SUMMARY
Optimal levels of ftsZ gene product are shown to be required for initiation of the cell division process in Mycobacterium tuberculosis. Here, we report that the ftsZ gene expression is sharply down-regulated during starvation and hypoxia, conditions that are believed to result in growth arrest, but is restored upon dilution of cultures into fresh oxygen-rich media. Primer extension analysis identified four transcriptional start sites, designated as P1, P2, P3 and P4 at nucleotide positions −43, −101, −263, and −787, respectively, in the immediate upstream flanking region of the ftsZ initiation codon. Promoter deletion and homologous recombination experiments revealed that ftsZ expression from the 101-bp region is sufficient for M. tuberculosis viability. All promoter strains had reduced FtsZ levels compared to wild-type, although the loss of P4 severely compromised FtsZ levels during both the active and stationary phases. We propose that ftsZ expression from all promoters is required for optimal intracellular FtsZ levels and that the activities of P4 and possibly other promoters are down-regulated during growth arrest conditions.
Keywords: FtsZ, Mycobacterium, cell division, transcription
1. Introduction
A hallmark of tuberculosis is latency, wherein Mycobacterium tuberculosis, the causative agent, is believed to remain in a non-proliferative persistent (NRP) state with limited bacterial turnover during its growth in granulomas1, 2. It is believed that Mtb encounters hypoxic conditions in granulomas3, 4. Mtb cultures exposed to oxygen-deprived conditions in nutrient broth attain NRP state, but upon resuspension in oxygen-rich media undergo a round of cell division prior to initiating new rounds of DNA replication2, 5. These results suggest that the cell division process is tightly regulated during hypoxia or NRP state.
FtsZ is an essential protein critical for the initiation of cell division in nearly all prokaryotes and in some mitochondria and chloroplasts6, 7. FtsZ protein polymerizes in a GTP dependent manner and forms a contractile cytokinetic ring or the Z-ring at a pre-determined septal site, usually at the mid-cell position6, 8, 9. Apart from recruiting as many as 13 additional cell division proteins to the division site, FtsZ is likely to provide the force for constriction of the septum in the dividing cell7, 10–13. FtsZ of M. tuberculosis (FtsZTB), although similar in structure and sequence to other prokaryotic FtsZ proteins, exhibits slow polymerization and weak GTPase activities14, 15. M. tuberculosis FtsZ protein levels decrease during stationary growth and optimal FtsZTB levels are needed to sustain cell division16–18. Investigations on the kinetics of FtsZ protein assembly dynamics revealed that the half-time for subunit turnover is 42 sec in vitro and 25 sec in vivo19. Together, these findings suggest that ftsZ expression is subject to regulation in M. tuberculosis and that characterization of ftsZ promoter region could provide clues in this direction.
The immediate upstream flanking genes of M. tuberculosis ftsZ include ftsQ and murA and the intergenic region between ftsZ and ftsQ is 174-bp. One earlier report identified several transcriptional start sites in the promoter region of ftsZ20. It is unknown if ftsZ expression from all these promoters is required for cell division and if there is a minimal promoter region needed to sustain cell division and growth. Furthermore, it is unknown if there is any correlation between the FtsZ levels and the expression from individual promoters. The present study addresses these questions.
2. Materials and methods
2.1. Bacterial growth conditions
Escherichia coli strains were grown in Luria–Bertani (LB) broth or agar supplemented with kanamycin (Km-50 μg ml−1) or hygromycin (hyg-50μg ml−1)17. M. tuberculosis strains were propagated in Middlebrook 7H9 broth supplemented with OADC (oleic acid, albumin, dextrose, catalase with sodium chloride) and plated on 7H10 plates containing appropriate antibiotics (Km at 25 μg ml−1; Hyg at 50 μg ml−1). Growth was monitored by absorbance at 600 nm and viability by determining colony forming units on Middlebrook 7H11 agar plates (Remel).
2.2. Construction of recombinant ftsZ promoter strains
The Hygr pJfr66 plasmid carrying ftsZ and its upstream 1 kb region contains four ftsZ promoters referred to as P1-P4 and was used as a template to generate DNA fragments containing the ftsZ coding region with different lengths of promoters21. The P1 fragment carries promoter 1, P2 carries promoters 1-2, P3 carries promoters 1-3 and P4 carries promoters 1-4. The promoter fragments were cloned individually into pMV306K, a Kmr integrating vector (Table 1), and confirmed by sequencing. The plasmids with the respective promoter fragments were named as pMK1, pMK2 and pMK3 (Table 1) and were used to exchange the resident pJFR66 in Mtb-66 by transformation as described21. Genomic DNA from all recombinant strains was extracted; the ftsZ region was amplified and verified by sequencing.
Table 1.
| Plasmids | Description | Reference |
|---|---|---|
| pMH66 | Mycobacterial integrating vector with luciferase gene, hygr | Park et al. |
| pEM43 | pMH66 with 1 kb 5’flanking region of ftsZtb, hygr | This study |
| pEM45 | 3 kb BamHI-ClaI fragment from pEM43 in pMV206, kmr | This study |
| pRM15 | 278-bp flanking 5’ftsZTB in pEM45, kmr | This study |
| pRM16 | 114-bp flanking 5’ ftsZTB in pEM45, kmr | This study |
| pRM17 | 804-bp flanking 5’ ftsZTB in pEM45, kmr | This study |
| pRM18 | 1068-bp flanking 5’ ftsZTB in pEM45, kmr | This study |
2.3. Western blotting
FtsZ levels were quantitated by immunoblotting and normalized to MtrA as the levels of the latter are shown to be constant under various growth conditions22.
2.4. RNA extraction, primer extension and quantitative real-time (QRT) PCR
Extraction of total RNA and determination of ftsZ transcript levels relative to house keeping 16S rRNA gene by quantitative real-time (qRTR-PCR) were essentially as described22, 23. The reactions were performed in triplicate on at least two independent RNA preparations. Primer extension was carried out using 10 to 20 µg total RNA, γ-32P end-labeled oligonucleotide primers specific to ftsZ upstream region and AMV reverse transcriptase utilizing the primer extension system from Promega. For each primer extension reaction a DNA sequencing ladder was generated with pJFR66 DNA as the template and the corresponding primer used for primer extension (not shown). DNA sequencing was done using the Sequenase Version 2.0 DNA sequencing kit from USB, the products were resolved on 10% denaturing acrylamide gels, and processed for autoradiography. Bands were visualized using the BIORAD Molecular Imager and analyzed by QuantityOne software.
2.5. Construction of ftsZ promoter–luciferase (lux) plasmids and measurement of lux activity
Different lengths of the 5’ flanking region of ftsZtb bearing appropriate promoters/transcriptional start sites were PCR amplified, cloned upstream of the promoter-less lux gene in pMV306K vector and transformed into M. tuberculosis. Luciferase activity was measured by mixing 100 µL of exponentially growing cultures with 100 µL of Beetle Luciferin (1mM; Promega) and measured in a luminometer (Turner Designs). Luciferase activity is expressed as lux units per O.D.600.
2.6. Microscopic analysis and cell length measurement
M. tuberculosis cells were visualized on a Nikon Eclipse 600 microscope equipped with a 100X (Nikon Plan Fluor) oil immersion objective with a numerical aperture of 1.4 and cell lengths were determined as described21.
3. Results and discussion
3.1. ftsZ expression is reduced during starvation and hypoxia
In an effort to understand if the ftsZ expression is altered under conditions that lead to growth arrest, we measured the ftsZ transcript levels relative to exponential growth phase by QRT PCR. The ftsZ transcript levels were reduced 10-fold during stationary phase and hypoxia relative to the exponential phase, but were restored to wild-type levels following resuspension of the hypoxic cultures in fresh broth (Fig. 1). Since hypoxic cultures are blocked at the cell division step, these results suggest that ftsZ gene expression is growth-phase dependent and that optimal ftsZ transcript levels are necessary for the initiation of cell division.
Figure 1.
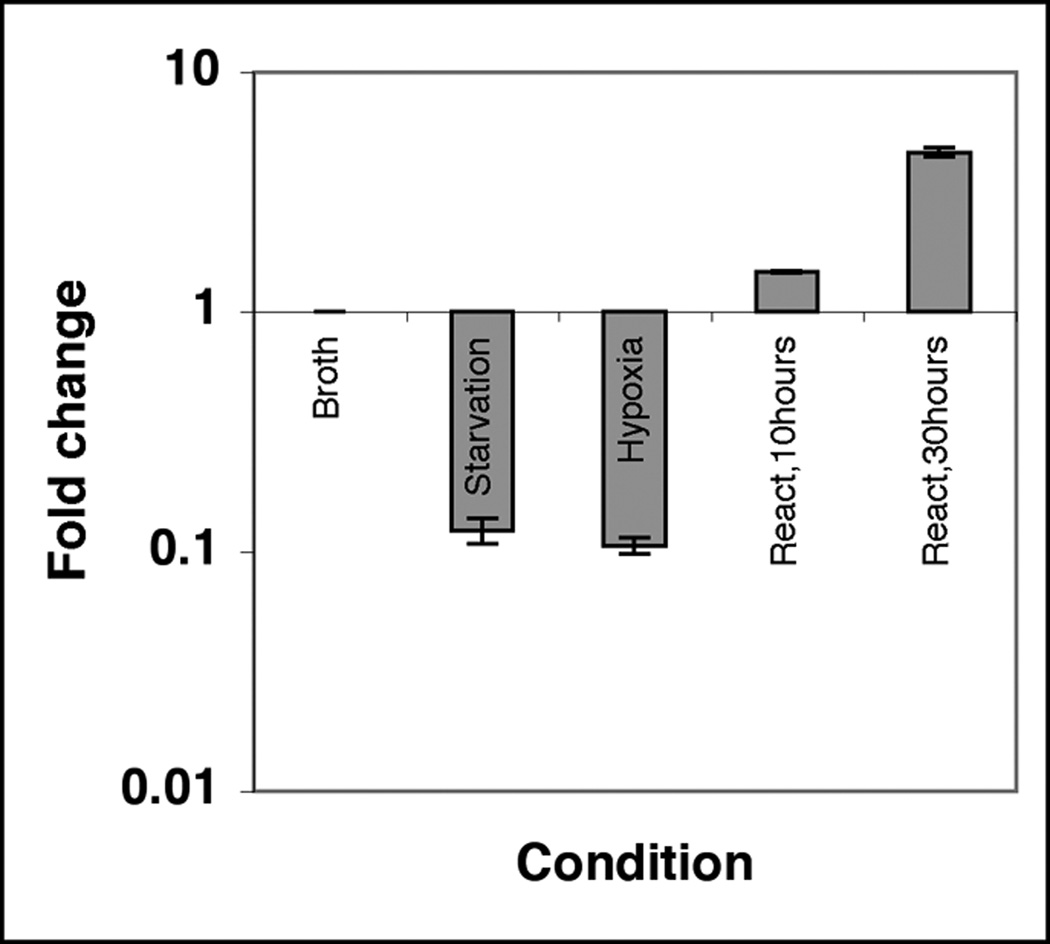
Expression levels of ftsZ under different growth conditions. M. tuberculosis was grown in broth under various indicated conditions as described. RNA was extracted and levels of ftsZ mRNA were measured by quantitative real-time PCR using Taqman chemistry (see Table 1). The expression levels of ftsZ mRNA were normalized to 16s mRNA levels. Mean ±SD from three independent experiments are shown.
3.2. Multiple ftsZ promoters contribute to ftsZ expression
To begin understanding how ftsZ transcription is regulated, we extracted total RNA and carried out primer extension using oligonucleotide primers specific to the 1 kb ftsZ upstream region. Four transcriptional start sites designated as P1, P2, P3 and P4 at −43, −101, −263 and −787 nucleotides, respectively, relative to the ftsZ start codon were identified (Table 1; Fig. 2A). The last 2 promoters, P3 and P4, were located in the ftsQ-coding region whereas those of the P1, and P2 were in the ftsQ-ftsZ intergenic region. While these results are in general agreement with those reported by Roy et al. (20), differences with respect to specific location of transcriptional start sites are noted (see Fig. 2A legend). These data, however, do not rule out the possibility of other regulatory elements or unidentified distant promoters present upstream of the 1 kb region that could affect ftsZ transcription.
Figure 2.
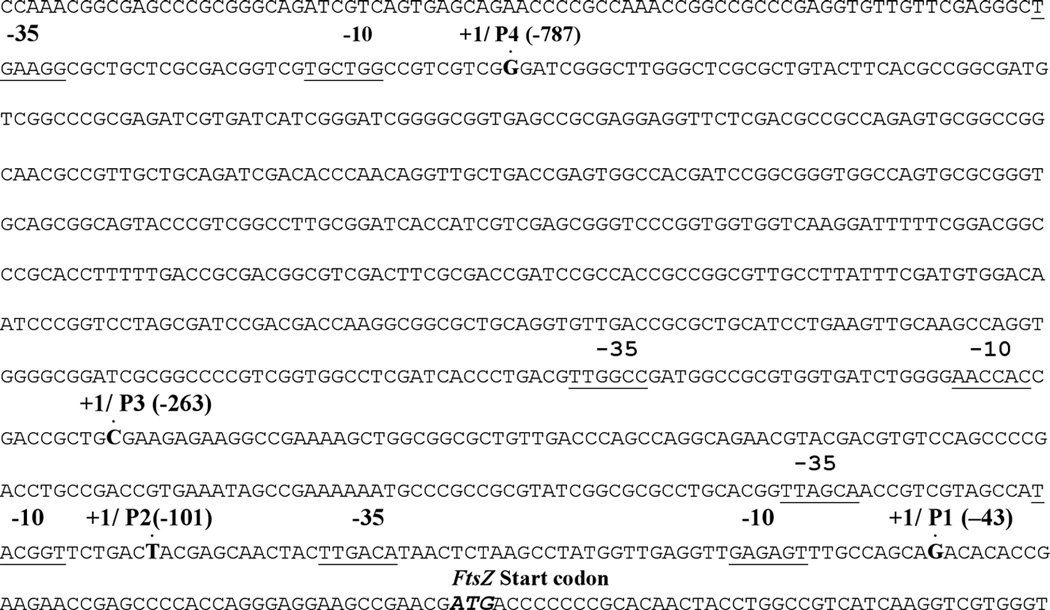
Putative transcription start sites for ftsZTB. Transcription start sites and the putative −35 and −10 regions in the 5’ flanking region of ftsZTB are shown. E. coli sigma 70 type −35 and −10 boxes, TTagCA and TAcggT, respectively (uppercase - identity with the consensus), identified upstream of the P2 promoter. Because of at least 3 matches to the s70 E. coli consensus, the P2 promoter was included in group A mycobacterial promoters. For promoters P1 and P4, distinct s70 type −35 regions were seen, but no identifiable −10 sequences were found. For promoter P3, no apparent −10 or −35 regions were seen. Therefore, these three promoters could belong to mycobacterial group C promoters and may contain recognition sequences for regulators other than σA or σB (33).
ftsZ promoter-luciferase fusion reporter gene expression analysis revealed that a construct bearing all four promoters showed the highest promoter activity whereas that containing P1 alone showed the least (Fig. 2B). DNA fragment containing the P2 and P1 promoters contributed to nearly 62% of total activity. The relative contribution of the four promoters was calculated to be: P1 - 5%, P2 - 57%, P3 - 14% and P4 - 23% with P2 being the most active promoter.
3.3. ftsZ expression from P2 promoter is sufficient for viability
M. tuberculosis FtsZ is an abundant protein with ~ 14, 000 molecules per cell17. To understand if ftsZ expression from all four promoters is needed for cell division and optimal FtsZ levels, we created Kmr plasmids expressing ftsZ from DNA fragments bearing P1 (pMK1), P1+P2 (pMK2), P1+P2+P3 (pMK3) and used them to exchange the integrated Hygr plasmid pJFR66 expressing ftsZ from P1-P4 promoters in M. tuberculosis ΔftsZ strain, Mtb-6616. Transformation of Mtb-66 with pMK2 and pMK3, but not that with pMK1, produced viable transformants indicating that ftsZ expression from a region containing P1 and P2 is sufficient to support growth and viability (not shown). DNA sequencing following PCR amplification confirmed that pMK2 and pMK3 had indeed replaced the resident plasmid pJFR66 in these experiments (not shown). One representative transformant each, designated as Mtb-pMK2 and Mtb-pMK3, respectively, was further characterized.
The recombinant strains Mtb-pMK2 and Mtb-pMK3, like the Mtb-66, showed reduced growth rate and viability as compared to the parent strain (Fig. 3, see inset). However, the growth rates of the Mtb-pMK2 and Mtb-MK3 were comparable to that of Mtb-66 (Fig. 3). Differences in the growth rate and viability between the wild-type and the recombinant promoter strains indicate that the sequences upstream of the 1-kb promoter region are necessary for optimal growth and viability. Alternatively, the location of ftsZ at the attB site may have affected the transcription of genes present downstream of ftsZ. Nonetheless, these data indicate that transcription from a fragment containing promoters 1 and 2 is sufficient for the viability of M. tuberculosis. Consistent with the slow growth rate, a modest 15% increase in average cell length of the recombinant strains was noted (data not shown).
Figure 3.
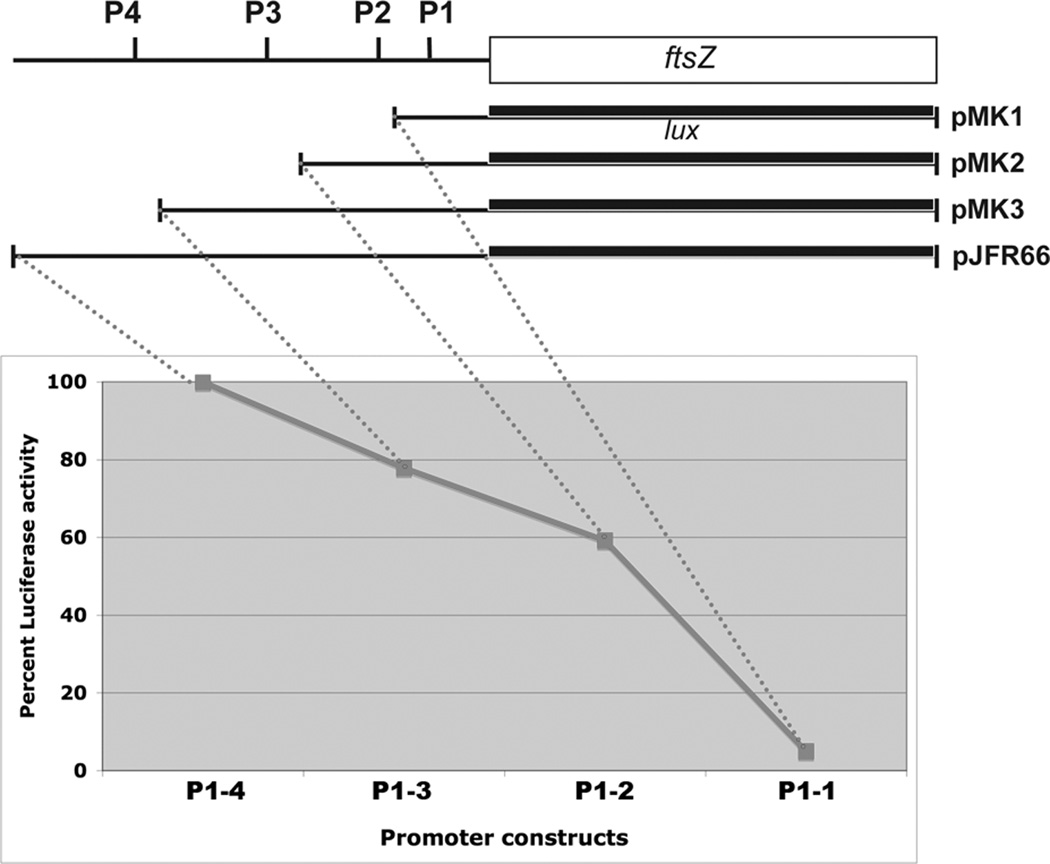
Transcription from ftsZTB promoters P1-P4. Transcriptional fusions bearing P1 to P4 promoters upstream of the lux gene in pEM45 (pRM15 – pRM18) were transformed to M. tuberculosis (upper panel) and luciferase assays were carried out using exponentially growing cultures of the recombinant strains. Lux units were determined and normalized to OD600 units (lower panel).
3.4. FtsZ levels and the contribution of promoters
Next, we examined the FtsZ levels in the promoter strains during exponential and the stationary growth phases (Fig. 4). The following observations were evident: (1) FtsZ levels were reduced in the stationary phase relative to the active phase in the wild type strain. These results are consistent with the mRNA expression (Fig. 1) and our earlier published data17. (2). Expression of ftsZ from attP site in the Mtb-66 strain led to a minor reduction in the FtsZ levels under both growth conditions (Fig. 4. compare P1-4 with WT, also see inset). (3) Removal of the P4 promoter led a significant reduction in the FtsZ levels (Fig. 4 and inset, compare P1-3 with WT, P1-4 and P1-2). Simultaneous removal of P4 and P3 promoters restored FtsZ levels, albeit partially (see P1-2 in Fig. 4 and inset).
Figure 4.
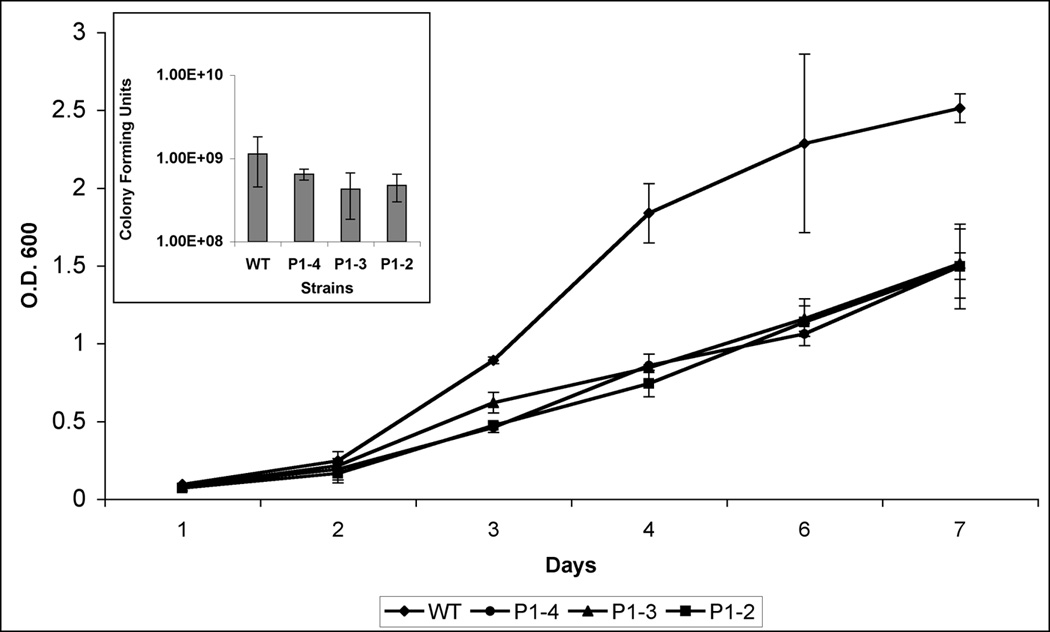
Growth and viability of M. tuberculosis ftsZ promoter strains. M. tuberculosis H37Ra strains, Mtb-66 (P1-4), Mtb-pMK3 (P1-3) and Mtb-pMK2 (P1-2) were grown in 7H9 broth for indicated time-points and OD600 was measured and plotted. Mean ±SD from three independent experiments are shown. Inset: Exponential cultures from each strain were plated on 7H10 agar and CFU determined after 3 weeks. WT – M. tuberculosis H37Ra strain.
The presence of multiple functional promoters in the upstream flanking region of the M. tuberculosis ftsZ, as in other bacteria, signals a complex regulation24–28. These results suggest that while hitherto unidentified regulatory elements present upstream of the 1-kb promoter region are important for optimal ftsZ expression, the absence of P4 promoter activity significantly reduces the intracellular FtsZ levels. It is possible that under the conditions that promote growth arrest, ftsZ expression from P4 and or more promoters is selectively prevented thereby resulting in decreased transcription and translation. Thus, decrease in ftsZ transcript levels during hypoxia and starvation and upregulation upon reactivation signals that ftsZ transcriptional regulatory mechanisms are at work under growth arrest conditions. We can also envision that intracellular FtsZ levels could be regulated at the protein level. For example, a recent report indicates that FtsZ is a substrate of FtsH protease in vitro29. Our preliminary data indicate that FtsZ levels are modulated in M. tuberculosis strains expressing altered levels of FtsH 30. Presumably, FtsH and other unrecognized proteases, could act to reduce FtsZ levels. Recent experiments to determine FtsZ dynamics by fluorescent recovery after photo-bleaching (FRAP) technique revealed that only ~30% of the available intracellular FtsZ is engaged in the formation of septal Z-rings at midcell sites indicating that cell division can proceed with reduced levels of FtsZ31, 32. We envision that a combination of transcriptional and translational regulation controls FtsZ levels and therefore Z-ring assembly and cell division during normal growth, NRP state2 and possibly during growth in granulomas.
Figure 5.
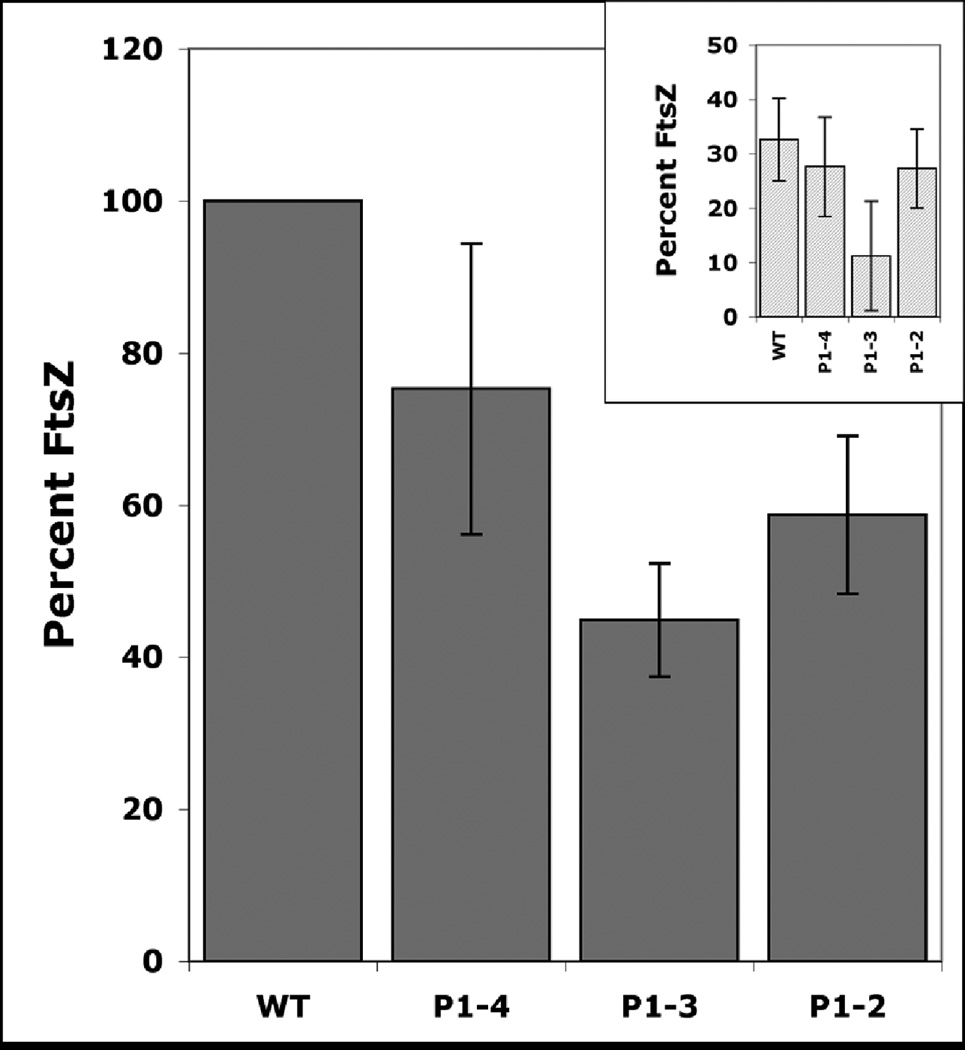
FtsZ levels in M. tuberculosis ftsZ promoter strains: Exponential or stationary phase cultures of Mtb-66 (P1-4), Mtb-pMK3 (P1-3) and Mtb-pMK2 (P1-2) were lysed and separated on SDS-PAGE as described in Methods. Proteins were transferred to nitrocellulose membranes and Western blot analysis was carried out as described in Methods. FtsZ levels were measured and normalized to MtrA22. FtsZ levels in various promoter strains are expressed with respect to exponential phase wild-type M. tuberculosis. Inset: FtsZ levels in stationary phase.
Table 2.
| Oligo name | Oligo sequence | Description |
|---|---|---|
| MVM535 | 5’- CCT TGA TGA CGG CCA GGT AG - 3’ | For mapping P1 by PE |
| MVM238 | 5’- GCG GAT CCG CTT CCT CCC TGG TGG GGC- 3’ | For mapping P2 by PE |
| MVM534 | 5’- CGG GGC TGG ACA CGT CGT ACG - 3’ | For mapping P3 by PE |
| MVM533 | 5’- CCT CCT CGC GGC TCA CCG CC - 3’ | For mapping P4 by PE |
| MVM542 | 5’- CGC GGA TCC GAC TAC GAG CAA CTA CTT GAC -3’ | For cloning P1 |
| MVM541 | 5’- CGC GGA TCC CCG CTG CGA AGA GAA GGC CG -3’ | For cloning P2 |
| MVM540 | 5’- CGC GGA TCC CCG TCG TCG GGA TCG GGC TTG -3’ | For cloning P3 |
| MVM461 | 5’- TGG CAA CAC TAG TAC GAG GAGGC GGT TAC GGA G -3’ | For cloning P4 |
Acknowledgments
This work is supported by RO1-AI48417, RO1-AI41406 and R56-AI073966.
References
- 1.Harries AD, Dye C. Tuberculosis. Ann Trop Med Parasitol. 2006;100:415–431. doi: 10.1179/136485906X91477. [DOI] [PubMed] [Google Scholar]
- 2.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 3.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE., 3rd Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehlers S. Lazy, Dynamic or Minimally Recrudescent? On the Elusive Nature and Location of the Mycobacterium Responsible for Latent Tuberculosis. Infection. 2009 doi: 10.1007/s15010-009-8450-7. [DOI] [PubMed] [Google Scholar]
- 5.Wayne LG. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 6.Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 7.Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson HP. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 9.Lutkenhaus J. Bacterial cytokinesis: let the light shine in. Curr Biol. 1997;7:R573–R575. doi: 10.1016/s0960-9822(06)00285-5. [DOI] [PubMed] [Google Scholar]
- 10.Goehring NW, Gueiros-Filho F, Beckwith J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 2005;19:127–137. doi: 10.1101/gad.1253805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan G, Wolgemuth CW, Sun SX. Z-ring force and cell shape during division in rod-like bacteria. Proc Natl Acad Sci U S A. 2007;104:16110–16115. doi: 10.1073/pnas.0702925104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopalan M, Atkinson MA, Lofton H, Chauhan A, Madiraju MV. Mutations in the GTP-binding and synergy loop domains of Mycobacterium tuberculosis ftsZ compromise its function in vitro and in vivo. Biochem Biophys Res Commun. 2005;331:1171–1177. doi: 10.1016/j.bbrc.2005.03.239. [DOI] [PubMed] [Google Scholar]
- 15.White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182:4028–4034. doi: 10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, Rajagopalan M. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol Microbiol. 2006;62:132–147. doi: 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- 17.Dziadek J, Madiraju MV, Rutherford SA, Atkinson MA, Rajagopalan M. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology. 2002;148:961–971. doi: 10.1099/00221287-148-4-961. [DOI] [PubMed] [Google Scholar]
- 18.Dziadek J, Rutherford SA, Madiraju MV, Atkinson MA, Rajagopalan M. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology. 2003;149:1593–1603. doi: 10.1099/mic.0.26023-0. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Anderson DE, Rajagopalan M, Erickson HP. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J Biol Chem. 2007;282:27736–27743. doi: 10.1074/jbc.M703788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy S, Ajitkumar P. Transcriptional analysis of the principal cell division gene, ftsZ, of Mycobacterium tuberculosis. J Bacteriol. 2005;187:2540–2550. doi: 10.1128/JB.187.7.2540-2550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006;188:1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair N, Dziedzic R, Greendyke R, Muniruzzaman S, Rajagopalan M, Madiraju MV. Synchronous replication initiation in novel Mycobacterium tuberculosis dnaA cold-sensitive mutants. Mol Microbiol. 2009;71:291–304. doi: 10.1111/j.1365-2958.2008.06523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MV, Rajagopalan M. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 24.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzy-Treboul G, Karmazyn-Campelli C, Stragier P. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J Mol Biol. 1992;224:967–979. doi: 10.1016/0022-2836(92)90463-t. [DOI] [PubMed] [Google Scholar]
- 26.Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sackett MJ, Kelly AJ, Brun YV. Ordered expression of ftsQA and ftsZ during the Caulobacter crescentus cell cycle. Mol Microbiol. 1998;28:421–434. doi: 10.1046/j.1365-2958.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- 28.Vicente M, Gomez MJ, Ayala JA. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell Mol Life Sci. 1998;54:317–324. doi: 10.1007/s000180050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anilkumar G, Srinivasan R, Anand SP, Ajitkumar P. Bacterial cell division protein FtsZ is a specific substrate for the AAA family protease FtsH. Microbiology. 2001;147:516–517. doi: 10.1099/00221287-147-3-516. [DOI] [PubMed] [Google Scholar]
- 30.Kiran M, Chauhan A, Dziedzic R, Maloney E, Mukherji S, Madiraju M, Rajagopalan M. Mycobacterium tuberculosis ftsH expression in response to stress and viability. Tuberculosis (Edinb) 2009 doi: 10.1016/S1472-9792(09)70016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez JE, Bishai WR. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc Natl Acad Sci U S A. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]


