Abstract
The CONNecticut Joint University Research (CONNJUR) team is a group of biochemical and software engineering researchers at multiple institutions. The vision of the team is to develop a comprehensive application that integrates a variety of existing analysis tools with workflow and data management to support the process of protein structure determination using Nuclear Magnetic Resonance (NMR). The use of multiple disparate tools and lack of data management, currently the norm in NMR data processing, provides strong motivation for such an integrated environment. This manuscript briefly describes the domain of NMR as used for protein structure determination and explains the formation of the CONNJUR team and its operation in developing the CONNJUR application. The manuscript also describes the evolution of the CONNJUR application through four prototypes and describes the challenges faced while developing the CONNJUR application and how those challenges were met.
Keywords: Bioinformatics, Integration, NMR, Protein
1. Introduction
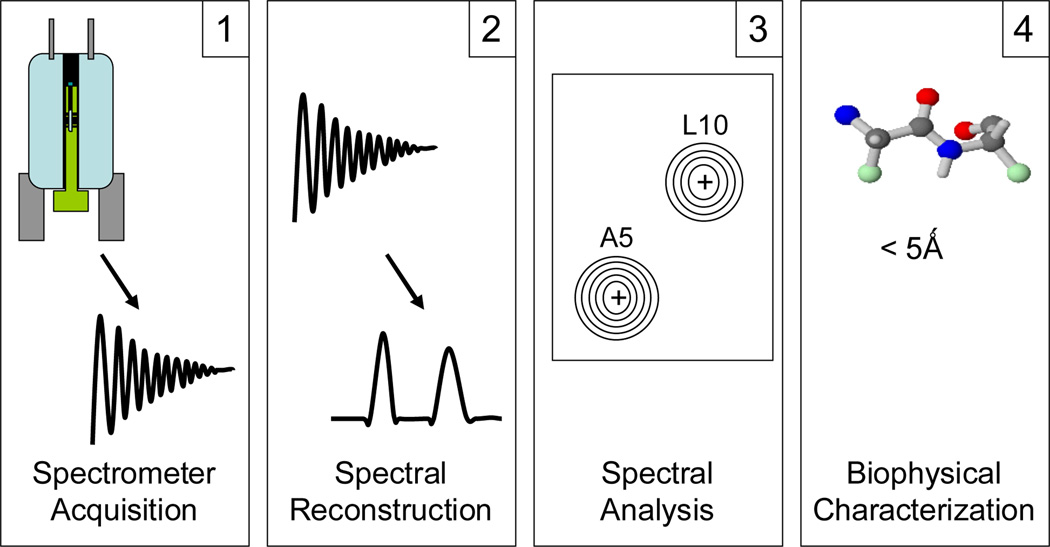
The current process of protein structure determination using NMR is lengthy and complex, relying on a variety of different software tools to process data in various forms with a large degree of user guidance. Figure 1 illustrates the four general phases of the NMR experiment process: 1. Spectrometer Acquisition: A protein sample in solution is probed using an NMR spectrometer over a period of hours or days, producing several binary files, each up to 100MB in size.
Figure 1.
NMR experiment process.
2. Spectral Reconstruction: This raw output is cleaned and processed to convert the output from time domain data to frequency domain data represented as frequency spectra.
3. Spectral Analysis: A series of multidimensional spectra representing the atoms and their relationship to each other are analyzed to associate which resonance frequencies correspond to which nuclei.
4. Biophysical Characterization: Lastly, the physical structure of the protein is calculated using distance and angular restraints inferred from the spectra.
Each of the four phases above may involve the iterative use of one or more software tools to refine the data and individual processing steps may be repeated if the results of a previous step are found to be insufficient. A single protein structure determination experiment may take weeks or months to complete and can generate up to several gigabytes of data in a variety of formats. Much, if not all, of this data is stored in flat files, in a variety of different formats, making the data difficult to manage. In the absence of a database, there is no simple way to query this experiment data. In addition, the flat format of the data differs for the various tools used throughout the experiment process.
Many researchers have voiced a desire to automate NMR structure determination. In fact, some NMR researchers have made attempts to write their own software to support this automation [1–4]. However, unlike many diagnostic machines (i.e. an ultrasound device) which have a single purpose, an NMR spectrometer is used for a wide range of tasks. In addition, individual user preferences vary significantly with respect to accomplishment of tasks. This results in the need for a variety of different data processing applications to support the range of purposes. Since the number of users of NMR spectrometers for the purpose of protein structure determination is low (in the thousands world-wide), there is little incentive for commercial companies to develop such data processing tools.
The need for modularity and flexibility in NMR analysis has resulted in the use of “scripts” as a common interface to NMR tools. One first step towards automating the NMR data analysis process is the use of a script generator (http://sbtools.uchc.edu). A typical script generator is a web-based interface to one or more NMR data processing tools. The application allows the NMR researcher to enter various characteristics of their experiment and generates an executable script that includes an appropriate set of parameters for use with one or more of the data analysis tools. While a step in the right direction, the script generator is only a partial solution to the problem of automating the NMR experiment process as it only eases the use of a small number of tools. Furthermore, the script generator approach cannot address the problems of data organization, lacks elements of workflow capture, nor can it support the querying of experiment data or easily support the iterative, cyclic nature of the NMR experiment process. To address the data and workflow issues in an NMR experiment, we have developed the CONNJUR (CONNecticut Joint University Research) application. The CONNJUR application is an integrated environment for biomolecular NMR data analysis.
2. The CONNJUR Project
The core CONNJUR team is made up of five individuals from three different institutions with a mix of backgrounds in computing, biochemistry and biology. The central idea for CONNJUR was first envisioned in 2001 by Dr. Michael Gryk, an Assistant Professor of Molecular, Microbial and Structural Biology at the University of Connecticut Health Center who has research interests in the area of protein structure determination. Dr. Gryk attended a database seminar held by Dr. Timothy Martyn at Rensselaer at Hartford and quickly grasped the benefits afforded by providing the NMR experimental process with database support and CONNJUR was born.
The team has an informal structure with each person responsible for their area of expertise. Dr. Gryk provides the domain understanding and is both an end user and developer. Dr. Gryk is adept in programming, having an understanding of several programming languages and databases, and was proficient in writing UNIX shell scripts to support his NMR experiments prior to the advent of the CONNJUR application. Dr. Martyn has extensive background in data modeling and database applications and provides data modeling support for the CONNJUR application and for the larger domain of NMR [5,6]. Dr. Heidi Ellis, currently from Western New England College, is responsible for the software engineering aspects of the project. Another key member (now a former member), Dr. Susan Fox-Erlich, has a B.S. in Biology, M.S. in Computer Science, and a Ph.D. in Physiology. Dr. Fox-Erlich’s unique background has resulted in important insights into the development of the CONNJUR application. The lead CONNJUR developer, Mr. Jay Vyas, has a M.S. in Computer Science and worked on the CONNJUR application during his computer science studies. Mr. Vyas is now working on a doctorate in biomedical sciences and has taken classes in biochemistry and NMR in order to broaden his understanding of the domain, while also working on other aspects of protein bioinformatics. In addition, the core team has had support from Dr. Mark Maciejewski, NMR facility manager at the University of Connecticut Health Center, and various graduate and undergraduate students.
The CONNJUR team faced several challenges when designing and developing software components necessary for the project. One typical problem faced when developing a scientific tool is the lack of common understanding between developers and domain experts. We have experienced this problem to some degree as the computing team members are not very familiar with the NMR domain while the NMR experts are only modestly familiar with software development. Since the group’s inception in 2001, Dr. Gryk has actively pursued an understanding of software and database design, including database normalization, which has allowed us to accurately and efficiently model the underlying data to support NMR experiments. Dr. Gryk has also gained understanding of computer science theory including regular expressions, finite state automata, object-oriented design, and process modeling. Dr. Gryk has demonstrated a facility for recognizing the way “relational” data management is implemented in an ad-hoc manner in legacy NMR tools, which store information in flat text files. In addition, we are heavily aided by the fact that Dr. Fox-Erlich has both computing and biochemical background and by the domain knowledge of Mr. Vyas which is continually being expanded.
One consequence of the emergent common knowledge-base among the CONNJUR team was the fast rate of change that occurred in the development of the CONNJUR application, especially in the early phases. At the outset, we had only a vague understanding of the requirements of the CONNJUR application; that it needed to wrap existing NMR data analysis tools and provide data management support. As the team gained a greater understanding of the NMR domain as well as software engineering and computing approaches, the requirements of a tool to support NMR data processing evolved and changed. The rate of change was rapid and sometimes work that had been done the previous week was discarded as a new approach was accepted. Handling such a high rate of change requires patience and flexibility on the part of all team members.
While there has been a fortuitous overlap of expertise by many of the team members, another critical mechanism we have used to create a common understanding has been abstraction. When we have encountered a problem, we have frequently abstracted the domain-specific issue to a higher level of abstraction. We then found a simpler analogous example that contains a similar issue and resolved the issue by applying the rules and heuristics that apply to the simpler approach to the domain-specific instance. For instance, in the process of modeling the complexity of atoms and bonds and their relationships, the problem was abstracted to use an example domain of cars and their components. The understanding gained from investigating this analogous domain allowed us to more clearly understand the correct modeling of the NMR domain. Minor inconsistencies may arise when resolving the problem at the domain level and only in rare instances may the problem may to be abstracted once again to correct for a fundamental misunderstanding of the system. These rarer occasions are often the most insightful!
Another challenge that we faced was the precision with which real world phenomena must be modeled. The first step in automating NMR data processing was modeling the data that supports protein structure determination using NMR. During the modeling process we realized that data modeling for the natural world provides fixed constraints and there is less flexibility in modeling as can occur for business data. The natural phenomena must be modeled completely and accurately in order to fully support automation of the NMR experiment process. It took a series of iterations over several months for Dr. Gryk and Dr. Martyn and other group members to construct a robust model of protein conformation.
The nature of the existing NMR data analysis tools provided yet another difficulty in automating the NMR experiment process. The current NMR data analysis tools were not typically developed to be scalable or maintainable and were not designed to be used in any integrated fashion. In addition, none of the data analysis tools support the storage and management of data. The characteristics of the existing tools provide NMR spectroscopists incentive to create their own helper tools and an abundance of scripts and script generators have been developed by NMR researchers. The CONNJUR application is intended to provide a unified environment for interacting with a variety of existing tools while providing data management capabilities, all in a flexible and scalable manner.
The development of the CONNJUR application has used a very agile approach with frequent team meetings to review progress and to plan. The core CONNJUR team began meeting regularly in the spring of 2003 and efforts were focused on creating a core data model of atoms and bonds from an NMR perspective. This work was completed in the spring of 2004 [5]. The utilities associated with various versions were able to be combined into several powerful tools enabling visualization and analysis of proteins in broader contexts[12]. The summer and fall of 2004 were spent learning the NMR processing tools and identifying requirements. During the fall of 2004, a graduate student researched and evaluated the possible architectures for the CONNJUR application. Below we describe the evolution of the CONNJUR application as we learned more about the requirements for automating the NMR experiment process to arrive at CONNJUR’s current workflow-driven design.
3. Tool-Oriented CONNJUR
After gaining an initial comprehension of the requirements for the CONNJUR application, we decided to take a tool-oriented approach. This decision was made based on the determination that it would be easier for NMR spectroscopists to have a direct view of the underlying tools that were being wrapped by the prototype. Development began in January 2005 with the goal of a prototype that could import raw data files from the NMR spectrometer and provide an interface for one data processing tool, NMRPipe [7], a tool which supports the spectral reconstruction phase of the NMR experimentation process.
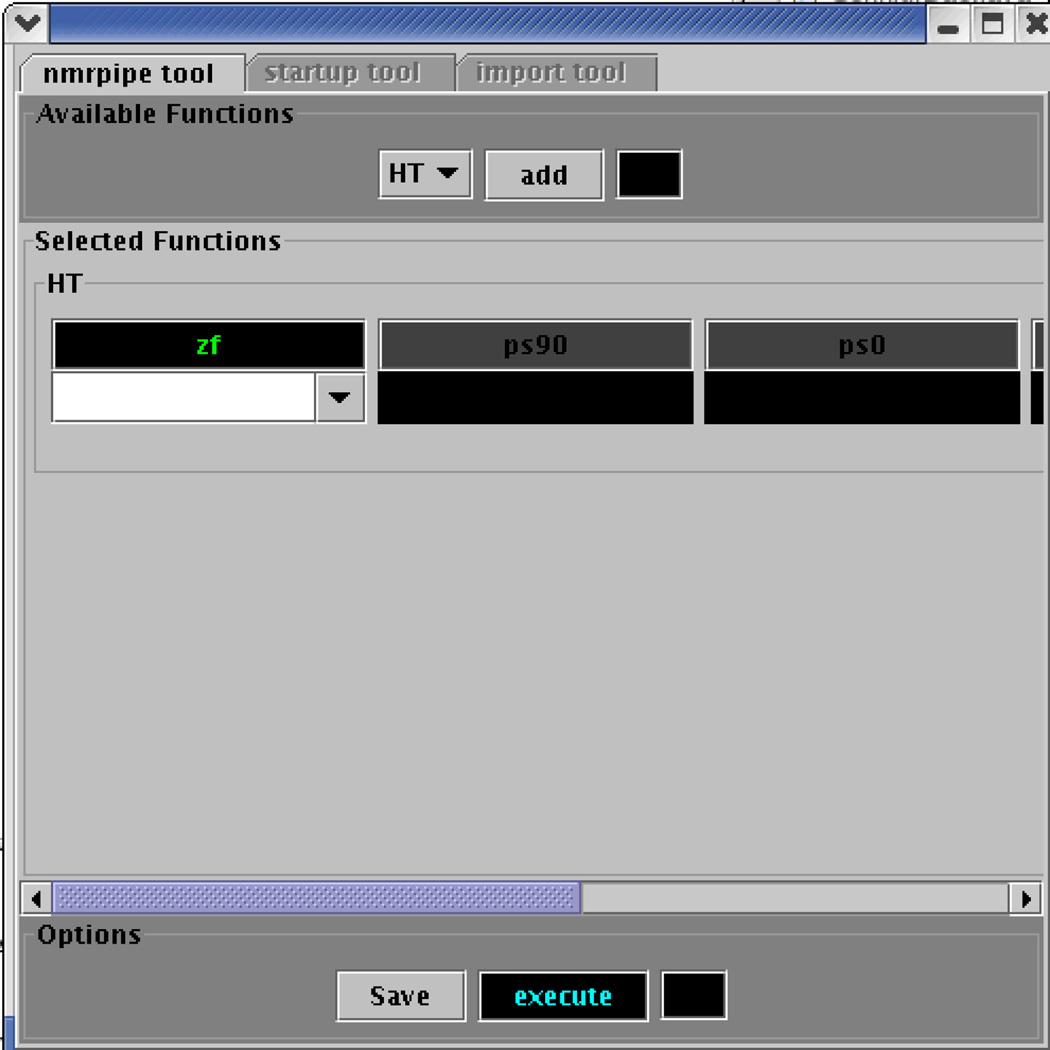
The basic interface to the first prototype, shown in Figure 2, included a series of panels which allowed for individual configuration of an NMRPipe function, execution of several functions at once, and the importation of new data as input to the system. Figure 2 shows the selection of the Hilbert Transform function of the NMRPipe tool.
Figure 2.
Tool-Oriented version of CONNJUR showing configuration choices for performing the Hilbert Transform Operation.
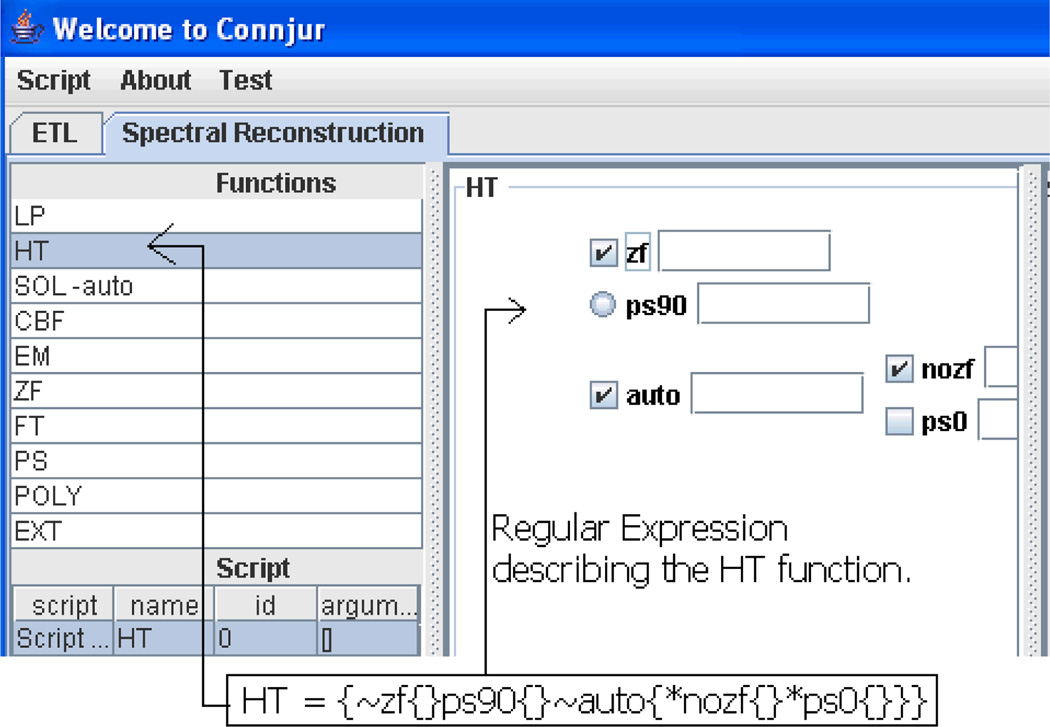
As the tool-oriented version of the CONNJUR application was being tested, it quickly became apparent that the application did not provide robust support for spectral analysis of data using NMRPipe. In particular, the user interface and data model supported only a highly oversimplified NMRPipe configuration process. NMRPipe has over 50 command-line functions each of which exhibits a variety of options, some of which may be individually parameterized. Some options depend on other options being activated, as shown in Figure 3. The prototype offered no support for such logic. The need to “merge” different processing schemes into one script for complicated spectral processing tasks was not addressed in this version, because it was not yet apparent. In addition, there were usability issues, such as the fact that the generated scripts were not “data-aware” - that is, they were not able to intelligently “reconfigure” parameterization according to the input data set.
Figure 3.
Data-oriented version of CONNJUR showing functionality for the Hilbert Transform with regular expressions guiding the function and parameter selection.
4. Data-Oriented CONNJUR
Development of the second version of the CONNJUR application started in September 2005 and was a modification of the tool-oriented version. The data-oriented version addressed the major drawback of the tool-oriented version by providing a more specific NMRPipe function configuration interface with accompanying changes in the database to support the relationships between specific functions and data in data sets. In this version, the experiment parameters for an individual NMRPipe function were automatically updated as functions were executed. The ability to automatically conform to such changes allowed for the creation of templates, i.e. generic combinations of processing functions which provide frequently reused functionality. The data-oriented version correctly modeled all of the functions and their parameters in the database using regular expressions. Having in-depth knowledge of NMRPipe and an understanding of regular expressions allowed Dr. Gryk to write all of the regular expressions which were used to provide guidance to the user in configuring the NMRPipe options. Figure 3 shows the configuring of the Hilbert Transform function and the underlying regular expression that supports the configuration.
The data-oriented version of the CONNJUR application provided the user with a more abstract view of the NMR experiment process. Instead of presenting the user with a full range of configuration options, this version allowed the user to configure their functions according to hierarchical pathways. Multiple templates could be combined for complex processing flows. Finally, the functions in this version were able to modify their parameterization depending on the input data set, which eased the user's burden of configuration. However, we recognized that as our understanding of the problem space became more clear, our user interface was growing in complexity. In the data-oriented version, individual NMRPipe functions were difficult to configure for anyone other than a domain expert, and then full blown complexity of the workflow was still not modelled. Just as the function options were dependent on one another, we learned that, ideally, the functions themselves might require higher logical ordering and branching.
5. Work-Flow-Oriented CONNJUR
The increase in complexity of the user interface in the data-oriented prototype of the CONNJUR application caused us to realize that we needed a greater understanding of the actual NMR experiment process. As a result, we spent the spring of 2006 using process modeling techniques to determine the workflow for a typical NMR experiment to determine a protein structure [8]. During the process of modeling the workflow, all group members including the domain expert gained a better understanding of the experiment process. In addition, we came to recognize how the codification of the experiment process could be used to make the process more rigorous as well as to instruct new NMR researchers about the process.
The workflow-oriented version of CONNJUR [9] solved the problem of user interface complexity by implementing an approach based on actors where individual functions were wrapped by actors. Each actor represents a domain-centric, object oriented implementation of a single function provided by a tool, which had unique execution and configuration properties.
The advantage to this approach is that “smart” actors can be built which have the capacity to guide the user through complex configuration tasks, automatically refactor themselves according to the data on which they are operating, and potentially even analyze or validate input data before running. In addition, the simple API for such actors would suggest that domain experts could easily write such actors and indeed, many actors were written by Dr. Gryk. The integration of actors and a workflow-oriented approach allowed users to be able to visualize their entire work process. The workflow oriented approach allowed for new functionality and coarse-grained, as well as fine-grained user configuration so that both novice and expert NMR researchers can accomplish their processing needs using a single, highly intuitive interface.
In addition to the use of actors, the workflow-oriented CONNJUR prototype also incorporated the Rowland NMR Toolkit [10,11], an alternative data processing tool similar to NMRPipe. The wrapping of two different tools supplied proof of concept of the interoperability provided by the CONNJUR application's layered, data driven architecture. It also allowed users to create heterogeneous workflows through a single user interface, introducing a new paradigm to the NMR data processing world.
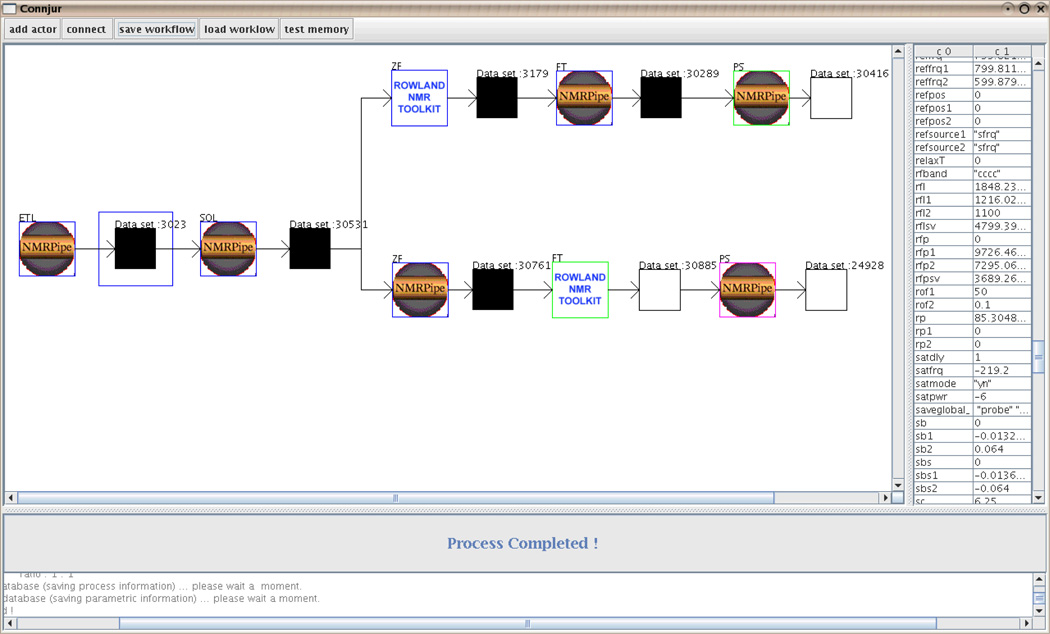
Figure 4 shows a typical NMR experiment workflow. Figure 4 shows branching workflows that contain actors for both the NMRPipe and Rowland NMR Toolkit tools. The workflow-oriented CONNJUR prototype supports a highly interactive interface where users can specify individual NMRPipe and Roland Toolkit functions as single atomic actions in a branched process flow.
Figure 4.
Workflow-oriented version of CONNJUR showing alternative branches of a typical data processing tree.
While the workflow-oriented CONNJUR prototype allowed the user much greater flexibility in creating NMR experiments, the combined effect of the actions carried out by the actors was not optimized. Execution of each actor individually resulted in a read and a write to disk of 1 MB or more for each actor, an unacceptable performance hit. In addition, the execution of each actor also required several mouse clicks by a user making the user interface unwieldy.
6. Composite-Actor CONNJUR
The current version of the CONNJUR application, the composite-actor version, solves the performance problem and simplifies modeling of workflows by allowing multiple actors to be combined into one larger, composite actor. A composite actor is an actor that is constructed of individual existing actors formed into a workflow. The workflow-based approach used in the workflow-oriented version of the CONNJUR application is maintained, and in addition a hierarchy of actors is supported.
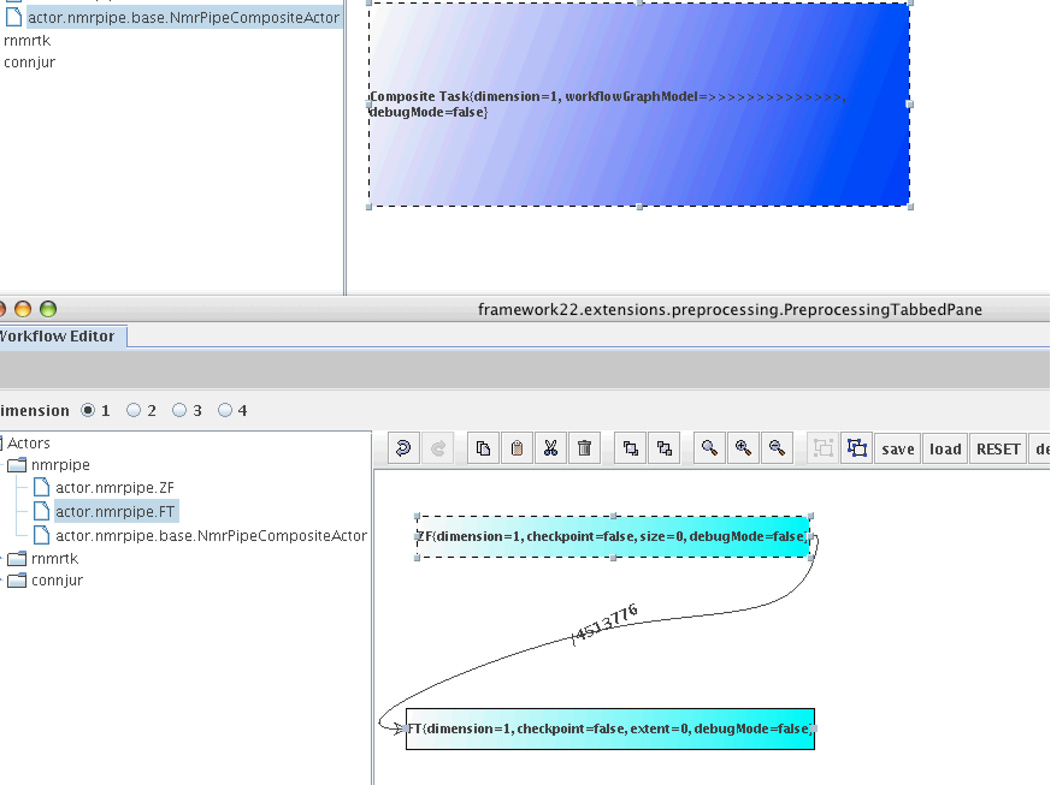
Figure 5 shows the construction of a composite actor which integrates two individual actors. The composite actor, NMRPipeCompositeActor, contains a zero fill actor followed by a Fourier transform actor (shown at the bottom of the figure).
Figure 5.
Composite-Actor Version of CONNJUR Showing Both Individual and Composite Actors.
The work-flow based approach used in the current version of the CONNJUR application has several benefits. First, the composite actor abstracts the underlying details of a lengthy and complex process from the workflow engine allowing users to design workflows in a platform independent, modular, and interactive fashion. Second, the combined actions of a composite actor can be optimized. For instance, one composite actor that we designed, the NMRPipe composite actor, strings together several NMRPipe atomic actors and the implementation of the CONNJUR application uses Unix pipes to optimize the data flow. A third benefit to our approach is that it is scalable. Atomic actor components are allowed to define parametric data about how a data set should be processed, while higher level composite actors are used to assemble atomic actors into optimized workflows which process data. As a result, this version is optimized for large workflows and supports higher resolution data such that the CONNJUR application will not take significantly longer to process data than the tools which it wraps.
7. From Prototype to Release
The summer of 2009 encompassed significant improvements as we revised our development pipeline, incorporating a greater degree of software engineering rigor. Build engineering, rigorous testing, and requirements analysis became core areas of focus. By differentiating research and engineering duties, we were able to confidently release our first software package (http://connjur.uchc.edu) to the general public. Unlike our previous, prototypical efforts, this application was designed primarily for public consumption and release, featuring robust testing protocols, code versioning, and automated build-release cycles. This release culminates an era of maturity in our engineering process which, in retrospect, will be essential for bringing our past prototypes to life.
8. Future Plans
Our aforementioned first release, like previous prototypes, is a utility which applies to the “Spectral Reconstruction” stage of the NMR workflow. In our current research efforts, in contrast, are continually spearheading a new wave of application prototypes which integrate data and applications in the later stages of this NMR structure calculation pipeline, such as “Analysis” and “Molecular Characterization” phases.
Consequently, our ability to differentiate “research” from “product” has been essential to our maturation as a team. We have adopted a two-pronged approach to software development : First, agile research development efforts produce prototypical software solutions, meanwhile, our engineering core follows behind, learning from research insights to produce robust, tested, software releases for public consumption.
This development culture should enable higher productivity, with new releases in the next few years, which we expect to be our most productive. We look forward to continue bridging research, engineering, and insight to ultimately enable a fully integrated, open-source environment for biomolecular NMR spectroscopy.
Acknowledgements
This research was funded by US National Institutes of Health grants EB-001496 and GM-083072. The authors wish to thank Gary Kesczewski for investigations of software architectures to support CONNJUR.
References
- 1.Baran MC, Huang YJ, Moseley HN, Montelione GT. Automated analysis of protein NMR assignments and structures. Chem Rev. 2004;vol 104:3541–3556. doi: 10.1021/cr030408p. [DOI] [PubMed] [Google Scholar]
- 2.Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;vol. 4:11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 3.Slupsky CM, Boyko RF, Booth VK, Sykes BD. Smartnotebook: a semi-automated approach to protein sequential NMR resonance assignments. J Biomol NMR. 2003;27:313–321. doi: 10.1023/a:1025870122182. [DOI] [PubMed] [Google Scholar]
- 4.Guntert P. Automated NMR protein structure calculation. Prog Nucl Magn Reson Spectrosc. 2003;vol. 43:105–125. [Google Scholar]
- 5.Fox-Erlich S, Martyn TO, Ellis HJC, Gryk MR. Delineation and Analysis of the Conceptual Data Model Implied by the IUPAC Recommendations for Biochemical Nomenclature. Protein Science. 2004;vol. 13:2559–2563. doi: 10.1110/ps.04810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis HJC, Fox-Erlich S, Martyn TO, Gryk MR. Development of an Integrated Framework for Protein Structure Determinations: A Logical Data Model for NMR Data Analysis. Proceedings of the Third International Conference on Information Technology : New Generations; ITNG; Apr. 2006; 2006. pp. 613–618.x. [Google Scholar]
- 7.Delaglio F, Grzesiek S, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;vol. 6(no. 3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 8.Verdi K, Ellis HJC, Gryk MR. Conceptual-level workflow modeling for NMR and other scientific experiments. BMC Bioinformatics. 2007 Jan.vol. 8(no. 31) doi: 10.1186/1471-2105-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gryk MR, Vyas J, Fox-Erlich S, Martyn TO, Ellis HJC. CONNJUR: An Open Source Integration Environment for Biomolecular NMR Data Analysis. 47th Experimental Nuclear Magnetic Resonance Conference; Pacific Grove; California. 2006. [Google Scholar]
- 10.Hoch JC, Stern AS. RNMR Toolkit. Version 3. 2005 [Google Scholar]
- 11.Rowland Institute at Harvard, Stern Allen. “Rowland NMR Toolkit Version 3”. [Online]. Available http://www.rowland.harvard.edu/rnmrtk/toolkit.htm. [Google Scholar]
- 12.Vyas Jay, Gryk Michael R, Schiller Martin R. VENN, a tool for titrating sequence conservation onto protein structures. Nucl. Acids Res. 2009 Jan.Vol. 37(no. 18) doi: 10.1093/nar/gkp616. [DOI] [PMC free article] [PubMed] [Google Scholar]