Abstract
Background and Aims
Although conservation biology has long focused on population dynamics and genetics, phenotypic plasticity is likely to play a significant role in population viability. Here, an investigation is made into the relative contribution of genetic diversity and phenotypic plasticity to the phenotypic variation in natural populations of Ranunculus nodiflorus, a rare annual plant inhabiting temporary puddles in the Fontainebleau forest (Paris region, France) and exhibiting metapopulation dynamics.
Methods
The genetic diversity and phenotypic plasticity of quantitative traits (morphological and fitness components) were measured in five populations, using a combination of field measurements, common garden experiments and genotyping at microsatellite loci.
Key Results
It is shown that populations exhibit almost undetectable genetic diversity at molecular markers, and that the variation in quantitative traits observed among populations is due to a high level of phenotypic plasticity. Despite the lack of genetic diversity, the natural population of R. nodiflorus exhibits large population sizes and does not appear threatened by extinction; this may be attributable to large phenotypic plasticity, enabling the production of numerous seeds under a wide range of environmental conditions.
Conclusions
Efficient conservation of the populations can only be based on habitat management, to favour the maintenance of microenvironmental variation and the resulting strong phenotypic plasticity. In contrast, classical actions aiming to improve genetic diversity are useless in the present case.
Key words: Metapopulation, Ranunculus nodiflorus, phenotypic plasticity, quantitative trait, genetic diversity, conservation
INTRODUCTION
The genetics of endangered species is of great interest to both evolutionary and conservation biologists (Avise and Hamrick, 1996; Young and Clarke, 2000; Hedrick, 2001; Reed and Frankham, 2003; Ellis et al., 2006), notably because the ability of a species to adapt to environmental changes (adaptive potential) is known to depend greatly on the genetic diversity available for traits subjected, or potentially subjected, to selection. Hence, the World Conservation Union (IUCN) has recommended the conservation of genetic diversity as one of three levels of biodiversity (McNeely et al., 1990). However, individuals can also respond plastically to environmental variations, and phenotypic plasticity should be considered a (potentially) major evolutionary force, especially in plants, which cannot escape their local environment. Phenotypic plasticity is sometimes adaptive (e.g. Thompson, 1991; Schmitt, 1993; Schmitt et al., 1995, 1999; Dudley and Schmitt, 1996; Price et al., 2003; Pigliucci et al., 2006; Valladares et al., 2006) and can be of great importance for conservation biology and conservation programmes, as it affects plant fitness and consequently population dynamics. For example, reintroduction or reinforcement programmes rely on the assessment of genetic variability among and within populations and aim to maximize genetic diversity in restored populations. A lot of effort is specifically put into finding an appropriate balance between inbreeding and outbreeding depression (Avise, 1992; Ellstrand, 1992; Fenster and Dudash, 2000), but large phenotypic plasticity could compensate local maladaptation or low genetic variability. Assessing the magnitude of phenotypic plasticity and its impact on plant fitness (Booy et al., 2000) could help design management programmes based on the control and/or manipulation of abiotic factors influencing the plastic response of individuals and their reproductive success.
Measuring the environmental and genetic diversity of (potentially) selected traits is, however, not straightforward. Some studies suggest that the diversity of molecular markers, e.g. microsatellites, could be a good indicator of the adaptive potential (e.g. Garten, 1976; Soulé and Wilcox, 1980; Beardmore, 1983; Allendorf and Leary, 1986; Houle, 1989; Merilä and Crnokrak, 2001), and conservation studies often infer adaptive potential from neutral molecular markers data (Hedrick, 2001). However, the relevance of this approach has recently been questioned (e.g. Hedrick and Miller, 1992; McKay et al., 2001; Reed and Frankham, 2001), notably because the association between neutral diversity and population fitness is highly variable across studies [e.g. Booy et al. (2000), Arden and Lambert (1997) and Calero et al. (1999) found no decreased fitness or lower population viability in populations with lower levels of genetic variation]. On average, the observed correlation between neutral and selected genetic variability remains, at best, weak (Reed and Frankham, 2001). In addition, molecular marker data carry no information regarding phenotypic plasticity. Hence, although genetic diversity and differentiation of neutral markers provide useful information on the demographic events that occurred in populations (bottlenecks, rapid expansion, migration events, etc.), estimation of adaptive potential and construction of reinforcement programmes should not rely exclusively on such markers, but also consider directly traits submitted to selection.
Most traits relevant to adaptation are quantitative traits that are coded by numerous, generally unidentified genes and are influenced by the environment. When some of the relevant genes are known, their genetic diversity can be measured directly by detection of quantitative trait loci (QTL), real time PCR or microarrays (e.g. Storfer, 1996; Kearsey and Farquhar, 1998; Korol et al., 1998; Gibson, 2002; Frary et al., 2003; Thomas and Klaper, 2004; Whitehead and Crawford, 2006). However, this remains difficult in wild species, particularly endangered ones (Falconer and McKay, 1989; Lynch and Walsh, 1998), as generally little is known regarding the genetic architecture of their traits. Thus, conservation biologists still depend upon quantitative genetics approaches to study phenotypic variation of endangered species and design efficient management programmes. Assessing the genetic and environmental parts of variation of morphological and fitness traits is a tedious task, as it requires knowledge of kinship and/or common garden experiments, but, as of now, it is a necessary step to study phenotypic plasticity.
Here, the respective roles of genetic diversity and phenotypic plasticity in the spatial differentiation of morphological and fitness traits of a rare endangered species, Ranunculus nodiflorus, are investigated and the consequences in terms of conservation are explored. The populations of this annual, essentially autogamous, species exhibit metapopulation dynamics and occur in a heterogeneous environment at a fine scale, both through space and time. In a previous study, significant differentiation of quantitative traits among natural (sub)populations of the Fontainebleau forest, France was shown (Noel et al., 2006). Here, the contributions of genetic and environmental components to the variability of morphological and reproductive traits are assessed by combining a common garden experiment and a molecular analysis using microsatellite markers. The following questions are addressed: (a) How is the variation of morphological characters and reproductive success of individuals spatially distributed among and within populations of the Fontainebleau forest? (b) What are the respective roles of genetic diversity and phenotypic plasticity in this spatial variation? (c) How can such information be used for the conservation of this species, in the Fontainebleau forest and at a national scale?
MATERIALS AND METHODS
Description of the metapopulation
Study species
Ranunculus nodiflorus L. (Ranunculaceae) is a rare and endangered annual plant inhabiting wet zones in Spain, Portugal and France. It has undergone a strong decline, caused by the drainage of wetlands and the reduction in grazing, during the last century (Danton and Baffray, 1995) and now appears as ‘vulnerable’ (VU) on the French Red List of threatened species (Olivier et al., 1995). The species has strict habitat requirements, growing only in puddles with thick soil and highly variable water levels. According to Kirchner et al. (2003), it reproduces mainly by selfing and produces small achenes. Ranunculus nodiflorus seedlings emerge biseasonally, in fall and spring (Noel et al., 2006). Flowering and seed production occur between April and May. The plants die rapidly after the last achene is mature. Ranunculus nodiflorus is unable to reproduce vegetatively and apomixis is unlikely (F. Noel et al., unpubl. res.).
Study area
The study was carried out in the Fontainebleau forest, 50 km south of Paris, in five sites where R. nodiflorus was previously observed (for details, see Noel et al., 2006): Coquibus (site 1), Meun (site 2), Couleuvreux (site 3), De Oliviera (site 4) and Belle-Croix (site 5). Each site is a ‘platière’, characterized by a sandstone ground and the occurrence of temporary puddles during rainy periods. Water levels, which depend chiefly on rainfall, are highly variable, so that puddles can transitorily be connected by water corridors. Previously each puddle occupied by the species as a population of R. nodiflorus had been considered. Networks were defined as a set of potentially connected puddles, i.e. puddles occasionally connected to one another via water corridors. Thus, in the terminology of metapopulation dynamics, a puddle corresponds to a deme and a network to a metapopulation. The site sizes vary from just only one puddle (one puddle in sites 3 and 4; three puddles in site 5) to around 200 puddles (sites 1 and 2); puddles may contain up to several thousands of individuals.
In all five sites, the respective roles of genetics and environment in total phenotypic variation of vegetative and reproductive traits were evaluated by conducting two series of measurements, in situ in two different years, and ex situ in a common garden.
In situ measurement of vegetative and reproductive traits
In situ measurements were conducted in 2004 and 2005 on individuals from 21 puddles: seven puddles from site 1 (3 networks), 11 from site 2 (5 networks), one from site 3 (1 network), one from site 4 (1 network) and one from site 5 (1 network). Puddles were chosen in such a way that (a) each site was sampled, (b) in sites 1 and 2, containing numerous puddles, isolated and connected puddles were sampled. Within these classes, puddles were sampled at random. One fixed square quadrat (30 cm × 30 cm) per sampled puddle was placed. Each quadrat was subdivided into 15 cells (each 6 cm × 5 cm).
Each year, 15 individuals per puddle were measured when available. If a puddle contained fewer than 15 individuals, all individuals were measured (e.g. only four individuals for the entire site 4). These 15 plants were sampled by generally measuring one plant, chosen at random, per cell, or, when some cells were empty, by randomly choosing additional plants in cells containing more than one individual. When quadrats contained fewer than 15 individuals, the sample was completed by measuring the closest individuals outside the quadrat [this occurred in 2005 for five quadrats only (24 %) and a total of 28 individuals (9·7 %)]. Measurements were made between 17 May and 20 May 2004 on adult plants. For each plant, the maximum height (Hind), stem diameter at soil level (Diam), number of leaves (Nleav), number of flowers (Nflw), number of achenes (Nak) and number of seeds per achene (Ns/a) were measured: the latter two traits yielded the number of seeds per individual (s/I). The number of individuals per quadrat was also recorded (dens). When possible, at least one achene per individual was collected in every quadrat. These seeds were used for the common garden experiment (see below). Finally, a leaf sample on each individual measured was collected for genetic analyses (see below).
The same measurements were performed on the same number of individuals (15 per quadrat) and in the same puddles between 24 May and 26 May 2005. Individuals measured can be considered as offspring of plants measured in 2004, although the soil seed bank has been shown to be a potential, though limited, source of new individuals in these populations (Noel et al., 2006).
Environmental data
To evaluate possible environmental variation between years among puddles, some characteristics of puddles were also recorded in May 2004 and May 2005: presence of water in puddles when plant measurements were conducted (water = humid or dried), presence (connect) and number (Ncorr) of potential corridors converging to a puddle, sunlight intensity (Sun = high, low and medium) describing canopy cover, ground vegetation cover (Veget = high, low and medium density), and indication of disturbance such as animal or human tracks (Tracks = 0 or 1).
Common garden experiment
A total of 60 seeds per quadrat was collected on the individuals measured in 2004 and were used for the common garden experiment. As the aim was to study population differentiation, all seeds from a given quadrat were pooled. Seeds were sown in pots 7 cm in diameter, containing one-quarter of Fontainebleau sand and three-quarters of mould, at two different densities: five seeds per pot (three replicates) and 15 seeds per pot (three replicates). The 126 pots were placed in a common environment (around 2 m2 of total space) in the gardens of the Conservatoire Botanique National du Bassin Parisien (CBNBP) in Paris. Pots were randomized once a week and watered regularly. Germination emergence was recorded once a week. The same traits as those recorded in situ were measured twice during the common garden experiment (between 16 June and 17 June and between 27 June and 28 June). These two records were performed to limit the effects of phenology differences when comparing in situ and ex situ trait values of individuals.
Genetic data
Seven microsatellite loci were previously isolated from R. nodiflorus (Noel et al., 2005). Preliminary analyses on 22 individuals showed no polymorphism in R. nodiflorus (for details, see Noel et al., 2005) but appreciable polymorphism in R. flammula, a closely related species, with a number of alleles per locus ranging from 2 to 7 and observed heterozygosities ranging from 0 to 0·261 (27 individuals tested). Here, these seven microsatellite loci were analysed on a total of 137 individuals (113 and 24 plants sampled in 2003 and 2004, respectively). The 113 individuals sampled in 2003 can be considered a representative sample of all puddles occupied in the five sites studied, with at least one individual per puddle (32 plants from 28 quadrats in site 1, 40 plants from 28 quadrats in site 2, 19 plants from one quadrat in site 3, four plants from one quadrat in site 4 and 18 plants from three quadrats in site 5). In 2004, one measured individual was sampled per quadrat, except in three quadrats where two individuals were sampled (i.e. a total of 21 + 3 individuals). The description of the protocol used for isolation and characterization of microsatellite markers is given in Noel et al. (2005).
Statistical analysis
Statistical analyses were performed with JMP.5·0·1·2, SAS 9·1 (SAS Institute Inc., Cary, NC, USA) and R2·1·1 (R Core Development Team, http://www.r-project.org./index.html).
Vegetative and reproductive traits analysis
To discriminate between the genetic and environmental contributions to the phenotypic variation of vegetative and reproductive traits observed, the ‘2004 in situ’, ‘2005 in situ’ and ‘2005 garden’ data (referred to as ‘experiment’) were compared via an analysis of variance on log-transformed trait values (to meet normality requirements), using the following model:
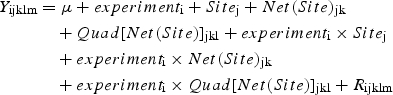 |
where Y is the log-transformed value of a trait (Nleav, Nflw, Nak, Diam, Hind and s/I), μ is the mean trait value, experiment is the effect of experimental set-up (i = 1, 2 or 3; three conditions under study, i.e. ‘2004 field’, ‘2005 field’ and ‘2005 garden’), Site is the site effect (j = 1…5; five sites under study), Net is the network effect (k = 1…5; up to five networks per site), Quad is the quadrat effect (l = 1…3; up to three quadrats per network) and R is an error term. All effects and their interactions were considered random, except the ‘experiment’ effect. The ‘Site’ effect was tested using the ‘Network’ mean square as the residual and the ‘Network’ effect was tested using the ‘Quadrat’ mean square as the residual. Because multiple tests were used to analyse phenotypic variation (one per trait), the level of significance was corrected using the Simes method (Simes, 1986), a Bonferroni-like method that is less conservative and more powerful than the classical Bonferroni procedure (Simes, 1986, Sarkar and Chang, 1997). After ordering the n P-values [P(1) ≤ … ≤ P(n)], the hypothesis H0 was rejected at the level of significance α if P(i) ≤ iα/n for at least one i.
To analyse each experiment in more detail, the effects of ‘Site’, ‘Quadrat’ and ‘Network’ were tested using a nested ANOVA for each year and site, with the following model:
where Y is the log-transformed value of a trait [Nleav, Nflw, Nak, Diam, Hind and s/I; for the 2005 garden data, the percentage of germination (%Ger.) was also tested]. Other abbreviations are as above.
Environmental data and correlation with plant traits
To test for a difference in environmental conditions for all the quadrats between 2004 and 2005, chi-square tests (for categorical variables, i.e. water, connect, Veget, Sun and Tracks) or Student's t-tests (for numerical variables, i.e. Dens and Ncorr) were performed to detect a possible ‘year’ effect (JMP.5·0·1·2).
In addition, an analysis of variance was performed on in situ data to test for possible effects of each environmental variable taken separately on morphological and fitness traits with the following model:
where Y is the log-transformed value of a trait (Nleav, Nflw, Nak, Diam, Hind and s/I), Env is an environmental variable (‘connect’, ‘Veget’, ‘Sun’, ‘Tracks’, ‘water’, ‘Ncorr’ and ‘dens’), Year is the time variable (‘2004’ and ‘2005’) and other abbreviations are as above. When the environmental variable was numerical (i.e. ‘Ncorr’ and ‘Dens’), the ‘Quadrat’ effect was not nested in the ‘Environment’ effect. Significance levels were again corrected using the Simes method.
Finally, density dependence and accumulation of deleterious mutations were explored in isolated puddles by testing the effect of density, connection status (isolated or connected), and their interaction on the probability of germination for the seeds sown in common garden. To achieve this, a mixed effects logistic regression (glmmPQL function in R, with a binomial error and a logit link) was performed with the model:
where connect is the connection status, dens is plant density in the common garden experiment, and other variables are as above. The effect of quadrat is considered random.
RESULTS
Vegetative and reproductive data
The total number of plants measured varied across years (315 in 2004 and 289 in 2005), due to small population sizes (<15 individuals) of some puddles and the extinction of one puddle in 2005. In the common garden experiment in 2005, 407 plants were measured. The mean and standard deviation over all sites are summarized in Fig. 1 for each trait.
Fig. 1.
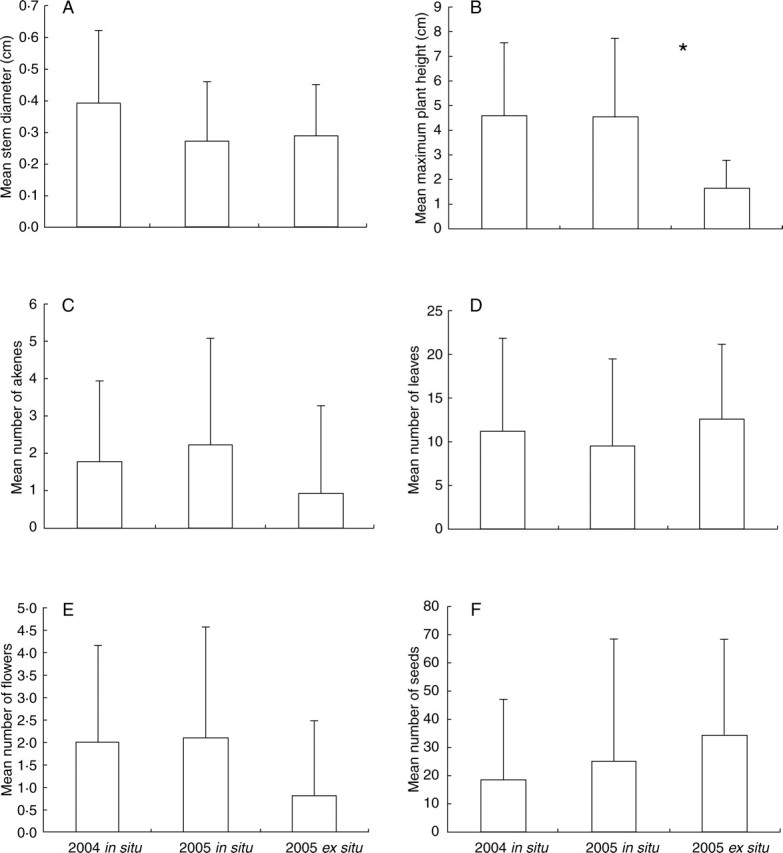
Mean and standard deviation of individual vegetative and reproductive traits in the three experimental set-ups: (A) stem diameter (cm); (B) maximum plant height (cm) (the asterisk indicates significant difference between ‘in situ’ and ‘ex situ’); (C) number of leaves; (D) number of achenes; (E) number of flowers; (F) number of seeds per individual.
In a comparison of the three experiments, vegetative and reproductive traits were highly variable within each experimental set-up, thus yielding no clear differences among the three experiments (Fig. 1 and Table 1). Only plant height exhibited significant variation across experimental set-ups (Table 1), with much smaller plants in the garden experiment than in the field (Fig. 1, asterisk). Although no other single effect was significant, the quadrat effect was highly variable across treatments for almost all traits (significant Quadrat × experiment effects for all but one trait; Table 1).
Table 1.
Results of an ANOVA on the full dataset (all three experimental set-ups)
| experiment | Site | Network (site) | Quadrat (network, site) | experiment × site | experiment × network (site) | experiment × quadrat (network, site) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | F | P > F | F | P > F | F | P > F | F | P > F | F | P > F | F | P > F | F | P > F |
| Nleav | 2·54 | 0·1392 | 0·42 | 0·7906 | 1·01 | 0·4892 | 1·85 | 0·1192 | 2·44 | 0·0412 | 0·91 | 0·5539 | 5·50 | <0·0001 |
| Nflw | 0·81 | 0·4730 | 1·18 | 0·3807 | 2·53 | 0·1046 | 1·79 | 0·1190 | 1·70 | 0·1534 | 1·20 | 0·3496 | 1·86 | 0·0144 |
| Nak | 0·46 | 0·6555 | 0·38 | 0·8204 | 0·68 | 0·6701 | 2·28 | 0·502 | 2·92 | 0·0371 | 1·22 | 0·3418 | 2·65 | 0·0005 |
| Hind | 17·93 | 0·0011 | 2·99 | 0·0728 | 1·20 | 0·3893 | 1·80 | 0·1290 | 3·42 | 0·0103 | 1·29 | 0·3012 | 5·78 | <0·0001 |
| Diam | 7·59 | 0·0137 | 0·96 | 0·4785 | 0·88 | 0·5453 | 1·68 | 0·1594 | 1·88 | 0·1145 | 1·33 | 0·2796 | 3·87 | <0·0001 |
| s/I | 1·02 | 0·4230 | 1·31 | 0·3364 | 0·71 | 0·6508 | 2·17 | 0·780 | 1·68 | 0·1812 | 0·96 | 0·5116 | 3·17 | <0·0001 |
Traits are abbreviated as follows: Diam, stem diameter; Height, maximum height of individuals; Nleav, number of leaves; Nak, number of akenes; Nflw, number of flowers; s/I, total number of seeds per individual.
The significant P-values after sequential Simes corrections at the significance level α (5 %) are in bold.
Results of the ANOVA on each dataset separately showed that, in 2004, most phenotypic variation observed in the field was due to differences among quadrats (Table 2). In addition, plant height also exhibited significant differentiation among sites. These patterns remained relatively unchanged during the second year of field measurements, with the exception of more widespread quadrat effects, and no significant site effect for plant height (Table 2). These results were consistent with previous measures performed in 2003 (Noel et al., 2006). In contrast, the individuals grown in a common garden environment did not display such a differentiation among quadrats or sites of origin: a significant quadrat effect was only detected on plant height (Table 2). Finally, the germination percentage observed in the common garden showed significant differences among quadrats of origin (Table 2).
Table 2.
Results of an ANOVA on each experimental set-up
| Year/experiment | Effects | ||||||
|---|---|---|---|---|---|---|---|
| Site | Network (site) | Quadrat (network, site) | |||||
| Variables | F | P > F | F | P > F | F | P > F | |
| 2004/in situ | Nleav | 3·31 | 0·0714 | 3·84 | 0·0299 | 1·89 | 0·0458 |
| Nflw | 2·60 | 0·0984 | 1·51 | 0·2700 | 3·03 | 0·0012 | |
| Nak | 1·35 | 0·3274 | 2·86 | 0·0713 | 3·45 | 0·0006 | |
| Hind | 17·51 | <0·0001 | 0·83 | 0·5697 | 8·22 | <0·0001 | |
| Diam | 5·66 | 0·0185 | 3·63 | 0·0355 | 1·66 | 0·0903 | |
| s/I | 2·59 | 0·1106 | 2·65 | 0·0830 | 3·53 | 0·0004 | |
| 2005/in situ | Nleav | 1·33 | 0·3064 | 0·59 | 0·7334 | 16·84 | <0·0001 |
| Nflw | 1·57 | 0·2594 | 1·75 | 0·2180 | 5·66 | <0·0001 | |
| Nak | 1·99 | 0·1936 | 2·07 | 0·1801 | 7·19 | <0·0001 | |
| Hind | 3·98 | 0·0337 | 1·32 | 0·3404 | 14·17 | <0·0001 | |
| Diam | 1·26 | 0·3367 | 0·86 | 0·5595 | 10·43 | <0·0001 | |
| s/I | 1·63 | 0·2530 | 1·50 | 0·3001 | 8·72 | <0·0001 | |
| 2005/ex situ | Nleav | 3·36 | 0·0379 | 0·63 | 0·7039 | 1·72 | 0·0737 |
| Nflw | 0·37 | 0·8265 | 1·72 | 0·1735 | 1·21 | 0·2947 | |
| Nak | 1·82 | 0·3675 | 0·67 | 0·6503 | 1·07 | 0·3963 | |
| Hind | 0·59 | 0·6796 | 2·24 | 0·1181 | 2·53 | 0·0059 | |
| Diam | 1·92 | 0·1787 | 0·92 | 0·5130 | 1·77 | 0·0638 | |
| s/I | 1·34 | 0·5614 | 0·29 | 0·9120 | 1·28 | 0·2685 | |
| %Ger | 2·44 | 0·0908 | 0·29 | 0·9308 | 2·99 | 0·0026 | |
Traits are abbreviated as follows: Diam, stem diameter; Height, maximum height of individuals; Nleav, number of leaves; Nak, number of achenes; Nflw, number of flowers; s/I, total number of seeds per individual and %Ger, percentage of germination per pot.
The significant P-values after sequential Simes corrections at the significance level α (5 %) are in bold.
Environmental data
Minor differences were detected across years for presence of water, connection status, number of potential corridors converging on the puddle, sunlight intensity, ground vegetation cover and disturbance status, but none of these were significant (Table 3). A significant difference in plant density was observed in the field between 2004 and 2005, with lower densities in 2005 (Student's t-test t = 3·403, P-value = 0·021).
Table 3.
Distribution of environmental variables across puddles in 2004 and 2005
| Environmental variables | 2004 | 2005 | Difference | ||||
|---|---|---|---|---|---|---|---|
| % connected puddles | 0·71 | 0·71 | n.s. | ||||
| % puddles with tracks | 0·76 | 0·80 | n.s. | ||||
| % dried puddles | 0·57 | 0·50 | n.s. | ||||
| Vegetation cover† | 0·43 | 0·52 | 0·05 | 0·55 | 0·40 | 0·05 | n.s. |
| Sunlight intensity‡ | 0 | 0·33 | 0·67 | 0·05 | 0·60 | 0·35 | n.s. |
| Mean number of connections§ | 3·8 ± 2·8 | 2·2 ± 0·4 | n.s. | ||||
| Mean plant density | 97·4 ± 73·2 | 52 ± 48·7 | * |
The environmental variables are the connection status (% of connected puddles), indication of animal or human disturbance (% of puddles with tracks), presence of water (% of dried puddles), ground vegetation cover, sunlight intensity, mean number of connections and mean plant density per quadrat (the asterisk indicates a significant difference between the 2 years, P < 0·05).
In 2005, one puddle went extinct and environmental data were not recorded.
† Distribution of vegetation cover across puddles, with three classes: low, medium and high.
‡ Distribution of sunlight intensity across puddles, with three classes: low, medium and high.
§ For connected puddles only.
None of the environmental variables measured had a significant effect on phenotypic trait means (results not shown), with a single exception: plants were taller under lower density (F = 6·68, corrected P-value = 0·01). The environmental effect was, however, variable across years for a number of traits, as indicated by significant year × env interactions (Veget and Nleav, Nak, Nflw, s/I; Sun and Hind; Tracks and Diam, Hind; water and Diam). In these analyses incorporating environmental variables, the quadrat effect remained highly significant for all traits (P < 0·0001)
Germination percentage in common garden
On average, 42 (±26) % of seeds germinated in the common garden experiment. The large differences in germination rates observed among quadrats (Table 2) were not explained by plant density or connection status of the puddles of origin (logistic regression, non-significant effects).
Genetic data
The genetic analysis of the 137 individuals showed a strictly identical genetic profile. All individuals exhibited a homozygous pattern at all seven microsatellite loci. No genetic diversity was detected at these molecular markers in the Parisian populations of R. nodiflorus.
DISCUSSION
Variation in morphological and fitness traits across puddles is mostly attributable to phenotypic plasticity
Measures of morphological characters (vegetative and reproductive traits) were performed on a large number of individuals of five populations of Ranunculus nodiflorus in Fontainebleau forest. The results showed a high variability of these traits among puddles in the field during the two years of the experiment.
This among-puddle variability in quantitative traits appeared to have little – if any – genetic component, as supported by the following observations: (a) the quadrat effect was highly significant in the field, but this effect was highly variable across the three experiments for almost all traits (Table 1), due to differences both between 2004 and 2005 in situ experiments and between in situ and ex situ 2005 experiments (not shown). This suggests that the observed variation in traits across puddles is due to local environmental variations rather than to genetic effects, which are expected to be more stable from one generation to the other; (b) almost all significant quadrat effects disappeared when individuals were grown in a common garden, with more homogeneous environmental conditions. Hence, most of the spatial differentiation of morphological and fitness traits observed in the field can be attributable to strong phenotypic plasticity for the majority of traits. Only for plant height and percentage of seed germination, did the quadrat effect remain significant in the common garden. For these traits, genetic effects or parental effects (i.e. differences arising from differences in the parental environment) could not be excluded (Andalo et al., 1999).
Despite the presumed strong phenotypic plasticity of morphological and fitness traits, differences of the quadrat effects between years (2004 and 2005 in situ experiments) were not apparently related to environmental changes. First, no among-quadrat differences could be detected for the environmental parameters measured during the two years of study (Table 3). In addition, there was generally no strong relationship between in situ trait means and environmental variables measured in the field (with the exception of a significant effect of plant density on plant height). Specifically, the quadrat effect remained highly significant despite incorporation of environmental effects into statistical analyses, i.e. the variability observed among quadrats for the morphological and fitness traits was not explained by the variability of the environmental variables measured. These unexpected results suggest that environmental measures may not have been performed at a relevant scale and that phenotypic plasticity is likely controlled by random environmental microvariations, both in space and time. This is supported the significant experiment × quadrat interaction for almost all traits (Table 1): although differences among quadrats were observed in situ in both years, mean trait values within quadrat varied across years, indicating fluctuating sources of variation.
Absence of or weak genetic differentiation among plant populations is often explained by (a) high migration rates that homogenize populations and prevent differentiation, (b) homogenizing selection or (c) low levels of genetic variability at the metapopulation level (Slatkin, 1987; Lande, 1992; Ellstrand and Elam, 1993; Linhart and Grant, 1996; Giles and Goudet, 1997). For R. nodiflorus, the latter hypothesis is supported by microsatellite data showing no polymorphism within and among populations of the Fontainebleau forest, although one should take into account that (a) neutral and selected genetic diversity are generally uncorrelated (Reed and Frankham, 2001) and (b) a relatively low number of loci was surveyed. As a consequence, differences among quadrats for plant height and germination percentage observed in the common garden are likely to be due to environmental parental effects, although genetic differences cannot be ruled out completely.
An attempt was made to explore further the potential influence of genetics on germination rate by examining the influence of isolation on germination. The differences in germination rate, observed in the common garden, among quadrats of origin, were not explained by quadrat connectivity or plant density, i.e. individuals from isolated or dense populations did not exhibit a significantly lower germination rate, a proxy for seed quality. This test was initially performed to detect a possible stronger inbreeding depression or accumulation of deleterious mutations in isolated puddles, which also exhibited the lower density populations (Noel et al, 2006). The present results suggest no differences in seed quality (and hence, fitness) between isolated versus connected puddles, regardless of seed density, which is consistent with the presumed low levels of genetic variation in the populations.
No genetic diversity in an endangered plant species
Variation in morphological characters appears to be mainly controlled by strong phenotypic plasticity and/or parental effects in the Parisian populations of R. nodiflorus. The within-population genetic diversity also appeared to be low; for the seven microsatellite markers typed in 137 individuals, absolutely no polymorphism was observed. These molecular data have shown a complete lack of genetic diversity for R. nodiflorus, a situation sometimes observed in plant species, especially endangered species (Arden and Lambert, 1997; Calero et al., 1999; Ibáñez et al., 1999). However, in the few studies mentioning monomorphic populations (Calero et al., 1999; Ibáñez et al., 1999; Meunier et al., 2001), monomorphism was restricted to the population scale, and genetic polymorphism was generally observed between populations. The complete absence of polymorphism observed here thus deserves further discussion, especially because (a) microsatellites were developed specifically from R. nodiflorus and (b) some of the Parisian populations have a large population size (e.g. site 1 contains around 100 000 individuals).
The first obvious explanation for the lack of genetic variation is a technical problem with molecular markers. Short (with few repetitions), interrupted or complex microsatellites, such as those isolated from R. nodiflorus (Noel et al., 2005), are known to be less variable than regular, long microsatellites (Primmer et al., 1996; Brinkmann et al., 1998). This potential technical problem is supported by the observation that populations from other regions (Corsica, Massif Central and Brittany for a total of 52 individuals from ten populations) showed the same complete absence of genetic diversity (not shown). Thus, no polymorphism was detected at a national scale (550 000 km2). Nevertheless, the same microsatellite markers analysed in a closely related species of R. nodiflorus, R. flammula, showed some genetic diversity among individuals; all markers were polymorphic with a number of alleles per locus varying from two to seven and observed heterozygosities ranging from 0 to 0·261 (Noel et al., 2005).
The complete absence of polymorphism in the Fontainebleau populations, some of which exhibit large population sizes, could appear unusual. Genetic diversity is nevertheless not systematically correlated with population size (Podolsky, 2001). Moreover, the mating system of R. nodiflorus (predominant selfing), its metapopulation dynamics and its limited number of populations could also be responsible for a low polymorphism. In selfing species, metapopulation dynamics could reinforce the genetic homogenization of individuals within populations (Amos and Harwood, 1998; Ingvarsson, 2002). In addition, recurrent extinctions and (re)colonizations could result in a strong decline in genetic diversity, especially for species where pollen migration is almost absent, such as R. nodiflorus. Thus, (re)colonization occurs only by seed migrations from patches of surrounding areas. Preliminary results on random amplified polymorphic loci (RAPD markers) between two individuals, one from the Parisian region and the other from Corsica, showed that a single marker among 20 exhibited a different pattern between the two individuals tested. The lack of polymorphism within R. nodiflorus thus seems to affect the whole genome. Nevertheless, more individuals and more RAPD primers should be analysed to confirm this result.
Consequences for the conservation of R. nodiflorus
Although conservation biology has long focused on demographic and genetic issues, the potential role of phenotypic plasticity has recently been considered for the management of endangered populations. Phenotypic plasticity is generally regarded as detrimental in conservation, notably because translocations (e.g. from captivity or gardens to natural populations, or between natural populations for reinforcement) expose individuals to new environments, and the changes in phenotype triggered by plasticity may decrease the survival or reproductive success of individuals (e.g. Lema and Nevitt, 2006). The potential beneficial role of phenotypic plasticity on population viability is, however, also recognized, as it creates phenotypic variation and therefore increases the probability that a population will persist in the face of environmental change (Watters et al., 2003). In R. nodiflorus, the high degree of phenotypic plasticity observed is likely to provide the opportunity for population persistence despite limited genetic variation. The phenotypic variation observed in natural populations of the Fontainebleau forest does not correspond solely to a response to poor environmental conditions, yielding a variety of unhealthy phenotypes: in fact, plants in natural populations tend to be taller and produce more flowers and achenes than plants grown in the common garden environment (Fig. 1), an a priori favourable environment with abundant nutrients and water. Overall, the absence of genetic diversity seems to be correctly compensated by phenotypic plasticity, enabling seed production despite sometimes severe environmental changes (e.g. major drought in summer 2003; Rebetez et al., 2006). Although the species is endangered at a national scale, local populations have large population sizes and demographic surveys suggest that they are not threatened in the immediate future (Noel et al., 2006).
In the present situation, genetics should not play a major role in the designing of management plans for R. nodiflorus. Despite the highly significant morphological differentiation among populations, transplantations among populations would be useless, as they would bring little, if any, genetic novelty to a declining population. Instead of maximizing genetic diversity, the most appropriate measure to enhance the viability of populations on longer time scales is manipulating the environment to help maintain an appreciable level of phenotypic plasticity (‘phenotypic management’; Watters et al., 2003). This requires first to preserve the habitat of R. nodiflorus via regular clearings, but also to maintain or even increase microenvironmental variation, which can be achieved, for example, by increasing the number of puddles occupied by the species. However, although phenotypic plasticity may favour the viability of R. nodiflorus in the short term by allowing the production of numerous seeds under a wide range of environmental conditions, the species survival may be compromised in the long term, as phenotypic plasticity without genetic variation is not sufficient to face large environmental changes (Potvin and Tousignant, 1996; Réale et al., 2003).
ACKNOWLEDGEMENTS
We thank G. Garcia, R. Masini, C. Griveau, A. Labat, J. Besco and P. Lherminier for their help during experimentation in garden and in the field, the Office National des Forêts for facilitating our work in the Fontainebleau forest, the Conservatoire Botanique National du Bassin Parisien for financial support and two anonymous reviewers for their helpful comments.
LITERATURE CITED
- Allendorf FW, Leary RF. Heterozygosity and fitness in natural populations of animals. In: Soulé ME, editor. Conservation biology: the science of scarcity and diversity. Sunderland, MA: Sinauer Associates; 1986. pp. 57–76. [Google Scholar]
- Amos W, Harwood J. Factors affecting levels of genetic diversity in natural populations. Philosophical Transactions of the Royal Society London B. 1998;353:177–186. doi: 10.1098/rstb.1998.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andalo C, Mazer SJ, Godelle B, Machon N. Parental environmental effects on life history traits in Arabidopsis thaliana (Brassicaceae) New Phytologist. 1999;142:173–184. [Google Scholar]
- Arden SL, Lambert DM. Is the black robin in genetic peril? Molecular Ecology. 1997;6:21–28. [Google Scholar]
- Avise JC. Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos. 1992;63:62–76. [Google Scholar]
- Avise JC, Hamrick JL. Conservation genetics: case histories from nature. New York, NY: Chapman and Hall; 1996. [Google Scholar]
- Beardmore J. Extinction, survival and genetic variation. In: Schonewald-Cox CM, Chambers SM, MacBryde S, Thomas L, editors. Genetics and conservation: a reference for managing wild animal and plant populations. Menlo Park, CA: Benjamin/Cummings; 1983. pp. 125–151. [Google Scholar]
- Booy D, Hendriks RJJ, Smulders MJM, Van Groenendael JM, Vosman B. Genetic diversity and the survival of the populations. Plant Biology Stuttgart. 2000;2:379–395. [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. American Journal of Human Genetics. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero C, Ibáñez O, Mayol M, Rosselló JA. Random amplified polymorphic DNA (RAPD) markers detect a single phenotype in Lysimachia minoricensis J.J.Rodr. (Primulaceae), a wild extinct plant. Molecular Ecology. 1999;8:2133–2136. doi: 10.1046/j.1365-294x.1999.00800.x. [DOI] [PubMed] [Google Scholar]
- Danton P, Baffray M. Inventaire des plantes protégées en France. Paris: Nathan; 1995. [Google Scholar]
- Dudley SA, Schmitt J. Testing the adaptive plasticity hypothesis: density-dependant selection on manipulated stem length in Impatiens capensis. The American Naturalist. 1996;147:445–465. [Google Scholar]
- Ellis JR, Pashley CH, Burke JM, McCauley DE. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Molecular Ecology. 2006;15:2345–2355. doi: 10.1111/j.1365-294X.2006.02937.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC. Gene flow by pollen: implications for plant conservation genetics. Oikos. 1992;63:77–86. [Google Scholar]
- Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics. 1993;24:217–242. [Google Scholar]
- Falconer DJ, Mackay TFC. Introduction to quantitative genetics. 3rd edn. Harlow: Longman Science and Technology; 1989. [Google Scholar]
- Fenster CB, Dudash MR. Genetic considerations for plant population restoration and conservation. In: Bowles ML, Whelan CJ, editors. Restoration of endangered species: conceptual issues, planning and implementation. Cambridge: Cambridge University Press; 2000. pp. 34–62. [Google Scholar]
- Frary A, Doganlar S, Daunay MC, Tanksley SD. QTL analysis of morphological traits in eggplant and implications for conservation of gene function during evolution of solanaceous species. Theoretical Applied Genetics. 2003;107:359–370. doi: 10.1007/s00122-003-1257-5. [DOI] [PubMed] [Google Scholar]
- Garten C., Jr Relationships between aggressive behaviour and genetic heterozygosity in the oldfield mouse, Peromyscus polionotus. Evolution. 1976;41:80–91. doi: 10.1111/j.1558-5646.1976.tb00881.x. [DOI] [PubMed] [Google Scholar]
- Gibson G. Microarrays in ecology and evolution: a preview. Molecular Ecology. 2002;11:17–24. doi: 10.1046/j.0962-1083.2001.01425.x. [DOI] [PubMed] [Google Scholar]
- Giles BE, Goudet J. Genetic differentiation in Silene dioica metapopulations: estimation of spatiotemporal effects in a successional plant species. The American Naturalist. 1997;149:507–526. [Google Scholar]
- Hedrick PW. Conservation genetics: where are we now? Trends in Ecology and Evolution. 2001;16:629–636. [Google Scholar]
- Hedrick PW, Miller P. Conservation genetics: techniques and fundamentals. Ecological Applications. 1992;2:30–46. doi: 10.2307/1941887. [DOI] [PubMed] [Google Scholar]
- Houle D. Allozyme associated heterosis in Drosophila melanogaster. Genetics. 1989;123:789–801. doi: 10.1093/genetics/123.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez O, Calero C, Mayol M, Rosselló JA. Isozyme uniformity in a wild extinct insular plant, Lysimachia minoricensis J.J.Rodr. (Primulaceae) Molecular Ecology. 1999;8:813–817. doi: 10.1046/j.1365-294x.1999.00800.x. [DOI] [PubMed] [Google Scholar]
- Ingvarsson K. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution. 2002;56:2368–2373. doi: 10.1111/j.0014-3820.2002.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Kearsey MJ, Farquhar AGL. QTL analysis in plants: where are we now? Heredity. 1998;80:137–142. doi: 10.1046/j.1365-2540.1998.00500.x. [DOI] [PubMed] [Google Scholar]
- Kirchner F, Ferdy JB, Andalo C, Colas B, Moret J. Role of corridors in plant dispersal: an example with the endangered Ranunculus nodiflorus. Conservation Biology. 2003;14:401–410. [Google Scholar]
- Korol AB, Ronin YI, Nevo E. Approximate analysis of QTL-environment interaction with no limits on the number of environments. Genetics. 1998;148:2015–2028. doi: 10.1093/genetics/148.4.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Neutral theory of quantitative genetic variance in an island model with extinction and colonization. Evolution. 1992;46:381–389. doi: 10.1111/j.1558-5646.1992.tb02046.x. [DOI] [PubMed] [Google Scholar]
- Lema SC, Nevitt GA. Testing an ecophysiological mechanism of morphological plasticity in pupfish and its relevance to conservation efforts for endangered Devils Hole pupfish. Journal of Experimental Biology. 2006;209:3499–3509. doi: 10.1242/jeb.02417. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Grant MC. Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics. 1996;27:237–277. [Google Scholar]
- Lynch M, Walsh B. Genetic analysis of quantitative traits. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- McKay JK, Bishop JG, Lin JZ, Richards JH, Sala A, Mitchell-Olds T. Local adaptation across a climatic gradient despite small effective population size in the rare sapphire rockcress. Proceedings of the Royal Society of London B. 2001;268:1715–1721. doi: 10.1098/rspb.2001.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely JA, Miller KR, Reid WV, Mittermeier RA, Werner TB. Conserving the world's biological diversity. Washington, DC: World Conservation Union, World Resources Institute, Conservation International, World Wildlife Fund-US and the World Bank; 1990. [Google Scholar]
- Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. Journal of Evolutionary Biology. 2001;14:892–903. [Google Scholar]
- Meunier C, Tirard C, Hurtez-Boussès S, Durand P, Bargues MP, Mas-Coma S, et al. Lack of molluscan host diversity and the transmission of an emerging parasitic disease in Bolivia. Molecular Ecology. 2001;10:1333–1340. doi: 10.1046/j.1365-294x.2001.01284.x. [DOI] [PubMed] [Google Scholar]
- Noel F, Boisselier-Dubayle MC, Lambourdière J, Machon N, Moret J, Samadi S. Characterization of seven polymorphic microsatellites for the study of two Ranunculaceae: Ranunculus nodiflorus L., a rare endangered species and Ranunculus flammula L., a common closely related species. Molecular Ecology Notes. 2005;5:827–829. [Google Scholar]
- Noel F, Porcher E, Moret J, Machon N. Connectivity, habitat heterogeneity, and population persistence in Ranunculus nodiflorus, an endangered species in France. New Phytologist. 2006;169:71–84. doi: 10.1111/j.1469-8137.2005.01572.x. [DOI] [PubMed] [Google Scholar]
- Olivier L, Galland JP, Maurin H, Roux JP. Livre rouge de la flore menacée de France. Paris: Muséum National d'Histoire Naturelle; 1995. [Google Scholar]
- Pigliucci M, Murren CJ, Schlichting CD. Phenotypic plasticity and evolution by genetic assimilation. Journal of Experimental Biology. 2006;209:2362–2367. doi: 10.1242/jeb.02070. [DOI] [PubMed] [Google Scholar]
- Podolsky RH. Genetic variation for morphological and allozyme variation in relation to population size in Clarkia dudleyana, an endemic annual. Conservation Biology. 2001;15:412–423. [Google Scholar]
- Potvin C, Tousignant D. Evolutionary consequences of simulated global change: genetic adaptation or adaptive phenotypic plasticity. Oecologia. 1996;108:683–693. doi: 10.1007/BF00329043. [DOI] [PubMed] [Google Scholar]
- Price TD, Qvarnström A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proceedings of the Royal Society of London B. 2003;270:1433–1440. doi: 10.1098/rspb.2003.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer CR, Ellegren H, Saino N, Moller AP. Directional evolution in germline microsatellite mutations. Nature Genetics. 1996;13:391–393. doi: 10.1038/ng0896-391. [DOI] [PubMed] [Google Scholar]
- Réale D, McAdam AG, Boutin S, Berteaux D. Genetic and plastic responses of a northern mammal to climate change. Proceedings of the Royal Society of London B. 2003;270:591–596. doi: 10.1098/rspb.2002.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebetez M, Mayer M, Dupont O, Schindler D, Gartner K, Kropp JP, et al. Heat and drought 2003 in Europe: a climate synthesis. Annals of Forest Science. 2006;63:569–577. [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conservation Biology. 2003;17:230–237. [Google Scholar]
- Sarkar SK, Chang CK. The Simes method for multiple hypothesis testing with positively dependant test statistics. Journal of the American Statistical Association. 1997;92:1601–1608. [Google Scholar]
- Schmitt J. Reaction norms of morphological and life-history traits to light availability in Impatiens capensis. Evolution. 1993;47:1654–1668. doi: 10.1111/j.1558-5646.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- Schmitt J, McCormac AC, Smith H. A test of the adaptive plasticity hypothesis using transgenic and mutant plant disabled phytochrome-mediated elongation responses to neighbors. The American Naturalist. 1995;146:937–953. [Google Scholar]
- Schmitt J, Dudley SA, Pigliucci M. Manipulation approaches to testing adaptive plasticity: phytochrome-mediated shade-avoidance responses in plants. The American Naturalist. 1999;154:S43–S54. doi: 10.1086/303282. [DOI] [PubMed] [Google Scholar]
- Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Soulé ME, Wilcox BA. Conservation biology: an evolutionary-ecological perspective. Sunderland, MA: Sinauer Associates; 1980. [Google Scholar]
- Storfer A. Quantitative genetics: a promising approach for the assessment of genetic variation in endangered species. Trends in Ecology and Evolution. 1996;11:343–347. doi: 10.1016/0169-5347(96)20051-5. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Klaper R. Genomics for the ecological toolbox. Trends in Ecology and Evolution. 2004;19:439–445. doi: 10.1016/j.tree.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Thompson JD. Phenotypic plasticity as a component of evolutionary change. Trends in Ecology and Evolution. 1991;6:246–249. doi: 10.1016/0169-5347(91)90070-E. [DOI] [PubMed] [Google Scholar]
- Valladares F, Sanchez-Gomez D, Zavala MA. Quantitative estimation of phenotypic plasticity: bridging the gap between the evolutionary concept and its ecological applications. Journal of Ecology. 2006;94:1103–1116. [Google Scholar]
- Watters JV, Lema SC, Nevitt GA. Phenotype management: a new approach to habitat restoration. Biological Conservation. 2003;112:435–445. [Google Scholar]
- Whitehead A, Crawford DL. Variation within and between species in gene expression: raw material for evolution. Molecular Ecology. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- Young AG, Clarke GM. Genetics, demography and viability of fragmented populations. Cambridge: Cambridge University Press; 2000. [Google Scholar]


