper chelator ammonium tetrathiomolybdate, and then the extents of growth of the wild type and sur7Δ cells were assessed after 4 h of infection. C. albicans cell size was scored as follows: S, small budding cells and cells with a germ tube smaller than the size of the mother cell; M, medium cells with a germ tube diameter less than that of the macrophage; and L, large cells that distorted the macrophage or lysed it. Results are the averages of results from three independent experiments. The decrease in the number of small cells and the increase in the numbers of medium and large cells caused by addition of the chelator were significant (P < 0.002). (B) Sensitivity of mutant strains to growth in the presence of CuSO4. The data represent the results of two independent experiments, each done in triplicate. (C) J774 cells were infected with the indicated C. albicans strains for 4 h, and then the extent of cell growth was recorded. Results represent the averages of results from three independent experiments. Strains used included the wild-type (DIC185), sur7Δ (YJA11), cup1Δ (YLD116-7), crp1Δ (YLD115-1), crp1Δ cup1Δ (KC25), and cup2Δ (YLD117-1) strains. Error bars indicate SD.
To determine if other copper-sensitive mutants showed a similar phenotype, we analyzed a previously reported crp1Δ cup1Δ double mutant that lacks both the Crp1 copper exporter and the Cup1 copper binding protein (42, 43). A cup2Δ mutant that lacks the Cup2 transcription factor, which induces the copper-responsive CRP1 and CUP1 genes, was also analyzed (44). All of the mutant cells that were not phagocytosed grew well outside the macrophages and formed large germ tubes in the culture medium (not shown). Interestingly, the crp1Δ cup1Δ mutant that is nearly 10,000-fold more sensitive to copper (Fig. 6B) grew poorly in macrophages, similar to what occurred with sur7Δ cells (Fig. 6C). This further indicates that copper resistance is important for efficient growth in macrophages. Although the cup2Δ mutant was ~2,000-fold more sensitive to copper than the wild type, like sur7Δ cells, it showed only a slight change in growth in macrophages. These results indicate that the poor growth of sur7Δ cells is likely due to a combination of factors that includes increased sensitivity to copper, oxidation, and cell wall stress.
Decreased virulence of the sur7Δ mutant in a mouse model systemic infection.
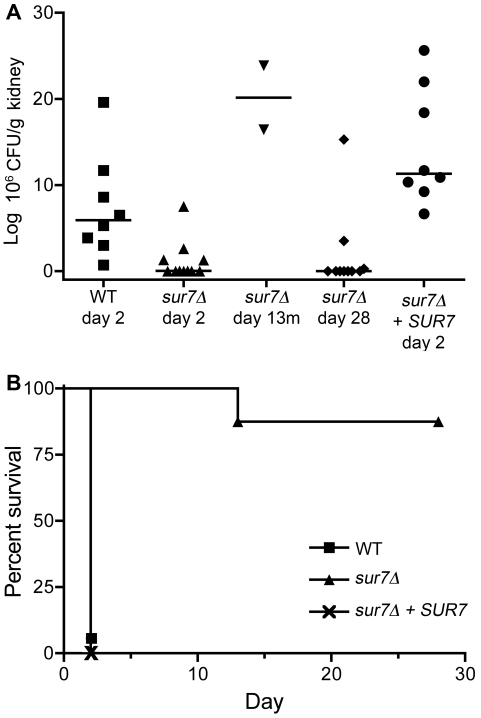
The virulence of the sur7Δ mutant was examined in a mouse model of hematogenously disseminated candidiasis in which BALB/c mice were infected via the lateral tail vein. To determine whether the sur7Δ mutant was defective in initiating an infection, the number of CFU/g of C. albicans in the kidneys was analyzed 2 days postinoculation. Mice infected with the wild-type strain contained high levels of C. albicans at day 2, with a median of 5.9 × 106 CFU/g kidney (Fig. 7A). In contrast, the median number of CFU/g kidney at day 2 after infection with the sur7Δ mutant was nearly 1,000-fold lower (7.6 × 103 CFU/g kidney). Surprisingly, the sur7Δ mutant showed a broad range of numbers of CFU/g, from 7.5 × 106 to 2.6 × 103. It is not clear why there was such a wide variation, but it is interesting to note that the highest CFU/g levels all resulted from four mice that were injected with the same batch of sur7Δ mutant cells. The other seven mice, which were infected with two other independent batches of sur7Δ cells, showed much lower levels of CFU/g kidney tissue at day 2. Thus, there may have been something distinct about the mice or the sur7Δ cells that were used in one of the three independent assays. Nonetheless, the sur7Δ mutation mutant showed significantly lower numbers of CFU/g at day 2 (P = 0.0053), indicating a defect in initiating an infection.
FIG 7 .
Virulence defect of the sur7Δ mutant in a mouse model of hematogenously disseminated candidiasis. BALB/c mice were injected via the tail vein with 106 cells of the indicated strain. (A) The number of CFU/gram of kidney tissue was determined at day 2 postinfection. Numbers of CFU/gram of kidney were also determined for two sur7Δ strain-infected mice that became moribund on day 13 (day 13m) and for the sur7Δ strain-infected mice that survived to the end of the experiment (day 28). (B) Survival curves for mice infected with the designated strains demonstrating the greatly reduced virulence of the sur7Δ mutant. Strains included the wild-type control (DIC185), the sur7Δ mutant (YJA11), and the complemented sur7Δ strain carrying a wild-type copy of SUR7 (YJA12).
The eight mice injected with the wild-type control strain, DIC185, became moribund after 2 days, as expected (Fig. 7B). Similar results were observed for mice infected with the complemented sur7Δ strain carrying a wild-type copy of SUR7. In contrast, only 2 out of 12 mice infected with the sur7Δ mutant succumbed to infection, and both of these took 11 days longer than wild-type-infected mice to succumb. These mice showed numbers of CFU/g kidney tissue that were greater than or equal to those of mice infected with wild-type C. albicans. The majority of the 10 mice infected with the sur7Δ mutant that remained viable up through the completion of the assay at day 28 contained very low numbers of or no detectable CFU/g kidney tissue (Fig. 7A). Seven of these surviving mice infected with the sur7Δ mutant lacked detectable CFU in their kidney tissue, indicating that they had cleared the infection (Fig. 7A). Thus, the sur7Δ mutant is very strongly defective in virulence. Interestingly, two mice infected with the sur7Δ mutant that did not display obvious symptoms of infection at day 28 had CFU/g levels in the kidney that were in the range of the values seen for moribund mice infected with the wild-type control strain. The diminished symptoms of these mice may be related to distinct patterns of growth of sur7Δ cells in vivo as described below.
Defects of sur7Δ cells in invasive growth and morphogenesis in vivo.
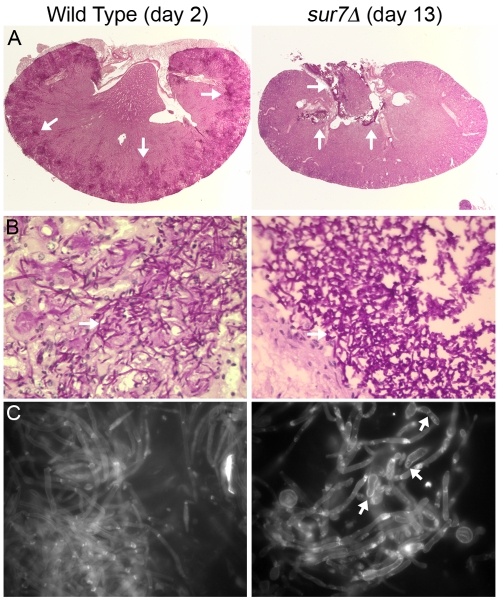
Kidneys from infected mice were fixed with formaldehyde, and then histopathological analysis was performed using periodic acid-Schiff (PAS) staining. As expected, the wild-type control strain caused widely disseminated infections in the kidneys, with large numbers of hyphal filaments emanating from multiple foci, which were found primarily in the outer cortex region of the organ (Fig. 8A). In contrast, kidneys from mice that succumbed to infection with the sur7Δ mutant at day 13 showed fungal staining primarily in the central renal space near the region that exits to the bladder. The sur7Δ cells were primarily in a large clump that did not invade the neighboring tissue (Fig. 8A and B). An interesting possibility is that the sur7Δ cells are better able to grow in the more open central renal space rather than in the solid outer cortex, since previous in vitro studies demonstrated that sur7Δ cells are defective in growing invasively into agar (27). Other mutants that display defects in invasive growth into agar have also been reported to grow predominantly in a cluster in the central renal space and cause less severe symptoms in mice (4, 45).
FIG 8 .
C. albicans growth in kidneys. (A) Kidneys were excised from a mouse infected with the wild-type strain (DIC185) for 2 days or from a mouse infected with the sur7Δ mutant strain (YJA11) that became moribund after 13 days of infection. Kidney sections were stained by the periodic acid-Schiff (PAS) staining method, which stains the C. albicans cells a dark magenta color (see the arrows for examples). Kidney tissue infected with the DIC185 wild-type strain demonstrated a high level of penetration by hyphal filaments emanating from multiple sites in the outer cortex. In contrast, sur7Δ mutant cells were mostly restricted to the central renal space of the kidney. (B) Higher magnification images of a region of the PAS-stained kidney sections taken with a 60× objective. (C) Kidney homogenates were stained with calcofluor white and then examined by fluorescence microscopy using a 100× objective. Arrows point to examples of sur7Δ cells with pseudohyphal morphology.
The morphogenesis of the sur7Δ cells grown in vivo was examined in more detail by staining kidney homogenates with calcofluor white, which preferentially stains fungal cell wall chitin but not mammalian tissues. As expected, the wild-type control cells were observed mainly as elongated filamentous hyphae, although some pseudohyphal cells could be observed (Fig. 8C). In contrast, the sur7Δ cells formed a broader range of cell morphologies that included wider hyphae and a variety of aberrant-looking pseudohypha-type cells.
DISCUSSION
Little is known about the organization of the plasma membrane or how it is affected by antifungal therapy. Thus, the recent identification of MCC enzymes/eisosomes as a novel type of membrane domain has provided important new insight into the lateral organization of the plasma membrane (11). A key role for these domains in C. albicans was suggested by previous studies showing that sur7Δ mutants mislocalized actin, septins, and cell wall growth (27). The abnormal cellular organization of sur7Δ mutants is linked to defects in morphogenesis, cell wall strength, and invasive growth into agar (27, 31, 32). Therefore, in this study, the virulence properties of a sur7Δ mutant were analyzed. The results revealed defects in intraphagosomal growth and virulence in a mouse model of systemic candidiasis that correlate with increased sensitivity to oxidation, copper, and cell wall stress.
Sur7 promotes resistance to a subset of stress conditions encountered in the macrophage phagosome.
Sur7 had opposing effects on the infection of macrophages in that sur7Δ cells were phagocytosed less efficiently than wild-type cells (Fig. 1) but grew poorly once they were internalized (Fig. 2). sur7Δ cells were not significantly altered in their responses to most conditions encountered in the phagolysosomal environment, such as low iron, nitric oxide, the antimicrobial peptide LL-37, or growth on poor carbon sources (Fig. 3). However, sur7Δ cells were more sensitive to a wide range of oxidants (Fig. 3 and 4). This correlates with a previous microarray analysis of gene expression, which suggested that sur7Δ cells are oxidatively stressed under normal growth conditions. Expression of the superoxide dismutase SOD4 was increased ~6-fold and SOD5 was increased ~12-fold in sur7Δ cells grown under standard conditions (27). Thus, sur7Δ cells are stressed under normal growth conditions and may therefore be more easily overwhelmed by exogenous sources of oxidation.
One likely contributing factor to the oxidative stress defect is that sur7Δ cells were also more sensitive to copper (Fig. 4 and 5). Copper catalyzes the conversion of hydrogen peroxide to more toxic hydroxyl radicals (36, 46). Consistent with this, sur7Δ cells were more sensitive than the wild type to a short-term 25-min exposure to a combination of copper and H2O2 that is expected to more closely mimic the conditions of a phagosomal oxidative burst.
Copper also has other antimicrobial properties, and recent studies have demonstrated that copper influx into macrophage phagosomes inhibits bacterial growth (40, 47, 48). Gamma interferon stimulates macrophages to take up copper by inducing the expression of the high-affinity copper importer CTR1. The ATP7A copper exporter is then stimulated to translocate from the Golgi apparatus to the phagosome, where it promotes an increase in copper concentration (40). Consistent with this, Escherichia coli and Salmonella enterica copA mutants that are more sensitive than wild-type strains to copper due to a defect in copper export showed decreased survival in macrophage phagosomes (40, 47). Thus, the 2,000-fold-increased copper sensitivity of sur7Δ cells indicates that increased copper in the phagosome may contribute to the growth defect of sur7Δ cells in macrophages. In further support of this, a C. albicans crp1Δ cup1Δ mutant that is ~10,000-fold more sensitive to copper also grew poorly in macrophages (Fig. 6). Since the concentration of copper in the phagosome is expected to increase over time, these results are consistent with observations that the growth initiated by many sur7Δ cells within the first few hours does not continue at later time points (Fig. 2). In contrast, wild-type C. albicans cells are highly resistant to copper and are probably not affected by copper influx into the phagosome. All together, these results identify a novel role for copper resistance in promoting the growth of C. albicans in macrophages. Interestingly, nonpathogenic S. cerevisiae lacks an ortholog of the CRP1 copper exporter (42, 43), whereas C. albicans and a diverse group of other pathogenic fungi, including Cryptococcus gattii, Aspergillus fumigatus, and Histoplasma capsulatum contain a CRP1 ortholog. This further suggests that resistance to copper is important for the survival of a broad range of fungal pathogens in the host.
Novel role for Sur7 in resistance to copper.
The CRP1 copper exporter has the most significant role in promoting resistance to copper of any known C. albicans gene (42, 43). Although Crp1 is a plasma membrane protein, its function does not seem to be altered in sur7Δ cells (Fig. 5). Crp1-GFP was induced by copper and localized to the plasma membrane as expected. Furthermore, the level of copper in the sur7Δ mutant was not significantly different than that in the wild type when cells were grown either in the presence or in the absence of added copper (Fig. 5D). Thus, sur7Δ cells do not accumulate high levels of copper but instead are more sensitive to copper. These new data indicate that another type of copper detoxification mechanism exists in C. albicans and that sur7Δ cells are deficient in this mechanism. One possibility for the increased sensitivity to copper comes from the results of high-throughput genetic studies of S. cerevisiae that suggest that excess copper is exported into endosomal vesicles and then accumulates in the vacuole (49). This mechanism could be impaired in C. albicans sur7Δ mutants, since they have a defect in the fusion of late endosomal vesicles with the vacuole (27).
Virulence defects of the sur7Δ strain.
The sur7Δ mutant was strongly defective in a mouse model of systemic candidiasis. Whereas all of the mice infected with the wild-type strain succumbed to infection by day 2, ~83% of the mice infected with the sur7Δ mutant survived to the end of the experiment (day 28) and 58% of the mice cleared the infection (Fig. 7). Part of the virulence defect was due to its reduced ability to initiate an infection; the median number of CFU/g of kidney at day 2 was almost 1,000-fold lower than for the wild-type control (Fig. 7). The decreased virulence is likely due to a combination of factors that include increased susceptibility to oxidation (Fig. 3 and 4), copper (Fig. 5 and 6), and cell wall stress (32). Furthermore, the sur7Δ cells showed reduced dissemination in vivo. sur7Δ cells were clumped in the central renal space and were not disseminated throughout the outer cortex of the kidney, as was seen for the wild type (Fig. 8). This phenotype is similar to that seen with other C. albicans mutants that are also defective in invasive growth into agar (4, 27, 45). However, other factors may also contribute to growth in the central renal space and reduced dissemination into kidney tissues. For example, it is potentially significant that the level of copper is about 30-fold lower in urine than in blood (50).
Implications for antifungal therapy.
The plasma membrane and its constituents are directly or indirectly targets of most of the commonly used antifungal drugs: fluconazole, amphotericin, and caspofungin (6). Studies on Sur7 and the MCC/eisosome domains are therefore likely to lead to a better understanding of the mechanisms by which these drugs act. For example, MCC domains preferentially stain with the ergosterol-binding drug filipin (51), suggesting that the related polyene drug amphotericin may bind and carry out its pore-forming function primarily at MCC domains. sur7Δ cells are ~2-fold more sensitive to caspofungin, which correlates with altered cell wall formation and decreased cell wall β-glucan (32). Although the primary target of caspofungin is β-glucan synthase, it is interesting that a related type of an echinocandin class drug can chemically cross-link to the MCC/eisosome proteins Pil1 and Lsp1 (52, 53). The cell wall defects of sur7Δ cells are also thought to account for the 8-fold-increased sensitivity to the drug cercosporamide, which inhibits Pkc1 from inducing cell wall repair genes (32). Interestingly, pkc1 mutants are more sensitive than the wild type to fluconazole (54), an inhibitor of ergosterol synthesis, suggesting that an impaired Pkc1 pathway in sur7Δ cells may contribute to the observed 5-fold-increased sensitivity of these cells to fluconazole (27).
Studies on Sur7 and other MCC/eisosome components also have important implications for the development of novel therapeutic approaches. An advantage of targeting MCC domains is that, rather than blocking one specific function, inhibitory drugs are expected to cause multiple defects, including increased sensitivity to cell wall stress, copper, oxidation, and the immune system, as seen in the sur7Δ mutant (Fig. 2 to 6). Disruption of Sur7 and MCC domains is also expected to cause defects in invasive growth and biofilm formation (27, 31). Sur7 is also a potential drug target in that it is not conserved in mammalian cells, aside from a short Cys-containing motif in extracellular loop 1 (27). Thus, further studies on MCC/eisosome domains will help to make current drug therapy more effective and to reveal new avenues for therapeutic intervention.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used in this study were derived from BWP17 or CAI4, and their genotypes are listed in Table 1. The construction of the homozygous sur7Δ deletion strain YJA11 (27), the complemented strain YJA12, in which a wild-type copy of SUR7 was reintroduced into the sur7Δ mutant (27), and the homozygous fmp45Δ deletion strain YHXW3 (32) was described previously. C. albicans strains expressing CRP1-GFP were created by homologous recombination of GFP sequences into the 3′ end of the CRP1 open reading frame, essentially as described previously (55). In brief, PCR primers containing ~70 bp of sequence homologous to the 3′ end of the CRP1 open reading frame was used to amplify a cassette containing a more photostable version of enhanced GFP (C. albicans GFPγ) and a URA3 selectable marker (55). This cassette was transformed into C. albicans, and the Ura+ colonies were screened to identify those that carry CRP1-GFP. Strains expressing GFP and RFP under the control of the ADH1 promoter were constructed as described previously (33). Cells were grown in rich YPD medium or synthetic medium lacking the nutrients indicated in the text (56).
TABLE 1 .
C. albicans strains used in this study
| Strain | Parent | Genotype |
|---|---|---|
| BWP17 | S. cerevisiae 5314 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| DIC185 | BWP17 | ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/ARG4 |
| YJA11 | YJA10 |
sur7Δ::ARG4/sur7Δ::HIS1 URA3/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA12 | YJA10 |
sur7Δ::ARG4/sur7Δ::HIS1 SUR7::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW3 | YHXW2 |
fmp45Δ::ARG4/fmp45Δ::HIS1 URA3/ura3D::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| SKY43 | BWP17 |
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG plus pADH1-GFP-URA3 |
| SKY66 | YJA10 |
sur7Δ::ARG4/sur7Δ::HIS1 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG plus pADH1-RFP-URA3 |
| YLD111-9 | CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 CRP1-GFPγ::URA3 |
| YHXW4 | BWP17 |
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG SUR7-GFPγ::URA3 |
| YHXW11 | CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 PMA1-GFPγ::URA3 |
| YLD116-7 | KC13 | ura3Δ::λimm434/URA3 cup1::hisG/cup1::hisG |
| YLD115-1 | KC16 | ura3Δ::λimm434/URA3 crp1::hisG/crp1::hisG |
| KC25 | CAF3 |
ura3Δ::λimm434/ura3Δ::λimm434 cup1::hisG/cup1::hisG crp1::hisG crp1::hisG-URA3-hisG |
| YLD117-1 | SN152 | cup2::HIS1/cup2::LEU2 his1/his1 leu2/leu2 ARG4/arg4 |
Competition assay for macrophage phagocytosis.
Competition assays testing the ability of C. albicans to be phagocytosed were carried out using the mouse macrophage-like cell line J774 and primary mouse bone marrow-derived macrophages, essentially as described previously (33). The bone marrow-derived macrophages were isolated out of cells harvested from the femurs of BALB/c mice as previously described (36, 41). C. albicans cells for the competition assays were grown in YPD plus 50 µg/ml uridine; the cells were harvested by centrifugation, washed once, and then resuspended in phosphate-buffered saline (PBS). The yeast cells were adjusted to a final concentration of 107 cells/ml using a hemocytometer. The C. albicans cells were added to J774 or primary macrophages at a multiplicity of infection (MOI) of 10. At 40 to 60 min after the addition of yeast, calcofluor white was added (1-µg/ml final concentration) directly to the culture medium for ~10 s and then the coverslips were immediately mounted on a glass slide for imaging. The images were captured using a Zeiss Axioskop fluorescence microscope equipped with a DAGE-MTI charge-coupled-device (CCD) camera, and then the number of cells expressing RFP or GFP was quantified.
Growth of C. albicans within macrophages.
The ability of C. albicans strains to grow within the mouse macrophage-like cell line J774 was assayed essentially as described previously (45). The J774 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum. The J774 cells were grown to near confluence and then seeded on coverslips in 24-well trays at 105 cells per well. After overnight growth, the J774 cells were washed with PBS, infected with C. albicans at an MOI of 1, and then incubated in DMEM with 10% fetal bovine serum at 37°C with 5% CO2. The C. albicans cells were prepared for the assay by harvesting a fresh overnight culture grown in YPD medium, washing it with PBS, and then resuspending the cells in serum-free DMEM. Coverslips removed at various times after infection were observed microscopically for the ability of C. albicans to form hyphae and lyse macrophages. The sur7Δ cells that grew poorly in macrophages were smaller and therefore more difficult to detect than larger hyphal cells, which could be observed in multiple focal planes. Thus, some of the variability in the quantitations of the extents of cell growth within macrophages was most likely caused by undercounting of the small cells. To examine the effects of copper chelation, the J774 cells were incubated with the concentration of ammonium tetrathiomolybdate (Sigma-Aldrich Co.) indicated in Fig. 6 for 30 min and then infected with C. albicans cells as described above. Images were captured using an Olympus BH2 microscope equipped with a Zeiss AxioCam camera.
Growth assays and sensitivity to inhibitors.
Growth under different conditions was assayed by spotting dilutions of cells onto agar plates containing the medium components or chemicals indicated in the text. The pH levels on agar plates were buffered to pH 3.5 with 100 mM citrate, to pH 5.5 with 150 mM succinate, to pH 7.5 with 150 mM HEPES, and to pH 8.5 with 150 mM bicine. Sensitivity to iron chelation, nitric oxide, and different oxidizing agents was also analyzed by broth dilution assays in 96-well plates. These assays made use of the iron chelator BPS (bathophenanthrolinedisulfonic acid; Sigma-Aldrich), the NO-generating compound DPTA NONOate (dipropylenetriamine NONOate; Cayman Chemical Co., Ann Arbor, MI), and the oxidizing agents hydrogen peroxide, menadione, diamide, cumene hydroperoxide, and cadmium chloride (Sigma-Aldrich). Wild-type and mutant cells were adjusted to 2 × 104 cells/ml, and then aliquots were placed in the wells of a 96-well plate. Serial dilutions of the chemicals were made, and then the plates were covered with an AeraSeal oxygen-permeable barrier (Research Products International Corp., Mount Prospect, IL) and incubated at 37°C for 2 days. The optical density at 600 nm (OD600) was then measured to determine the extent of cell growth. Susceptibility to the antimicrobial host defense peptide LL-37 (Sigma-Aldrich Co.) was tested with strains grown overnight in YPD at 30°C, washed two times in 1 mM potassium phosphate buffer, pH 7, and then diluted to 2.5 × 105 cells/ml. Twenty microliters (5,000 cells) was then combined with 1 mM potassium phosphate buffer, pH 7, and LL-37 (Sigma-Aldrich Co.) in low-retention tubes, to yield a 40-µl final volume. Reaction mixtures were incubated for 90 min at 37°C with shaking at 250 rpm, followed by dilution with 360 µl 1 mM potassium phosphate buffer. Thirty microliters was then spread onto a YPD plate and incubated at 30°C for 48 h. The number of colonies was assessed to determine the effects of LL-37 on viability.
Analysis of copper responses.
Strains carrying the CRP1-GFPγ fusion gene were grown in synthetic medium with different concentrations of CuSO4 for 2 h. Cells were then analyzed by fluorescence microscopy using an Olympus BH2 microscope or a Zeiss Axiovert 200M microscope equipped with an AxioCam HRm camera and Zeiss AxioVision software for deconvoluting images. The fluorescence intensity of Crp1-GFP in wild-type and mutant cells was quantified using AxioVision software. Copper levels were analyzed using an inductively coupled plasma mass spectrometer (ICP-MS) at the Stony Brook University Trace Element Laboratory. Cells were grown to log phase in synthetic medium (containing <1 µM copper) and then grown for an additional 4 h in the absence or presence of 100 µM CuSO4. Cell pellets were harvested, dissolved in nitric acid, and then analyzed by ICP-MS.
Virulence assays.
C. albicans strains were grown overnight at 30°C in YPD medium with 80 mg/liter uridine, reinoculated into fresh medium the next day, and incubated again overnight at 30°C. Cultures of sur7Δ cells were sonicated for 5 s to break up aggregates, washed twice in PBS, counted in a hemocytometer, and diluted to 1 × 107 cells/ml with PBS. Cell density determinations were confirmed by plating dilutions of cells onto YPD agar plates. Female BALB/c mice were injected in the lateral tail vein with 1 × 106 cells. Mice were considered to be moribund if they could no longer reach food and water and were then euthanized humanely. Statistical analysis of the survival curves was performed using Prism 4 (GraphPad Software Inc., La Jolla, CA) to carry out a log rank test (Mantel-Haenszel test). To quantify the fungal burden, kidneys were disrupted for 30 s with a tissue homogenizer (PRO Scientific, Inc., Oxford, CT) and then serial dilutions of the homogenate were plated onto YPD plates. After incubation at 30°C for 2 days, the number of CFU per gram of kidney tissue was determined. Statistical analysis of the CFU data was carried out with Prism software using one-way ANOVA with the nonparametric Kruskal-Wallis test and Dunn’s post hoc test.
Periodic acid-Schiff staining of formaldehyde-fixed kidneys from infected mice was carried out by McLain Laboratories (Smithtown, NY). Homogenized kidney tissue was stained with calcofluor white (20 ng/ml) for 10 min, pelleted by centrifugation, resuspended in 1% KOH, incubated for 5 min at room temperature, and then examined by fluorescence microscopy (57).
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI-47837 awarded to J.B.K. from the National Institute of Allergy and Infectious Diseases.
We thank the members of our labs for their helpful advice and comments on the manuscript and gratefully acknowledge Francisco Javier Alvarez, Scott Filler, Aaron Mitchell, and Daniel Kornitzer for providing strains and plasmids and the Fungal Genetics Stock Center for providing strains deposited by Suzanne Noble. We also thank the lab of James Bliska for sharing expertise and equipment, Qingzhi Zhu for help with ICP-MS analysis of copper, and Patricio Mentaboada for assistance with experimental procedures.
Footnotes
Citation Douglas LM, Wang HX, Keppler-Ross S, Dean N, Konopka JB. 2012. Sur7 promotes plasma membrane organization and is needed for resistance to stressful conditions and to the invasive growth and virulence of Candida albicans. mBio 3(1):e00254-11. doi:10.1128/mBio.00254-11.
REFERENCES
- 1. Heitman J, Filler SG, Edwards JEJ, Mitchell AP. 2006. Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 2. Odds FC. 1988. Candida and candidosis. Bailliere; Tindall, Philadelphia, PA [Google Scholar]
- 3. Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 4. Douglas LM, Martin SW, Konopka JB. 2009. BAR domain proteins Rvs161 and Rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence. Infect. Immun. 77:4150–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klis FM, de Groot P, Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl 1):1–8 [PubMed] [Google Scholar]
- 6. Odds FC, Brown AJ, Gow NA. 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11:272–279 [DOI] [PubMed] [Google Scholar]
- 7. Munro S. 2003. Lipid rafts: elusive or illusive? Cell 115:377–388 [DOI] [PubMed] [Google Scholar]
- 8. Strádalová V, et al. 2009. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 122:2887–2894 [DOI] [PubMed] [Google Scholar]
- 9. Malínská K, Malínský J, Opekarová M, Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14:4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malinska K, Malinsky J, Opekarova M, Tanner W. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 117:6031–6041 [DOI] [PubMed] [Google Scholar]
- 11. Malinsky J, Opekarová M, Tanner W. 2010. The lateral compartmentation of the yeast plasma membrane. Yeast 27:473–478 [DOI] [PubMed] [Google Scholar]
- 12. Young ME, et al. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berchtold D, Walther TC. 2009. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 20:1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fröhlich F, et al. 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 185:1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grossmann G, et al. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183:1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshikawa K, et al. 2009. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 9:32–44 [DOI] [PubMed] [Google Scholar]
- 17. Hosiner D, et al. 2011. Pun1p is a metal ion-inducible, calcineurin/Crz1p-regulated plasma membrane protein required for cell wall integrity. Biochim. Biophys. Acta 1808:1108–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martinez MJ, et al. 2004. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell 15:5295–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu T, et al. 2010. A profile of differentially abundant proteins at the yeast cell periphery during pseudohyphal growth. J. Biol. Chem. 285:15476–15488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walther TC, et al. 2006. Eisosomes mark static sites of endocytosis. Nature 439:998–1003 [DOI] [PubMed] [Google Scholar]
- 21. Olivera-Couto A, Grana M, Harispe L, Aguilar PS. 2011. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell 22:2360–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ziolkowska NE, Karotki L, Rehman M, Huiskonen JT, Walther TC. 2011. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 18:854–856 [DOI] [PubMed] [Google Scholar]
- 23. Dickson RC, Sumanasekera C, Lester RL. 2006. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 45:447–465 [DOI] [PubMed] [Google Scholar]
- 24. Luo G, Gruhler A, Liu Y, Jensen ON, Dickson RC. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283:10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walther TC, et al. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alvarez FJ, Douglas LM, Konopka JB. 2009. The Sur7 protein resides in punctate membrane subdomains and mediates spatial regulation of cell wall synthesis in Candida albicans. Commun. Integr. Biol. 2:76–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alvarez FJ, Douglas LM, Rosebrock A, Konopka JB. 2008. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol. Biol. Cell 19:5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabeche R, Baldissard S, Hammond J, Howard L, Moseley JB. 2011. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol. Biol. Cell 22:4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seger S, Rischatsch R, Philippsen P. 2011. Formation and stability of eisosomes in the filamentous fungus Ashbya gossypii. J. Cell Sci. 124:1629–1634 [DOI] [PubMed] [Google Scholar]
- 30. Vangelatos I, et al. 2010. Eisosome organization in the filamentous ascomycete Aspergillus nidulans. Eukaryot. Cell 9:1441–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernardo SM, Lee SA. 2010. Candida albicans Sur7 contributes to secretion, biofilm formation, and macrophage killing. BMC Microbiol. 10:133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang HX, Douglas LM, Aimanianda V, Latgé JP, Konopka JB. 2011. The Candida albicans Sur7 protein is needed for proper synthesis of the fibrillar component of the cell wall that confers strength. Eukaryot. Cell 10:72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keppler-Ross S, Douglas L, Konopka JB, Dean N. 2010. Recognition of yeast by murine macrophages requires mannan but not glucan. Eukaryot. Cell 9:1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seider K, Heyken A, Lüttich A, Miramón P, Hube B. 2010. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr. Opin. Microbiol. 13:392–400 [DOI] [PubMed] [Google Scholar]
- 35. Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roos D, Winterbourn CC. 2002. Immunology. Lethal weapons. Science 296:669–671 [DOI] [PubMed] [Google Scholar]
- 37. Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. U. S. A. 101:6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gardarin A, et al. 2010. Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol. Microbiol. 76:1034–1048 [DOI] [PubMed] [Google Scholar]
- 39. Jin YH, et al. 2008. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 4:e1000053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284:33949–33956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elzanowska H, Wolcott RG, Hannum DM, Hurst JK. 1995. Bactericidal properties of hydrogen peroxide and copper or iron-containing complex ions in relation to leukocyte function. Free Radic. Biol. Med. 18:437–449 [DOI] [PubMed] [Google Scholar]
- 42. Riggle PJ, Kumamoto CA. 2000. Role of a Candida albicans P1-type ATPase in resistance to copper and silver ion toxicity. J. Bacteriol. 182:4899–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weissman Z, Berdicevsky I, Cavari BZ, Kornitzer D. 2000. The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 97:3520–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Warenda AJ, Kauffman S, Sherrill TP, Becker JM, Konopka JB. 2003. Candida albicans septin mutants are defective for invasive growth and virulence. Infect. Immun. 71:4045–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu X, Zweier JL. 2001. A real-time electrochemical technique for measurement of cellular hydrogen peroxide generation and consumption: evaluation in human polymorphonuclear leukocytes. Free Radic. Biol. Med. 31:894–901 [DOI] [PubMed] [Google Scholar]
- 47. Osman D, et al. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285:25259–25268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wolschendorf F, et al. 2011. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 108:1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jo WJ, et al. 2008. Identification of genes involved in the toxic response of Saccharomyces cerevisiae against iron and copper overload by parallel analysis of deletion mutants. Toxicol. Sci. 101:140–151 [DOI] [PubMed] [Google Scholar]
- 50. Lech T, Sadlik JK. 2007. Contribution to the data on copper concentration in blood and urine in patients with Wilson’s disease and in normal subjects. Biol. Trace Elem. Res. 118:16–20 [DOI] [PubMed] [Google Scholar]
- 51. Grossmann G, Opekarová M, Malinsky J, Weig-Meckl I, Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edlind TD, Katiyar SK. 2004. The echinocandin “target” identified by cross-linking is a homolog of Pil1 and Lsp1, sphingolipid-dependent regulators of cell wall integrity signaling. Antimicrob. Agents Chemother. 48:4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Radding JA, Heidler SA, Turner WW. 1998. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob. Agents Chemother. 42:1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. LaFayette SL, et al. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:e1001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C, Konopka JB. 2010. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot. Cell 9:224–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sherman F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41 [DOI] [PubMed] [Google Scholar]
- 57. Pringle JR. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194:732–735 [DOI] [PubMed] [Google Scholar]