Abstract
Recent studies of several species have reported a latitudinal cline in the circadian clock gene, Clock, which influences rhythms in both physiology and behavior. Latitudinal variation in this gene may hence reflect local adaptation to seasonal variation. In some bird populations, there is also an among-individual association between Clock poly-Q genotype and clutch initiation date and incubation period. We examined Clock poly-Q allele variation in the Barn Swallow (Hirundo rustica), a species with a cosmopolitan geographic distribution and considerable variation in life-history traits that may be influenced by the circadian clock. We genotyped Barn Swallows from five populations (from three subspecies) and compared variation at the Clock locus to that at microsatellite loci and mitochondrial DNA (mtDNA). We found very low variation in the Clock poly-Q region, as >96% of individuals were homozygous, and the two other alleles at this locus were globally rare. Genetic differentiation based on the Clock poly-Q locus was not correlated with genetic differentiation based on either microsatellite loci or mtDNA sequences. Our results show that high diversity in Clock poly-Q is not general across avian species. The low Clock variation in the background of heterogeneity in microsatellite and mtDNA loci in Barn Swallows may be an outcome of stabilizing selection on the Clock locus.
Introduction
By providing the opportunity to explore the association between genetic variation and adaptive phenotypic differentiation, candidate genes contribute to our understanding of the genetic basis of traits that are of ecological importance [1], [2]. The circadian clock, which generates endogenous ∼24 hr biochemical, physiological and behavioral rhythms, has been fully characterized at the molecular genetics level (reviewed by [3]). The circadian clock gene, Clock, has been sequenced from hundreds of taxa, and in vertebrates it codes for a protein (CLOCK) that serves as a transcription factor (as a heterodimer with BMAL1) in the core circadian clock oscillator [4]–[6]. The Clock gene is highly conserved among vertebrates throughout most of its sequence; however, its C-terminal region contains a variable poly-glutamine (poly-Q) motif with variability in the number of glutamine repeats both among and within species [7]–[9]. This region is known to influence the transcription activating potential of the protein, and therefore it can affect both behavior and physiology related to circadian rhythms [10], [11].
Clock poly-Q variation provides an opportunity to examine its association with life-history traits that may be related to circadian clock. Studies on various taxa have reported an association between Clock genotype and breeding latitude [9], [12], [13]. In birds, latitudinal variation in Clock genotypes have been detected in the Blue Tit (Cyanistes caeruleus) but not for the Bluethroat (Luscinia svecica) [9] or Tachycineta swallows [14], therefore the generality of this pattern in birds and its possible relationship with adaptive phenotypic differentiation remains unclear. Additional within-population analyses showed an association between Clock genotype and breeding phenology (clutch initiation date and incubation duration) in a Blue Tit population [15] but not in a sympatric Great Tit (Parus major) population, in which Clock poly-Q variation was low [16]. Thus, further comparisons are required to assess patterns of association between Clock variation and life-history traits.
The Barn Swallow (Hirundo rustica) is one of the world's most widely distributed passerines, with a broad geographic distribution throughout most of the northern hemisphere. The six described subspecies [17]–[20] exhibit substantial geographic variation in morphometric characteristics (such as tail streamer length and ventral coloration) and in life-history traits such as migration patterns and time of breeding [19], [21], [22]. This species' wide geographic distribution combined with life-history variation make it a good candidate in which to examine the generality of the association between Clock poly-Q and life-history traits related to the timing of seasonal reproductive behavior.
In this study we examined Clock poly-Q allelic length variation in five breeding populations of Barn Swallows (from three subspecies). These populations differ in several life-history traits related to breeding latitude, migratory behavior and breeding phenology (Table 1). We sampled three populations of the New World subspecies, H. r. erythrogaster, from New York, California and from a recently established breeding population in Argentina [23], and two populations from the Old World: a non-migratory population from Israel (H. r. transitiva) and a population from the United Kingdom (H. r. rustica). The Old World and New World clades are divergent in mtDNA with ∼1.6% sequence difference [24], [25]. Our main objectives were to (a) characterize the Clock poly-Q region in Barn Swallows, (b) examine genetic variation among pairs of five populations sampled from three subspecies in relation to differences in their life history traits and (c) compare genetic differentiation at the Clock locus to that of presumably neutral loci (mitochondrial DNA haplotypes and microsatellite loci). Comparisons among the Clock locus and loci that are selectively neutral provide an important background to examine whether the Clock locus is under selection or exhibits patterns of neutral evolution.
Table 1. Characteristics of the Barn Swallow populations used in this study with sample sizes for the genetic markers used in this study.
| Population | Subspecies | Breeding latitude | Migratory behavior | Time of breeding | Breeding cycles1 | # of samples analyzed for | ||
| Clock | ND2 | Microsatellites | ||||||
| Israel | transitiva | 32°55′N | No2 | Feb–Jun | 2 (3) | 49 | 31 | 178 |
| United Kingdom | rustica | 50°42′N | Yes | Apr–Sep | 2 | 49 | 30 | 62 |
| New York | erythrogaster | 42°30′N | Yes | May–Aug | 2 | 49 | 31 | 41 |
| California | erythrogaster | 38°N | Yes | May–Jul | 2 | 31 | 19 | 23 |
| Argentina | erythrogaster | 38°30′S | Yes | Oct–Feb | 2 (3) | 52 | 63 | 63 |
Typical number of breeding cycles with maximum breeding cycles in parentheses.
Barn Swallows from Israel only move locally outside the breeding season.
Methods
Study populations and sample collection
We studied five breeding populations of Barn Swallows representing three of the six described subspecies. In each of these breeding populations we collected blood samples from adults during the breeding season and stored them in lysis buffer until DNA was extracted. Population characteristics and sample sizes used in this study are presented in Table 1.
Analysis of Clock poly-Q alleles
Genomic DNA was extracted from blood using the E-Z 96 Tissue DNA kit (Omega Bio-Tek, Norcross, GA) or using the DNAeasy blood Extraction kit (Qiagen, Valencia, CA). In order to examine variability in Barn Swallow Clock poly-Q region and to verify the genetic sequence, we first amplified this region (corresponding to human Clock gene exon 20 [26]) from 3–5 individuals from each of the Barn Swallow populations using the sequencing primers developed by [9]. 10 µL PCR amplifications included 1 µL undiluted DNA, 10 µM Tris-HCl , 50 µM KCl, 4 mM MgCl2, 0.25 mM of each nucleotide, 0.25 mM from each primer, and 0.025 U jumpstart Taq polymerase (Sigma-Aldrich, St. Louis,Missouri). PCR amplification conditions were: initial denaturation at 95°C for 4 min 30 s; 30 cycles of denaturating at 95°C for 1 min, annealing at 64°C for 1 min, and extension at 72°C for 2 min; and a final extension at 72°C for 4 min 30 s. PCR products were purified using Exonuclease and Shrimp Alkaline Phosphatase enzymatic reactions (United States Biochemical, Cleveland, OH). Purified products were cycle sequenced in both directions using amplification primers and ABI BigDye Terminator chemistry. Sequencing products were cleaned using Sephadex columns and electrophoresed in an ABI 3730 Automated DNA Analyzer (Applied Biosystems, Foster City, CA). We aligned forward and reverse strands for each specimen and checked them using Sequencher 4.7 (Gene Codes Corp., Ann Arbor, MI). The amplified sequence generated for all Barn Swallows matched the expected sequence for this gene for birds. In total, we sequenced the Clock poly-Q region for 17 Barn Swallows (GenBank accession numbers JN986726-JN986747). All Barn Swallow sequences were identical to each other and differed only in the number of poly-Q repeats (Fig. 1). The first and last glutamine amino acids in the poly-Q repeat were coded by CAA codons, whereas the middle codons were exclusively CAG. Thus, the only variation was in the number of CAG codons.
Figure 1. Amino acid alignment of avian CLOCK polyQ alleles.
Alignment includes sequences from Barn Swallows (Hirundo rustica; Hr) together with published Blue Tit (Cyanistes caeruleus; Cc), Great Tit (Parus Major; Pm), Bluethroat (Luscinia svecica; Ls) and Tree Swallow (Tachycineta bicolor; Tb). For each sequence the species name and number of Clock poly-Q repeats are shown. The predicted protein sequences of Barn Swallow Clock poly-Q repeats only differ in the number of CAG codon (coded by Q) repeats (The first and last glutamine (Q) amino acids in the poly-Q repeat were coded by CAA codons). Q residues coded by CAA are underlined and lower-case Qs are within-population polymorphic site encoded by either CAA or CAG. Asterisk indicates identical amino acids in the poly-Q flanking region between the all sequences.
We then screened 49 adults from Israel, 49 from the United Kingdom, 49 from New York, 31 from California and 52 from Argentina for length polymorphism in the Clock poly-Q region using the genotyping primer set developed by [9] in which the forward primer was labeled at the 5′ end with 6-FAM fluorescent dye. The PCR protocol was similar to the one used for sequencing (see above). PCR products were genotyped on an ABI 3100 Genetic Analyzer (Applied Biosystems) with GeneScan-500 LIZ (Applied Biosystems) as the molecular size standard. Allele sizes were estimated using Genemapper v3.7 (Applied Biosystems) together with control samples with known repeat numbers determined by sequencing. We were able to successfully genotype all Barn Swallow samples (N = 230).
Analysis of mitochondrial sequences and microsatellite loci
Mitochondrial and microsatellite loci for the Israeli and UK populations were analyzed as part of a population genetics study which focus on the Old World samples [27], while loci for the three New World populations were analyzed as part of a population genetics study which focus on the New world samples [28]. The same individuals were used across all markers in all populations. Sample sizes for the Clock marker and the two neutral markers (microsatellites and mtDNA) are not equal due to sample and data availability. However, similar analyses with a subset of individuals that were analyzed for all three markers (for all five populations) generated similar results. Therefore, we report here only results using all available samples.
We amplified and sequenced the mitochondrial protein-coding gene nicotinamide adenine dinucleotide dehydrogenase subunit 2 (ND2) for a subset of each population sample (Table 1). We used primers METb and TRPc [29]. 10 µL PCR amplifications included 1 µL undiluted DNA, 10 µM Tris-HCl , 50 µM KCl, 3–4 mM MgCl2, 0.25 mM of each nucleotide, 0.25 µM from each primer, and 0.025 U jumpstart Taq polymerase (Sigma-Aldrich). PCR amplification conditions were as follows: initial denaturation at 95°C for 4 min 30 s; 30–35 cycles of denaturating at 95°C for 1 min, annealing at 54°C or 62°C for 1 min, and extension at 72°C for 2 min; and a final extension at 72°C for 4 min 30 s. PCR products were purified using Exonuclease and Shrimp Alkaline Phosphatase enzymatic reactions (United States Biochemical). Purified reactions were cycle sequenced in both directions using amplification primers and ABI BigDye Terminator. Sequencing products were cleaned using Sephadex columns and processed with an ABI 3730 Automated DNA Analyzer (Applied Biosystems). We aligned forward and reverse strands for each specimen and checked them using Sequencher 4.7 (Gene Codes Corp.). All sequence data were deposited in GenBank (Accession Nos. JN642465-JN642524 and HQ333550-HQ333663).
A total of six nuclear microsatellite loci previously developed for H. rustica or other birds were analyzed for all populations: Escµ6 [30], Ltr6 [31], POCC6 [32], Hir11, Hir19 and Hir20 [33]. Forward primers were labeled at the 5′ end with fluorescent tags (PET, 6-FAM, VIC or NED; Applied Biosystems). Individual PCR amplifications were combined into multiplex mixes to decrease the number of reactions needed per individual. 10 µL PCR amplifications included 10–100 ng of genomic DNA, 10 µM Tris-HCl , 50 µM KCl, 1.5–3.25 mM MgCl2, 0.25 mM of each nucleotide, 0.12–0.24 µM from each primer, and 0.025 U jumpstart Taq polymerase (Sigma-Aldrich). PCR amplification conditions were as following: initial denaturation at 95°C for 5 min; 35 cycles of denaturating at 95°C for 30 s, annealing at 50°C or 58°C for 30 s (specific per multiplex PCR mix), and extension at 72°C for 30 s; and a final extension at 72°C for 30 min. PCR products were genotyped on an ABI 3100 Genetic Analyzer (Applied Biosystems) with GeneScan-500 LIZ (Applied Biosystems) as molecular size standard. Allele sizes were estimated using Genemapper v3.7 (Applied Biosystems) and verified and amended by eye to ensure standardized data collection.
Statistical analysis
Observed and expected heterozygosities for the Clock poly-Q region for each population were calculated using ARLEQUIN v3.11 [34]. We tested for departures from Hardy-Weinberg equilibrium (HWE) per locus and population and for deviation from linkage equilibrium (LD) for all microsatellite loci combinations using GENEPOP v4 [35], [36] with parameters of 10,000 dememorization, 10,000 batches, and 10,000 iterations. We estimated overall genetic differentiation between populations using FST values in ARLEQUIN v3.11 [34]. We applied a sequential Bonferroni correction [37] to these test results.
We used Pearson correlation matrix in order to examine the relationship between pairwise genetic differentiation values (FST) between populations estimated from the various genetic markers.
Statistical analysis was done using Statistica 7.0 (Statsoft, Inc., Tulsa, OK). Results are presented as mean ± SE unless stated otherwise.
Ethics Statement
This study was conducted under the permits of Israel Nature Reserve Authority (permit numbers 28234-2007 and 31345-2008), UK Home Office (project license number 30/2740), Dirección de Administración de Areas Protegidas y Conservación de la Biodiversidad, Provincia Buenos Aires, Argentina (collecting permit number 6607) and the U.S. Fish and Wildlife Service (Federal Bird Banding Permit number 20576).
Results
Clock poly-Q variation in Barn Swallows
We genotyped a total of 230 Barn Swallows from five populations representing three subspecies. Overall, we found only three different length-variant alleles, ClkpolyQ6, 7, 8, corresponding to 6–8 poly-Q repeats coded by CAG codons (Table 2). However, one allele, ClkpolyQ7, accounted for 98% of the overall allelic diversity and 96.5% of individuals were homozygous for this allele. We found all three alleles in only one population (Israel), whereas all individuals from one population (Argentina) were homozygous to the ClkpolyQ7 allele. Mean observed heterozygosity (H = 0.030) was correspondingly low and ranged from 0.000 for the Argentina population to 0.082 for the Israeli population.
Table 2. Clock poly-Q allele frequencies, number of alleles (k), mean allele size (with standard errors) and observed heterozygosities (H) for the five Barn Swallow populations used in this study.
| Population | Subspecies | N | k | Mean allele size (s.e.) | Allele proportion | H | ||
| Q6 | Q7 | Q8 | ||||||
| Israel | transitiva | 49 | 3 | 6.96 (0.03) | 0.051 | 0.939 | 0.010 | 0.082 |
| United Kingdom | rustica | 49 | 2 | 6.99 (0.01) | 0.010 | 0.990 | 0.000 | 0.020 |
| New York | erythrogaster | 49 | 2 | 7.01 (0.01) | 0.000 | 0.990 | 0.010 | 0.020 |
| California | erythrogaster | 31 | 2 | 7.02 (0.02) | 0.000 | 0.984 | 0.016 | 0.032 |
| Argentina | erythrogaster | 52 | 1 | 7.00 (0.00) | 0.000 | 1.000 | 0.000 | 0.000 |
| Total | 230 | 3 | 6.99 (0.01) | 0.013 | 0.980 | 0.007 | 0.030 | |
Allele frequencies did not deviate from Hardy-Weinberg equilibrium for all populations (P>0.05).
Owing to the low variation in Clock poly-Q alleles, we could test for deviation from HWE only in the Israeli population, which did not show deviation from HWE (P = 0.150).
Population frequencies of microsatellite alleles
We genotyped Barn Swallows from the five populations at six polymorphic microsatellite loci. Mean number of alleles for all loci was 11.83 (±4.50 SD) and ranged from 5.40 for Ltr6 to for 14.80 for Escu6 (minimum number of alleles was 4 for Ltr6 in California and maximum was 21 for Hir20 in Israel). Most of the populations did not deviate from HWE; however we detected deviation from HWE for the Argentina population in Hir11 and for the New York population in Escu6 and Hir19 (after sequential Bonferroni correction). We found no evidence for significant LD for any pair of microsatellite loci.
Molecular characterization of mitochondrial haplotypes
We aligned most of the ND2 gene (1023 bp) for 174 individuals from the five populations and detected 45 unique haplotypes: 21 for the Old World populations and 24 for the New World populations, which represented two discrete clades (minimum of 13 nucleotide changes between them). Out of the 21 Old World haplotypes, two common haplotypes were shared by individuals from the Israeli and UK populations. Three New World haplotypes were shared by individuals from all three New World populations, while two haplotypes were shared by individuals from the New York and Argentina populations.
Genetic differentiation
We examined genetic differentiation between all pairs of populations based on the different genetic markers. Mean genetic differentiation based on the Clock poly-Q locus (FST = 0.0095±0.005) was much lower (and not significant) compared to genetic differentiation based on six microsatellite loci (FST = 0.038±0.005, all statistically significant) or on mitochondrial haplotypes (FST = 0.578±0.115, all statistically significant). Analysis of genetic differentiation for each of the microsatellite loci separately showed that only Hir20 (FST = 0.0082±0.003) had similarly low values to the Clock poly-Q locus and the other five loci had higher FST values (range: 0.021–0.078).
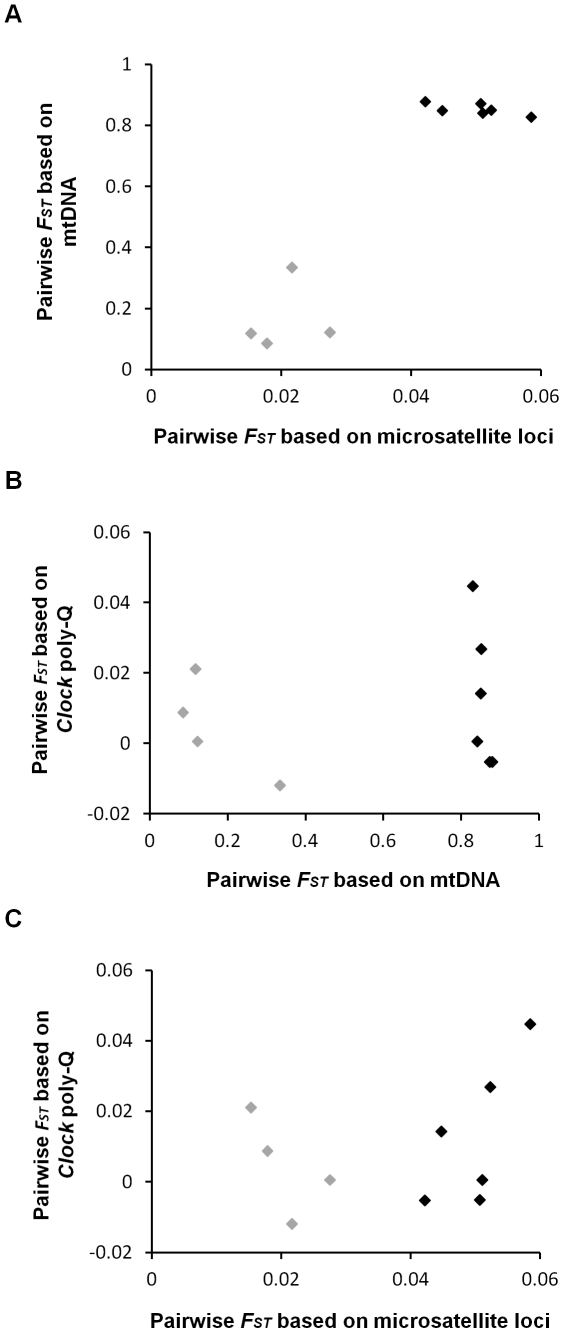
Pairwise genetic differentiation values based on mitochondrial haplotypes and microsatellite loci were highly correlated (Fig. 2A; N = 10, r2 = 0.858, t = 6.691, P = 0.0001), while similar comparisons between Clock poly-Q locus pairwise FST values and either mitochondrial haplotypes (Fig. 2B; N = 10, r2 = 0.020, t = 0.407, P = 0.694) or microsatellite loci (Fig. 2C; N = 10, r2 = 0.110, t = 0.995, P = 0.349) pairwise FST values were not correlated. Separate analyses for each of the microsatellite loci showed that pairwise FST values based on these loci were positively correlated with mitochondrial haplotype pairwise FST values (range of Pearson correlation coefficient: r = 0.237–0.864), although only correlations for Escu6, Ltr6 and Hir11 were significant. Parallel correlations of pairwise FST values based on these microsatellite loci to Clock poly-Q pairwise FST values were not significant and lower than the correlations between pairwise FST values of these microsatellite loci and mitochondrial haplotypes (range of Pearson correlation coefficient: r = −0.177–0.734), except for Hir19 which showed a significant correlation with the Clock poly-Q pairwise FST values.
Figure 2. Between-populations pairwise genetic differentiation values (FST) comparisons.
(A) Comparisons between mitochondrial haplotypes and microsatellite loci, (B) between Clock poly-Q locus and mitochondrial haplotypes and (C) between Clock poly-Q locus and microsatellite loci. Points in black represent comparisons between the erythrogaster subspecies to either the rustica or transitiva subspecies, while points in grey represent comparison within the erythrogaster subspecies and between the rustica and transitiva subspecies (which are mitochondrially intermixed).
The low variation in Clock poly-Q in Barn Swallows precluded additional analysis of the statistical association between Clock poly-Q variation and life history traits among the Barn Swallow populations or subspecies.
Discussion
Variation within the circadian clock core oscillator genes is generally low, but the Clock gene poly-Q region shows a unique polymorphism both among and within bird species. This polymorphism is associated with both breeding latitude and breeding phenology in Blue Tits, suggesting that this locus might be involved in local adaptation to seasonal environments, possibly through the response to photoperiod [9], [15]. However, this association has not been detected in several other bird species [9], [16], leaving the question regarding the generality of this relationship unresolved.
Barn Swallows demonstrate a great amount of variation in morphological and life-history traits throughout the Holarctic, including trait diversity that might be related to circadian rhythms such as variation in breeding latitude, time of breeding and migratory patterns. In this study we surveyed five populations of Barn Swallows in order to examine Clock poly-Q variation in the species and compare it among Barn Swallow populations that exhibit variation in life-history traits. We identified three Clock poly-Q length variants in Barn Swallows, however the variation was extremely low with one allele (Q7 – corresponding to 7 poly-Q repeats) accounting for 98% of the total alleles (and two other very rare alleles – Q6 and Q8), and a mean observed heterozygosity of only 0.030 (range: H = 0.000–0.082). This variation is much lower than similar values reported for the Blue Tit (9 alleles; Q9-Q17; H = 0.489), Bluethroat (9 alleles; Q10-Q16; H = 0.213) [9] or even a low-variability Great Tit population (5 alleles; Q11-Q15; H = 0.077) [16] and five species of Tachycineta swallows (2–4 alleles; Q5-Q9; H = 0.047–0.472) [14]. This low variation in Clock poly-Q in Barn Swallows was also reflected in the genetic differentiation analysis that showed no differentiation among populations based on this locus, while similar analyses based on mtDNA haplotypes or microsatellite loci showed significant and correlated differentiation between these populations (Fig. 2).
Comparison of population genetic differentiation based on the Clock poly-Q locus and presumably neutral loci (mtDNA haplotypes and microsatellite loci) showed contrasting findings. While neutral loci reflected, as expected, greater divergence between the Old World and New World clades and less structure within these clades [25], [27] genetic differentiation based on Clock poly-Q locus did not follow such a pattern (Fig. 2). This low variation in the Clock poly-Q locus precluded us from associating it with any of the populations' life-history traits (breeding latitude, time of breeding, migration). Still, the sole non-migratory population (H. r. transitiva from Israel) showed the highest genetic variation (Table 2). Genetic variation in the non-migratory Blue Tit was also higher compared to the migratory Bluethroat [9]. Yet, a non-migratory Great Tit population showed lower variation than both [16]. Therefore, we cannot assume any general relationship between migratory patterns and genetic variation in the Clock poly-Q locus based on the available data.
Why is Clock poly-Q variation in Barn Swallows so low compared to other bird species? Clock poly-Q allele size in Barn Swallows was shorter compared to those of the other species reported (Q9–17 in Blue tit, Q11–15 in Great Tit and Q10–16 in Bluethroat). Longer tandem repeats are more likely to be more polymorphic, therefore the lower variation observed for Barn Swallows may be related to their relatively shorter Clock poly-Q repeats. However, despite long Clock poly-Q repeats in a Great Tit population (Q11–15), polymorphism was relatively low [16]. Liedvogel and Sheldon [16] suggested that this might be due to the fact that CAG-repeat variation in this locus is tightly regulated and that selection in Great Tit acts in favor of the Clock poly-Q14 allele. Clock poly-Q variation in Barn Swallows in based on CAG-repeats as well, which may be also tightly regulated. However, CAG-repeat polymorphism exists also in the much more polymorphic Blue Tit and Bluethroat [9]; therefore it is less likely to assume that CAG-repeats will be more regulated in one species than the other.
Selection for a specific allele could explain the low variation observed in Barn Swallows (and Great Tits). It is possible that the Q7 allele is under positive selection in Barn Swallows (similarly to the Q14 allele in Great Tits). However, Johnsen et al. [9] have suggested that the fact that selection has not stabilized the Clock poly CAG repeat at a single length in several species is at least suggestive that the variation is itself selectively advantageous. Our results demonstrate homogeneity in the locus in a background of heterogeneity of presumably selectively neutral loci. This scenario suggests that the Clock poly-Q variation does reflect the demographic processes among the Barn Swallow population, and is more expected from a selection operating on the Clock locus to maintain the low variation while neutral processes, such as genetic drift, may be contribute to the variation in unrelated microsatellite and mtDNA loci [38].
Alternatively, this low variation in the Clock poly-Q locus in Barn Swallows might represent a holdover from an ancestral population that itself had low variation in the Clock locus. Examination of the Clock poly-Q locus in a few individuals of other swallow species in the genus Hirundo (H. angolensis, H. aethiopica and H. albigularis) has shown that they are also homozygotes for the Q7 allele (Dor R., unpublished data). In addition, the Barn Swallow population from Argentina, which showed no polymorphism in the Clock poly-Q locus, was established only in the 1980's, most likely from North American populations [23], [25], [28], thus in this population at least low variation in Clock poly-Q may stem from a founder effect. However, even in this newly founded population, presumably selectively neutral loci (mtDNA haplotypes and microsatellite loci) displayed much more variation than the Clock poly-Q locus. We cannot exclude the possibility that these presumably selectively neutral loci were already more variable in the ancestral population. However, it is more likely that the Clock poly-Q locus in Barn Swallows is not entirely neutral and that some type of selection is operating to reduce variation in the poly-Q region.
The timing of the central clock can be adjusted by several environmental cues. Photoperiod cues have been studied extensively; however the effects of non-photic cues on reproduction have been demonstrated as well [39]. These non-photic cues may include social cues, food and temperature (reviewed in [39]). The mechanisms that enable these non-photic cues to influence circadian rhythms are unknown, however they most likely utilize different genetic pathways. Species and populations may differ in the primacy of each of these cues as the most important cue to adjust reproductive phenology, depending on their ecology. Thus, we might expect genetic differences in species or population reflective of the cues used to adjust their circadian rhythm. For example, if the Clock gene is associated with photoperiod, then the nature of selection for this gene (e.g., balancing or positive) could vary depending on whether individuals in that species or population had increased reproductive success relying on non-photic or photoperiod cues. Social cues (associated with mate choice preferences) are very important in determining reproductive success in Barn Swallows [40]–[43], thus it is possible that the lack of Clock gene variation reflects social cues primacy over photoperiod cues in this species. Examination of this hypothesis warrants future work.
Acknowledgments
We thank Amanda Talaba and Laura Stenzler for laboratory assistance. We thank two anonymous reviewers for their comments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funded by the Cornell Lab of Ornithology and NSF grants IOS-0717421 to RJS and OISE-0730180 to DWW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- 2.Steinmeyer C, Mueller JC, Kempenaers B. Search for informative polymorphisms in candidate genes: clock genes and circadian behaviour in blue tits. Genetica. 2009;136:109–117. doi: 10.1007/s10709-008-9318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 5.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 6.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 7.Saleem Q, Anand A, Jain S, Brahmachari SK. The polyglutamine motif is highly conserved at the Clock locus in various organisms and is not polymorphic in humans. Hum Genet. 2001;109:136–142. doi: 10.1007/s004390100550. [DOI] [PubMed] [Google Scholar]
- 8.Fidler AE, Gwinner E. Comparative analysis of Avian BMALI and CLOCK protein sequences: a search for features associated with owl nocturnal behaviour. Comp Biochem Physiol Biochem Mol Biol. 2003;136:861–874. doi: 10.1016/s1096-4959(03)00276-8. [DOI] [PubMed] [Google Scholar]
- 9.Johnsen A, Fidler AE, Kuhn S, Carter KL, Hoffmann A, et al. Avian Clock gene polymorphism: evidence for a latitudinal cline in allele frequencies. Mol Ecol. 2007;16:4867–4880. doi: 10.1111/j.1365-294X.2007.03552.x. [DOI] [PubMed] [Google Scholar]
- 10.Avivi A, Albrecht U, Oster H, Joel A, Beiles A, et al. Biological clock in total darkness: The Clock/MOP3 circadian system of the blind subterranean mole rat. Proc Natl Acad Sci U S A. 2001;98:13751–13756. doi: 10.1073/pnas.181484498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayasaka N, LaRue SI, Green CB. In vivo disruption of Xenopus CLOCK in the retinal photoreceptor cells abolishes circadian melatonin rhythmicity without affecting its production levels. J Neurosci. 2002;22:1600–1607. doi: 10.1523/JNEUROSCI.22-05-01600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauber E, Kyriacou CP. Molecular evolution and population genetics of circadian clock genes. Methods Enzymol. 2005;393:797–817. doi: 10.1016/S0076-6879(05)93042-5. [DOI] [PubMed] [Google Scholar]
- 13.O'Malley KG, Banks MA. A latitudinal cline in the Chinook salmon (Oncorhynchus tshawytscha) Clock gene: evidence for selection on PolyQ length variants. Proc R Soc Lond B Biol Sci. 2008;275:2813–2821. doi: 10.1098/rspb.2008.0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dor R, Cooper CB, Lovette IJ, Massoni V, Bulit F, et al. Clock gene variation in Tachycineta Swallows. 2011. In press, Ecology and Evolution. doi: 10.1002/ece3.73. [DOI] [PMC free article] [PubMed]
- 15.Liedvogel M, Szulkin M, Knowles SCL, Wood MJ, Sheldon BC. Phenotypic correlates of Clock gene variation in a wild blue tit population: evidence for a role in seasonal timing of reproduction. Mol Ecol. 2009;18:2444–2456. doi: 10.1111/j.1365-294X.2009.04204.x. [DOI] [PubMed] [Google Scholar]
- 16.Liedvogel M, Sheldon BC. Low variability and absence of phenotypic correlates of Clock gene variation in a great tit Parus major population. J Avian Biol. 2010;41:543–550. [Google Scholar]
- 17.Peters JL. Family Hirundinidae. In: Mayr E, Greenway JC, editors. Check-list of the Birds of the World, vol IX. Cambridge, MA: Museum of Comparative Zoology; 1960. pp. 80–129. [Google Scholar]
- 18.Cramp S. Handbook of the Birds of Europe, the Middle East and North Africa, vol. 5. Oxford, UK: Oxford University Press; 1988. [Google Scholar]
- 19.Turner AT, Rose C. Swallows and Martins: An Identifcation Guide and Handbook. Boston, MA: Houghton Mifflin; 1989. [Google Scholar]
- 20.Dickinson EC, Dekker RWRJ. Systematic notes on Asian birds. 13. A preliminary review of the Hirundinidae. Zool Verh. 2001;335:85–126. [Google Scholar]
- 21.Turner A. Family Hirundinidae. In: Del Hoyo J, Elliott A, Christie D, editors. The birds of the World, vol 9. Barcelona, Spain: Lynx Edicions; 2004. pp. 602–685. [Google Scholar]
- 22.Turner A. The Barn Swallow. London, UK: T&AD Poyser; 2006. [Google Scholar]
- 23.Martínez MM. NidiWcacion de Hirundo rustica erythrogaster (Boddaert) en la Argentina (Aves, Hirundinidae). Neotropica. 1983;29:83–86. [Google Scholar]
- 24.Sheldon FH, Whittingham LA, Moyle RG, Slikas B, Winkler DW. Phylogeny of swallows (Aves : Hirundinidae) estimated from nuclear and mitochondrial DNA sequences. Mol Phylogenet Evol. 2005;35:254–270. doi: 10.1016/j.ympev.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Dor R, Safran RJ, Sheldon FH, Winkler DW, Lovette IJ. Phylogeny of the genus Hirundo and the Barn Swallow subspecies complex. Mol Phylogenet Evol. 2010;56:409–418. doi: 10.1016/j.ympev.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Steeves TDL, King DP, Zhao YL, Sangoram AM, Du FH, et al. Molecular cloning and characterization of the human CLOCK gene: Expression in the suprachiasmatic nuclei. Genomics. 1999;57:189–200. doi: 10.1006/geno.1998.5675. [DOI] [PubMed] [Google Scholar]
- 27.Dor R, Safran RJ, Vortman Y, Lotem A, McGowen A, et al. Population genetics and morphological comparisons of migratory European (Hirundo rustica rustica) and sedentary East-Mediterranean (H. r. transitiva) Barn Swallows. J Hered. Published. 2011 doi: 10.1093/jhered/esr114. online November 9, 2011. doi: 10.1093/jhered/ESR114. [DOI] [PubMed] [Google Scholar]
- 28.Billerman SM, Huber GH, Winkler DW, Safran RJ, Lovette IJ. Population genetics of a recent transcontinental colonization of South America by breeding Barn Swallows (Hirundo rustica). Auk. 2011;128:506–513. [Google Scholar]
- 29.Hunt JS, Bermingham E, Ricklefs RE. Molecular systematics and biogeography of Antillean thrashers, tremblers, and mockingbirds (Aves : Mimidae). Auk. 2001;118:35–55. [Google Scholar]
- 30.Hanotte O, Zanon C, Pugh A, Greig C, Dixon A, et al. Isolation and characterization of microsatellite loci in a passerine bird - the Reed Bunting Emberiza schoeniclus. Mol Ecol. 1994;3:529–530. doi: 10.1111/j.1365-294x.1994.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 31.McDonald DB, Potts WK. Cooperative display and relatedness among males in a lek-mating bird. Science. 1994;266:1030–1032. doi: 10.1126/science.7973654. [DOI] [PubMed] [Google Scholar]
- 32.Bensch S, Price T, Kohn J. Isolation and characterization of microsatellite loci in a Phylloscopus warbler. Mol Ecol. 1997;6:91–92. doi: 10.1046/j.1365-294x.1997.00150.x. [DOI] [PubMed] [Google Scholar]
- 33.Tsyusko OV, Peters MB, Hagen C, Tuberville TD, Mousseau TA, et al. Microsatellite markers isolated from barn swallows (Hirundo rustica). Mol Ecol Notes. 2007;7:833–835. [Google Scholar]
- 34.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 35.Raymond M, Rousset F. GENEPOP (version-1.2) - population-genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 36.Rousset F. GENEPOP' 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 37.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 38.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 39.Visser ME, Caro SP, van Oers K, Schaper SV, Helm B. Phenology, seasonal timing and circannual rhythms: towards a unified framework. Philos Trans R Soc Lond B Biol Sci. 2010;365:3113–3127. doi: 10.1098/rstb.2010.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Møller AP. Female choice selects for male sexual tail ornaments in the monogamous swallow. Nature. 1988;332:640–642. [Google Scholar]
- 41.Møller AP. Sexual Selection and the Barn Swallow. Oxford, UK: Oxford University Press; 1994. [Google Scholar]
- 42.Safran RJ, Neuman CR, McGraw KJ, Lovette IJ. Dynamic paternity allocation as a function of male plumage color in barn swallows. Science. 2005;309:2210–2212. doi: 10.1126/science.1115090. [DOI] [PubMed] [Google Scholar]
- 43.Vortman Y, Lotem A, Dor R, Lovette IJ, Safran RJ. The sexual signals of the East-Mediterranean barn swallow: a different swallow tale. Behav Ecol. 2011;22:1344–1352. [Google Scholar]