Abstract
Thiopurine methyltransferase (Tpmt) is the primary enzyme responsible for deactivating thiopurine drugs. Thiopurine drugs (i.e., thioguanine [TG], mercaptopurine, azathioprine) are commonly used for the treatment of cancer, organ transplant, and autoimmune disorders. Chronic thiopurine therapy has been linked to the development of brain cancer (most commonly astrocytomas), and Tpmt status has been associated with this risk. Therefore, we investigated whether the level of Tpmt protein activity could predict TG-associated cytotoxicity and DNA damage in astrocytic cells. We found that TG induced cytotoxicity in a dose-dependent manner in Tpmt+/+, Tpmt+/− and Tpmt−/− primary mouse astrocytes and that a low Tpmt phenotype predicted significantly higher sensitivity to TG than did a high Tpmt phenotype. We also found that TG exposure induced significantly more DNA damage in the form of single strand breaks (SSBs) and double strand breaks (DSBs) in primary astrocytes with low Tpmt versus high Tpmt. More interestingly, we found that Tpmt+/− astrocytes had the highest degree of cytotoxicity and genotoxicity (i.e., IC50, SSBs and DSBs) after TG exposure. We then used human glioma cell lines as model astroglial cells to represent high (T98) and low (A172) Tpmt expressers and found that A172 had the highest degree of cytoxicity and SSBs after TG exposure. When we over-expressed Tpmt in the A172 cell line, we found that TG IC50 was significantly higher and SSB's were significantly lower as compared to mock transfected cells. This study shows that low Tpmt can lead to greater sensitivity to thiopurine therapy in astroglial cells. When Tpmt deactivation at the germ-line is considered, this study also suggests that heterozygosity may be subject to the greatest genotoxic effects of thiopurine therapy.
Introduction
Thiopurine methyltransferase (Tpmt) is a cytoplasmic enzyme responsible for catalyzing the S-methylation of aromatic and heterocyclic compounds and is the primary deactivating enzyme for thiopurine drugs [1]. Thiopurines are antimetabolite pro-drugs that require intracellular conversion to the active metabolites to exert their cytotoxic effects (Fig. 1) [2]. Alternatively, thiopurine activation is in competition with deactivation pathways primarily involving Tpmt. Tpmt has been studied extensively because of inter-patient variability in Tpmt protein expression owing to a few common genetic polymorphisms [3], [4], [5]. Deactivating polymorphisms in Tpmt can result in a trimodal population distribution in enzymatic activity. Inheritance of two functional alleles results in high protein activity (∼90% of individuals), heterozygosity leads to intermediate activity (∼10% of individuals), and inheritance of two dysfunctional alleles results in practically no protein activity (<1% of individuals) [6], [7]. Importantly, low Tpmt has been associated with life-threatening toxicities including cancer in patients receiving thiopurine therapy [8], [9], [10], [11], [12].
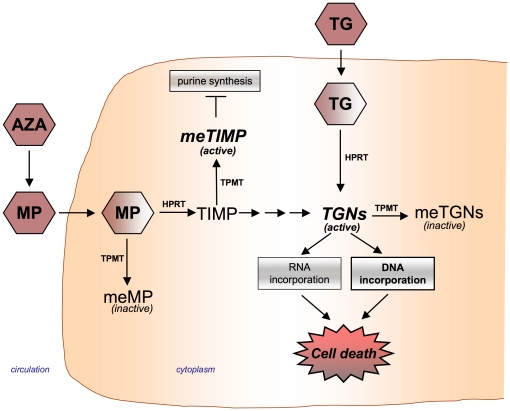
Figure 1. Thiopurine drug metabolism pathway.
Azathioprine (AZA) is a prodrug of mercaptopurine (MP). Thioguanine (TG) and MP can be converted by hypoxanthine phosphoribosyl transferase (HPRT) to thioguanine nucleotide (TGNs) metabolites. MP can also be converted to the methylthioinosine monophosphate (meTIMP) by thiopurine methyltransferase (Tpmt) which inhibits purine synthesis; however, thioguanine bypasses the conversion to this metabolite. Thiopurines can be converted to inactive metabolites [i.e. methyl-mercaptopurine (meMP); methyl-thioguanine (meTG); and methyl-thioguanine nucleotides (meTGNs)] by Tpmt. TGN metabolites are incorporated into DNA and RNA leading to cell death. However, DNA incorporation is believed to be the primary mode of cytotoxicty.
Over the past several decades, thiopurines [e.g. thioguanine (TG), mercaptopurine, and azathioprine] have been commonly used in the treatment of cancer, autoimmune disorders, and in organ transplant. In each of these patient populations there have been a number of reports of cancer development after thiopurine therapy to include acute myeloid leukemias, lymphomas, and cancers of the skin and brain [8], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]; although, the relevance of Tpmt status is not clear. However, in a study by Relling et al it was found that when patients were treated with antimetabolites (i.e. mercaptopurine, methotrexate) and prophylactic cranial irradiation as part of their antileukemic therapy, whether they developed a brain tumor depended on their Tpmt status [8], [24]. Patients who had a low Tpmt phenotype were more likely to develop a brain tumor as a late complication than those who had high Tpmt (42.9% versus 15.8% 10-year cumulative incidence, respectively) [8].
Tpmt is constitutively expressed in several tissues including blood, kidney, liver, and brain [25], [26], [27], [28], [29]. The association of Tpmt status with brain tumor risk after thiopurine exposure prompts the question of whether Tpmt genotypes can predict thiopurine drug phenotypes in astroglial cells. The relevance of Tpmt status to thiopurine-associated phenotypes in important brain cell populations has not been studied. Indeed, the first step to address the question of whether Tpmt plays any role in brain cancer risk after thiopurines is to determine whether Tpmt phenotypes in the brain are similar to those in other tissues. Thiopurine drug-associated genotoxicity has been linked to mutagenesis and transformative events [30], [31], [32]. We hypothesize that Tpmt deficiency can lead to greater genotoxicity (i.e. DNA damage) and cytotoxicity in astroglial cells after thiopurine exposure. To address this hypothesis we used primary astrocytes isolated from transgenic mice of each Tpmt genotype and performed in vitro studies to compare thiopurine-induced cytotoxicity and DNA damage between Tpmt genotypes. We also used established human glioma cell lines as model astroglial cells to validate the findings observed in primary mouse astrocytes.
Results
Characterization of Tpmt+/+, Tpmt+/−, and Tpmt−/− primary mouse astrocyte cultures
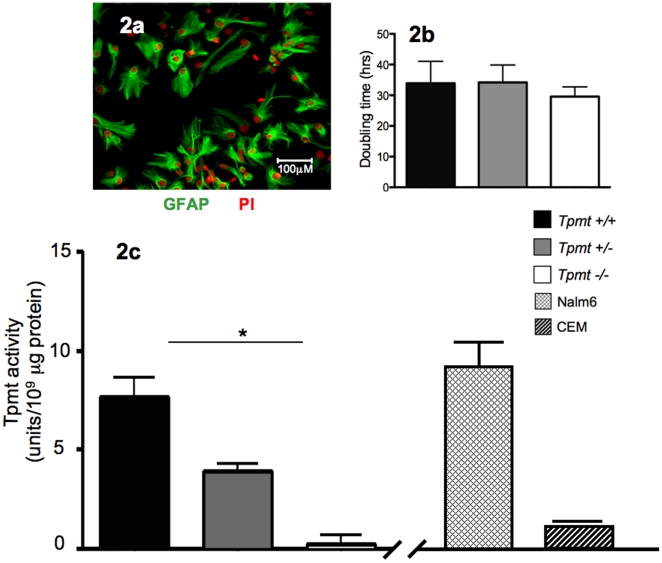
The primary mechanism of thiopurine cytoxicity is believed to be through incorportation of TG nucleotide metabolites into DNA [2], [31], [33](Fig. 1), and therefore is dependent on the rate of cell proliferation. Hence, we first compared the doubling rates between Tpmt+/+, Tpmt+/−, and Tpmt−/− primary astrocyte cultures. Primary cultures were established from mouse cortices of each Tpmt genotype and astrocyte purity was confirmed by GFAP (glial fibrillary acid protein, an astrocyte marker) fluorescent staining (Fig. 2a). We found that there was no significant difference in growth rates between cultures of different Tpmt genotypes (Fig. 2b). The mean doubling rates for Tpmt+/+, Tpmt+/− and Tpmt−/− cultures were 33.9, 34.2, and 29.6 hours, respectively (p = 0.89). Next, we measured the level of Tpmt protein activity using a well-established radiochemical assay that measures the conversion of 6-mercaptopurine to 6-methylmercaptopurine using [14C] S-adenosyl-L-methionine (SAM) as a methyl donor [34]. In agreement with previous studies comparing protein activity in hematological cells of different Tpmt genotypes [5], [29], [35], protein activities in astrocytes were significantly different and were predicted by Tpmt genotype (Fig. 2c). Tpmt+/+, Tpmt+/−, and Tpmt−/− primary astrocytes displayed high (7.6 unit/109 µg protein), intermediate (3.9 unit/109 µg protein), and low (0.25 unit/109 µg protein) levels of protein activity, respectively (p = 0.018). Based on these data, we next determined whether Tpmt could modulate the cytotoxic effects of TG in primary astrocytes.
Figure 2. Characterization of Tpmt+/+, Tpmt+/−, and Tpmt−/− primary astrocyte cultures.
(a) Micrograph image (200×) of primary astrocytes showing high astrocyte purity determined by GFAP (green) and propidium iodide (red) fluorescent staining. (b) There was no significant difference in growth rates between astrocyte cultures of each Tpmt genotype (p = 0.89). (c) The level of Tpmt protein activity was significantly different between the three Tpmt genotypes (p = 0.018); and levels were comparable to Nalm6 and CEM control lymphoblastoid cell lines. Bars represent the means; whiskers represent the standard deviation (±); * p<0.05.
Comparison of thioguanine-associated cytotoxicity in Tpmt+/+, Tpmt+/−, and Tpmt−/− primary astrocyte cultures
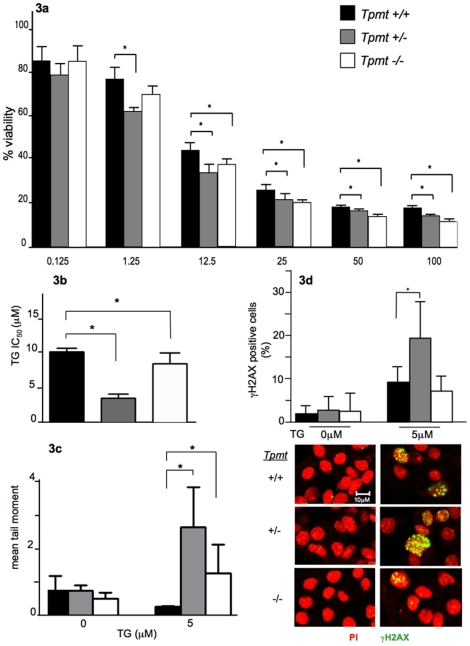
To determine whether Tpmt could predict thiopurine cytotoxicity in primary astrocytes, the MTT colorimetric assay was employed to compare TG sensitivity between astrocytes of each Tpmt genotype. This assay measures cell viability by quantitating the extent at which metabolically active cells can reduce tetrazolium salt (MTT) to form purple formazan crystals. Astrocytes were exposed to escalating concentrations of TG (0.125, 1.25, 12.5, 25, 50, and 100 µM) for five days and then subjected to the MTT assay. Cell viability data at each TG concentration was used to calculate IC50 values. As expected, TG treatment resulted in cytotoxicity in a dose dependant manner (Figure 3a). Cell viability was significantly lower in Tpmt+/− and Tpmt−/− cultures at four concentrations of TG (12.5, 25, 50, and 100 µM) when compared to Tpmt+/+ cultures (p<0.04). Interestingly, Tpmt+/− but not Tpmt−/− astrocytes were significantly more sensitive at 1.25 µM of TG than Tpmt+/+ astrocytes (p = 0.004); and this finding was reflected in corresponding IC50 values. There was a 2.9 fold reduction in IC50 in Tpmt+/− (3.5 µM TG) versus a 1.2 fold reduction in Tpmt−/− (8.6 µM TG) when compared Tpmt+/+ astrocytes (10.3 µM TG); and the Tpmt+/− and Tpmt−/− IC50 values were significantly different from Tpmt+/+ (p<0.05) (Fig. 3b). Studies have shown that genotoxicity (i.e. DNA strand breaks) correlates with cytotoxicity after TG exposure [33], [36]. Hence, we investigated whether the degree of TG-induced DNA strand breaks was associated with the level of cytotoxicity in primary astrocytes.
Figure 3. Comparison of thioguanine phenotypes in primary astrocyte cultures of each Tpmt genotype.
(a) Primary astrocytes of each Tpmt genotype were exposed to thioguanine (TG) and cell viability was determined using the MTT assay. Tpmt+/− and Tpmt−/− astrocytes were significantly more sensitive to TG (12.5–100 µM) than Tpmt+/+ (p<0.04). (b) IC50 values were significantly different between the three genotypes (p = 0.027) and Tpmt+/− astrocytes were the most sensitive. (c) The alkaline comet assay was used to compare TG-induced DNA damage between genotypes. Tpmt+/− and Tpmt−/− astrocytes had significantly higher mean comet tail moments than Tpmt+/+ astrocytes (p = 0.033). (d) Immunoflourescence staining for γH2AX confirms IC50 and comet data. Tpmt+/− astrocytes had greater damage than Tpmt+/+(p = 0.03). Below are representative images of γH2AX staining for control treated (left) and TG treated (right) astrocytes. γH2AX foci stained in green; nuclei stained in red (Zeiss LSM 710 Laser Scanning Microscopes, 400× magnification). Bars represent the means; whiskers represent the standard deviation (±); * p<0.05.
Comparison of the extent of DNA damage between Tpmt+/+, Tpmt+/−, and Tpmt−/− primary astrocytes
To address the question of whether the level of TG-induced cytotoxicity is correlated with DNA damage, we employed the alkaline comet assay. This assay allows for the quantitation of DNA damage [i.e. single strand breaks (SSBs), double strand breaks (DSBs) and alkali labile sites] by using fluorescent microscopy to visualize the extent of DNA migration from the nucleus of individual cells embedded in agarose [37]. We exposed astrocytes to 5 µM of TG for 72 hours. IC50 data was used to select a dose that would allow us to capture damage in each genotype. 72 hour time-point was selected to allow at least two cell doublings for TG incorporation and to capture cell damage prior to cell death. We observed a significant difference in DNA damage between the three genotypes as measured by mean comet tail moment. Both Tpmt+/− and Tpmt−/− astrocytes had a significantly greater degree of DNA damage than Tpmt+/+ astrocytes (mean comet tail moment = 2.64, 1.26, and 0.4, respectively; p = 0.034) (Fig. 3c). In agreement with MTT studies, Tpmt+/− astrocytes were more sensitive to TG, as evidenced by a greater degree of DNA damage, than were Tpmt−/− astrocytes. Although the difference in DNA damage between Tpmt+/− and Tpmt−/− astrocytes was not significantly different (p = 0.083). There was no difference in DNA damage between Tpmt genotypes of control treated astrocytes.
Studies have shown that the predominant DNA lesions caused by TG are SSBs although some DSB lesions are formed [33]. Therefore, we then compared γH2AX (an established DSB marker) foci staining after TG exposure to determine the extent of the DNA lesions that are in the form of DSBs and whether this damage is different between Tpmt genotypes. Astrocytes were seeded on glass coverslips and exposed to either 0 or 5 µM of TG. We found that Tpmt+/− astrocytes had the greatest degree of foci staining (19.4%) compared to Tpmt+/+ (9.3%), and Tpmt−/− (7.22%) (Fig. 3d). In agreement with comet data, Tpmt+/− was the most sensitive genotype. There was a significant difference between Tpmt+/− and Tpmt+/+ (p = 0.03) but no significant difference was observed between Tpmt−/− and Tpmt+/+ cells.
Comparison of thioguanine phenotypes between human glioma cell lines with different Tpmt protein activities
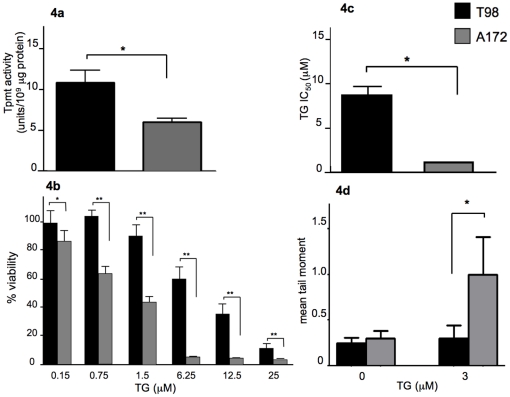
To determine whether our findings could be recapitulated in human cell lines, we used established human glioma cell lines as model astroglia and conducted MTT and comet experiments. We first identified cell lines with high and low Tpmt activity and compared TG-induced cytotoxicity and DNA damage between them. First, we measured Tpmt protein activity in four human glioma cell lines (T98, A172, U87, and LN18) to identify a high and low Tpmt expresser. We chose T98 (10.48 unit/109 µg protein) and A172 (6.15 unit/109 µg protein) as the high and low expressers, respectively (Fig. 4a). We then subjected T98 and A172 to the MTT assay to measure TG-induced cytotoxicity. Both cell lines were exposed to escalating concentrations of TG (0.15, 0.75, 1.5, 6.25, 12.5 and 25 µM) for five days. The cell viability was determined for each concentration and these data were used to calculate IC50 values. As predicted by Tpmt activity and in agreement with MTT studies performed in primary astrocytes, A172 had significantly lower cell viabilities at each TG concentration as compared to T98 (p<0.05) (Fig. 4b). The higher TG sensitivity of A172 was also reflected in the IC50 values. A172 displayed a 6.9 fold lower IC50 (1.2 µM) when compared to T98 (8.3 µM) (Fig. 4c).
Figure 4. Assessment of Tpmt-associated phenotypes in model astroglial cell lines.
Tpmt protein activity was measured in T98 and A172 glioma cell lines. (a) T98 displayed a significantly higher level of protein activity than A172 (p<0.05). (b) As observed in primary astrocyte cultures, low Tpmt protein activity is associated with greater cytotoxicity as determined by the MTT assay; (c) and reflected in IC50 values. A172 had a significantly more sensitive at all TG concentrations and lower IC50 than T98 (p<0.05). (d) The alkaline comet assay was used to compare TG-induced DNA damage. A172 displayed a significantly greater degree of TG-induced DNA strand breaks than T98 (p = 0.02) as measured by mean comet tail moment. Bars represent the means; whiskers represent the standard deviation (±); * p<0.05; ** p<0.005.
Next, we employed the comet assay to compare DNA damage between T98 and A172 after exposing cells to 3 µM TG for 72 hours. In agreement with primary astrocyte data, a low level of Tpmt activity led to significantly more DNA damage compared to high activity (mean comet tail moment: A172 = 1, T98 = 0.3; p = 0.02) (Fig. 4d). DNA damage in control treated cells was not significantly different between the two cell lines.
Comparison of thioguanine phenotypes between A172mock and A172Tpmt isogenic cell lines
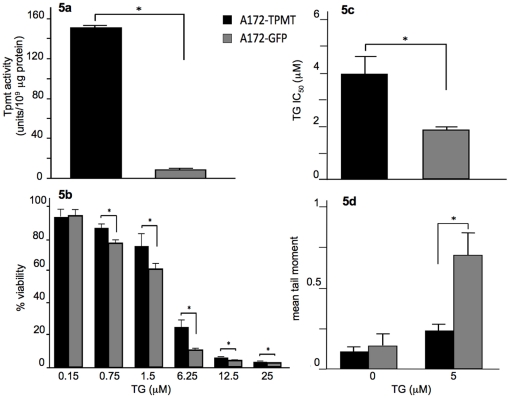
To further validate our findings, TG phenotypes were compared using isogenic cell lines. A172 was the lowest Tpmt expresser, hence we over-expressed Tpmt in this cell line and compared TG-induced cytotoxicity and DNA damage between the mock transduced (A172mock) and Tpmt transduced (A172Tpmt) cell lines. We performed growth rate studies and found that there were no differences in doubling times between A172Tpmt and A172mock (28.5 and 29.6 hours, respectively; p = 0.38). The level of Tpmt protein activity in A172Tpmt was significantly higher as compared to A172mock (151.23 unit/109 µg protein versus 9.47 unit/109 µg protein, respectively; p<0.05) (Fig. 5a). When A172mock and A172Tpmt were exposed to escalating concentrations of TG for five days we found that A172mock had significantly lower cell viabilities at 0.75, 1.5, 6.25, 12.5 and 25 µM of TG as compared to A172Tpmt (p<0.005) (Fig. 5b). These findings were reflected in the A172Tpmt IC50 value that was approximately two-fold higher than A172mock (3.9 µM versus 1.9 µM, respectively; p<0.05)(Fig. 5c).
Figure 5. Comparison of Tpmt-associated phenotypes in A172 isogenic cells.
Tpmt protein activity was measured in A172mock and A172Tpmt cell lines. (a) Tpmt protein activity was significantly higher in A172Tpmt (p<0.05); additionally, (b) A172Tpmt showed a significantly higher resistance to TG-induced toxicity when compared to A172mock as determined by the MTT cell viability data (p<0.005) and (c) by IC50 values (p<0.05). (d) The alkaline comet assay was used to compare TG-induced DNA damage. A172mock displayed a significantly higher DNA damage than A172Tpmt (p = 0.03) as measured by mean comet tail moment. Bars represent the means; whiskers represent the standard deviation (±). * p<0.05.
Next, we exposed A172Tpmt and A172mock to 5 µM of TG for 72 hours and compared the extent of DNA damage using the comet assay. We selected 5 µM based on the IC50 value of A172Tpmt. As expected, A172mock had a significantly greater degree of DNA damage when compared to A172Tpmt (mean comet tail moment: A172mock = 0.7, A172Tpmt = 0.24; p = 0.03) (Fig. 5d). DNA damage in control treated cells was not significantly different between the two cell lines (p = 0.35). These data support our findings observed in both the primary mouse astrocytes and the glioma cell lines (T98 and wild-type A172) that when Tpmt protein activity is low a greater degree of cytotoxicity and DNA damage can occur after TG exposure.
Discussion
Tpmt is a cytoplasmic enzyme responsible for deactivating thiopurine drugs and a low Tpmt phenotype can lead to life-threatening hematological toxicities when thiopurines are given. Indeed, chronic administration of thiopurines has been linked to an increased risk of cancer including cancers of the brain [8], [15], [17], [22], [24]. Unfortunately, not all patient populations that are treated with thiopurines are evaluated for their Tpmt status prior to initiating thiopurine therapy; hence, the importance of Tpmt to thiopurine-associated cancer risk is not clear. This fact coupled with the fact that Tpmt deficiency is relatively rare across populations, makes it difficult to assess the importance of Tpmt status to thiopurine therapy-related cancer risk. However, in a report by Relling et al whether patients developed a brain tumor as a late complication post-antileukemic therapy depended on their Tpmt status [24]. In this report, patients who were determined to be at high risk for treatment relapse received prophylactic cranial irradiation concurrent with antimetabolite therapy; and those patients with a low Tpmt had a surprisingly high incidence of brain tumors as a late complication. Indeed, ionizing radiation in itself is a risk factor for brain tumorigenesis. However, in this report, patients with low Tpmt had a 42.9% incidence of brain tumors compared to 15.8% for those with high Tpmt.
Astrocytomas are the most common brain tumor type that has been reported in patients after thiopurine therapy [8], [15], [22], [24]. The importance of Tpmt status to thiopurine-associated phenotypes in astroglial cells has never been studied. We assessed Tpmt and TG phenotypes in astroglial cells using primary mouse astrocytes of each Tpmt genotype and human glioma cells of different Tpmt phenotypes. In this study we have shown that TG induces cytotoxicity in a dose-dependent manner in astroglial cells; and cells with a low Tpmt phenotype (Tpmt+/− and Tpmt−/− astrocytes and A172mock) were significantly more sensitive. We also showed that TG exposure induced more DNA damage in the form of SSBs in astroglial cells with low Tpmt than in cells with high Tpmt. DNA damage in the form of DSBs was significantly different between astroglial cells with low and high Tpmt. SSBs have been shown to be the primary DNA lesion induced by thiopurine drugs [33].
Interestingly, we found that low but not the complete absence of Tpmt sensitized primary astrocytes to a higher degree of cytotoxicity and genotoxicity (i.e. IC50, SSBs and DSBs) after TG exposure. This discordance could possibly be explained by the induction of a compensatory mechanism that deactivates thiopurines with the complete loss of Tpmt. Another possible explanation could be a defect in the mismatch repair (MMR) system's ability to recognize DNA-TG nucleotide lesions. The MMR system is responsible for recognizing DNA mis-pairing caused by DNA-TG nucleotides and the subsequent signaling for cell death [38], [39], [40], [41]. The latter possibility suggests that astroglial cells with a defective Tpmt could have DNA lesions persisting in the genome that could potentially lead to mutagenesis and cancer. Indeed, some studies suggest that heterozygous children develop brain tumors and other cancers more often than children with mutant Tpmt. However, the frequency of heterozygous or mutant Tpmt is very low making it is difficult to make any definitive conclusions [24], [42]. It would be interesting to investigate whether Tpmt status influences the extent of TG nucleotide incorporation into DNA and damage recognition by the MMR system as well as whether ionizing irradiation alone or combined with thiopurines results in similar findings.
In summary, chronic thiopurine therapy has been associated with the development of brain cancer and Tpmt status has been linked to this risk. We found that Tpmt is an important factor for thiopurine drug-induced cytotoxicity and genotoxicity in astroglial cells. Even though glioma cells can exhibit a number of genetic alternations that could contribute to the phenotypes observed, our study shows that Tpmt is indeed an important determinant of thiopurine toxicity. Low Tpmt can increase the level of TG-induced genotoxic damage in both normal and in neoplastic astroglial cells. Indeed, the observations that glioma cells can have variable degrees of Tpmt protein activity and that low protein activity can cause cells to be more sensitive to TG poses the question of whether thiopurines may potentially have a therapeutic role in low Tpmt expressing glial tumors. In vitro studies have shown that thiopurine drugs are associated with mutagenic events in a variety of cell lines [30], [36]. It is possible that the DNA damage caused by low Tpmt function can ultimately contribute to transformative events over time. One study found that Tpmt was not an important factor for cancer risk after thiopurine therapy [42]; however in this study only Tpmt genotype was determined which may have excluded patients with discordance between Tpmt genotype and phenotype. Individuals who are wild-type for Tpmt can display a wide variation in their Tpmt phenotype [43]. Further studies are needed to understand the importance of Tpmt in the brain when thiopurines are given. Whether theses findings can be replicated in vivo and whether factors important to DNA repair mechanisms are responsible for the TG phenotypes described in this study should also be investigated.
Materials and Methods
Animals
The Tpmt knockout mice were a generous gift of Dr. Mary Relling (St. Jude Children's Research Hospital) and the generation of these mice has been described previously [35]. The Tpmt+/−, and Tpmt−/− mice were indistinguishable from Tpmt+/+ mice by appearance, life span, and organ histology (i.e. brain, liver, spleen, intestine, thymus, and lymph nodes). Animals were maintained in accordance to NIH guidelines for the care and use of laboratory animals and were housed in an AAALAC accredited facility. The University of Tennessee Health Science Center Institutional Animal Care and Use Committee approved all animal procedures.
Primary astrocyte cultures and cell lines
Astrocyte cultures were established from 2–5 day old pups of each Tpmt genotype. The DNA extracted from tails snips was used to confirm Tpmt genotype. A dissecting microscope was used to remove the meninges and hippocampus and the cortices were mechanically dissociated. Subsequently, cultures were established and maintained in Dulbecco's modified Eagle's medium/Ham's F-12 50/50 mix (Cellgro) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 IU/ml penicillin, 100 µg/ml streptomycin and 20 ng/ml epidermal growth factor (Millipore) and grown in Primaria flasks, (BD biosciences). Cells were refed with 20 ng/ml epidermal growth factor after three days of incubation, and the media was changed after five days. At passage 2, three individual primary cultures of the same genotype were pooled and seeded for experiments. Only cultures with greater than 80% astrocyte purity as determined by GFAP immunohistochemistry were included in experiments.
The T98 and A172 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). T98 was grown in Minimum Essential Medium Eagle (Cellgro), supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 IU/ml penicillin, and 100 µg/ml streptomycin. A172 was grown in Dulbecco's Modified Eagle's Medium (Cellgro), supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 IU/ml penicillin, and 100 µg/ml streptomycin. All cell cultures were maintained at 37°C in a 5% CO2 humidified atmosphere.
Lentiviral transduction
Human Tpmt cDNA was obtained by digesting pCMV6-Tpmt plasmid (Origene) at BamHI and EcoRI sites. Tpmt cDNA was then amplified by PCR, purified through agarose gel electrophoresis, and subcloned into BamHI and EcoRI sites of pLenti-CMV-pgk-puro vector (Viral vector core, UTHSC). The plenti-Tpmt-pgk-puro and plenti-GFP-pgk-puro lentivial vectors were produced by packaging in 293T cells [44]. The lentiviral vectors were used to transduce the A172 cell line in the presence of 6 µg/mL polybrene (Sigma). Transduction efficiency was monitored by GFP expression using fluorescent microscope. The mock- (A172mock) and Tpmt-transduced (A172Tpmt) cell lines were seeded for experiments at comparable passage numbers and Tpmt activity was measured prior to the experiments.
Cell proliferation comparison
The doubling rates for primary astrocyte cultures were determined and used as an index of cell proliferation. At passage two, 75,000 cells/well were seeded in duplicate 6-well plates. Every 24 hours the cells from one well of each plate were harvested using 0.05% Trypsin-EDTA (GIBCO) and viable cells were counted by trypan blue exclusion for four consecutive days. Doubling rates were calculated using the following formula: Doubling time (hours) = (T2-T1)/[log2×(Log N-Log N0)]. Where, T2 = Harvesting time, T1 = Initial time, N = Final cell concentration, and N0 = Initial cell concentration. The mean doubling rate was calculated from replicate experiments using two different pooled cultures established on different days.
TPMT activity assay
Primary mouse astrocyte and human glioma cell lines were sonicated using a probe sonicator (Masonix XL-2000 series) prior to performing the Tpmt activity assay. The level of Tpmt activity was determined using a pre-established non-chelated radiochemical assay [26], [34] with slight modifications. Tpmt activity was quantitated by measuring the extent of conversion of 6-mercaptopurine (Sigma) to radioactively labeled 6-methylmercaptopurine with [14C] S-adenosyl-L-methionine (SAM) (Perkin-Elmer). The protein concentration of each lysate was measured prior to performing the assay and was used to calculate Tpmt activity. The mean activities for each genotype were calculated using data from triplicate inter-day studies of three separate pooled cultures per Tpmt genotype.
MTT assay
Cell viability was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma). Briefly, 1000 cells were seeded per well in 96-well plates. Plates were pre-coated with poly-L-ornithine (Sigma) and Laminin (Invitrogen) for primary astrocytes to facilitate cell attachment. A fifteen millimolar stock solution of TG (Sigma) was prepared by dissolving TG in 0.1 N NaOH. Cells were allowed to attach over night and TG or vehicle control was added to the appropriate wells. After five days of drug exposure, MTT was added and allowed to incubate for 3 hours. Afterwards, the media was carefully aspirated and the purple formazan crystals were dissolved in 100 µl DMSO (Fisher). Absorbance was measured at wavelength of 570/690 nm using a FLx800 fluorescence microplate reader (BioTek Instruments, Inc.).
Alkaline comet assay
Techniques used to perform the comet assay were adopted from the laboratory of Dr. Peter Mckinnon (St. Jude Children's Research Hospital) [45]. Briefly, 50,000 primary astrocyte or 30,000 human astroglial cells were seeded in Primaria 24-well plates (BD Biosciences) or in Falcon 24-well plates (BD Biosciences), respectively. Cells were allowed to attach overnight and then exposed to TG for three days. After three days of drug treatment, cells were harvested and counted to prepare a 3×105 cells/ml suspension in PBS. The cell suspension was mixed with 1.2% ultra-pure low melting point agarose (Invitrogen) and casted onto chilled fully frosted glass slides (Fisher) pre-coated with 0.6% agarose (Bio-Rad). The cells were then lysed for 1.5 hours at 4°C, in lysis buffer (100 µM EDTA, 2.5 M NaCl, 10 mM Tris, 1.3% triton X-100 and 3.3% DMSO). The slides were washed twice with distilled water and incubated for 45 minutes at 4°C in electrophoresis buffer (1 mM EDTA, 50 mM NaOH and 1% DMSO). Electrophoresis was conducted at 12 V and ∼90 mA at 4°C for 25 minutes. Slides were incubated for one hour in 400 mM Tri-HCl neutralization buffer followed by a 20 minute incubation in SYBR green I (Sigma). Images were captured using a Nikon eclipse TE300 fluorescent microscope. All experiments were replicated using two different sets of pooled cultures. The extent of DNA damage was expressed as the comet tail moment (the amount and distribution of DNA in the tail). The comet tail moment was measured in a minimum of 60 cells using Cometscore version 1.5 (Tri Tek corp).
Immunohistochemistry
Primary astrocytes were grown on poly-D-lysine chamber slides (BD Biocoat) for GFAP staining (to confirm astrocyte purity) or on glass coverslips and treated with 0 or 5 µM of TG for γH2AX staining. Cells were fixed with 4% PFA/PBS for 10 minutes, permeabilized with 0.5% Triton X-100/PBS for 5 minutes. Cells were then washed with PBS and blocked with 3% BSA/PBS for one hour. Cells were then immunostained with either anti-GFAP (1∶500 in PBS, Sigma) for two hours or anti-γH2AX (1∶1000 in 3%BSA, Milipore) for one hour. Subsequently, cells were washed with PBS and incubated for one hour with in Cy2-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch, 1∶250 in PBS containing 2% donkey serum and 0.1% Triton X-100) for GFAP or with donkey anti-mouse secondary antibody labeled with Alexa 488 (In vitrogen, 1∶800 in 3% BSA) for one hour. Finally, vectashield mounting media (Vectorlabs) containing propidium iodide was used to stain nuclei. GFAP slides were visualized and images were captured using Nikon eclipse TE300 fluorescent microscope. Images were captured for γH2AX stained slides using a LSM-710 confocal microscope (Zeiss). Images were analyzed for γH2AX foci by counting a minimum of 100 cells per genotype. Cells containing fewer than five foci were excluded to normalize for baseline staining observed in control treated cells. Cells with “pan-staining” were considered to have more than 5 foci per cell. Hence, the percent of cells with γH2AX foci staining represents the percent of cells with greater than five foci per cell.
Statistical analysis
Differences in cytotoxicity and DNA damage among all three groups were compared using Kruskal-Wallis analysis of ranks. Mann-Whitney U test was used when only two groups were compared. Significance was declared when p<0.05. All statistical analyses were performed using SAS 9.2 (SAS Institute Inc., Cary, NC).
Acknowledgments
We thank Dr. Sachin Katyal, Dr. Qhong Zhou, Zorica Janjetovic, and Christian Muenyi for their technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was funded by the Center for Translational Science Institute, University of Tennessee Health Science Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Remy CN. Metabolism of thiopyrimidines and thiopurines. S-Methylation with S-adenosylmethionine transmethylase and catabolism in mammalian tissues. J Biol Chem. 1963;238:1078–1084. [PubMed] [Google Scholar]
- 2.Tidd DM, Paterson AR. A biochemical mechanism for the delayed cytotoxic reaction of 6-mercaptopurine. Cancer Res. 1974;34:738–746. [PubMed] [Google Scholar]
- 3.Krynetski EY, Tai HL, Yates CR, Fessing MY, Loennechen T, et al. Genetic polymorphism of thiopurine S-methyltransferase: clinical importance and molecular mechanisms. Pharmacogenetics. 1996;6:279–290. doi: 10.1097/00008571-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, et al. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 5.Otterness D, Szumlanski C, Lennard L, Klemetsdal B, Aarbakke J, et al. Human thiopurine methyltransferase pharmacogenetics: gene sequence polymorphisms. Clin Pharmacol Ther. 1997;62:60–73. doi: 10.1016/S0009-9236(97)90152-1. [DOI] [PubMed] [Google Scholar]
- 6.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 7.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 8.Hijiya N, Hudson MM, Lensing S, Zacher M, Onciu M, et al. Cumulative incidence of secondary neoplasms as a first event after childhood acute lymphoblastic leukemia. Jama. 2007;297:1207–1215. doi: 10.1001/jama.297.11.1207. [DOI] [PubMed] [Google Scholar]
- 9.Tassaneeyakul W, Srimarthpirom S, Reungjui S, Chansung K, Romphruk A. Azathioprine-induced fatal myelosuppression in a renal-transplant recipient who carried heterozygous TPMT*1/*3C. Transplantation. 2003;76:265–266. doi: 10.1097/01.TP.0000070521.22024.F1. [DOI] [PubMed] [Google Scholar]
- 10.Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J Pediatr. 1991;119:985–989. doi: 10.1016/s0022-3476(05)83063-x. [DOI] [PubMed] [Google Scholar]
- 11.Schutz E, Gummert J, Mohr F, Oellerich M. Azathioprine-induced myelosuppression in thiopurine methyltransferase deficient heart transplant recipient. Lancet. 1993;341:436. doi: 10.1016/0140-6736(93)93028-y. [DOI] [PubMed] [Google Scholar]
- 12.Evans WE, Hon YY, Bomgaars L, Coutre S, Holdsworth M, et al. Preponderance of thiopurine S-methyltransferase deficiency and heterozygosity among patients intolerant to mercaptopurine or azathioprine. J Clin Oncol. 2001;19:2293–2301. doi: 10.1200/JCO.2001.19.8.2293. [DOI] [PubMed] [Google Scholar]
- 13.Smith MA, Irving PM, Marinaki AM, Sanderson JD. Review article: malignancy on thiopurine treatment with special reference to inflammatory bowel disease. Aliment Pharmacol Ther. 2010;32:119–130. doi: 10.1111/j.1365-2036.2010.04330.x. [DOI] [PubMed] [Google Scholar]
- 14.Kandiel A, Fraser AG, Korelitz BI, Brensinger C, Lewis JD. Increased risk of lymphoma among inflammatory bowel disease patients treated with azathioprine and 6-mercaptopurine. Gut. 2005;54:1121–1125. doi: 10.1136/gut.2004.049460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiesse C, Larue JR, Kriaa F, Blanchet P, Bellamy J, et al. Incidence and type of malignancies occurring after renal transplantation in conventionally and in cyclosporine-treated recipients: single-center analysis of a 20-year period in 1600 patients. Transplant Proc. 1995;27:2450–2451. [PubMed] [Google Scholar]
- 16.Guenova E, Lichte V, Hoetzenecker W, Woelbing F, Moehrle M, et al. Nodular malignant melanoma and multiple cutaneous neoplasms under immunosuppression with azathioprine. Melanoma Res. 2009;19:271–273. doi: 10.1097/CMR.0b013e32831bc44a. [DOI] [PubMed] [Google Scholar]
- 17.Snanoudj R, Durrbach A, Leblond V, Caillard S, Hurault De Ligny B, et al. Primary brain lymphomas after kidney transplantation: presentation and outcome. Transplantation. 2003;76:930–937. doi: 10.1097/01.TP.0000079253.06061.52. [DOI] [PubMed] [Google Scholar]
- 18.Penn I. Incidence and treatment of neoplasia after transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 1993;12:S328–336. [PubMed] [Google Scholar]
- 19.Walter AW, Hancock ML, Pui CH, Hudson MM, Ochs JS, et al. Secondary brain tumors in children treated for acute lymphoblastic leukemia at St Jude Children's Research Hospital. J Clin Oncol. 1998;16:3761–3767. doi: 10.1200/JCO.1998.16.12.3761. [DOI] [PubMed] [Google Scholar]
- 20.Rosso P, Terracini B, Fears TR, Jankovic M, Fossati Bellani F, et al. Second malignant tumors after elective end of therapy for a first cancer in childhood: a multicenter study in Italy. Int J Cancer. 1994;59:451–456. doi: 10.1002/ijc.2910590402. [DOI] [PubMed] [Google Scholar]
- 21.Nygaard R, Garwicz S, Haldorsen T, Hertz H, Jonmundsson GK, et al. Second malignant neoplasms in patients treated for childhood leukemia. A population-based cohort study from the Nordic countries. The Nordic Society of Pediatric Oncology and Hematology (NOPHO). Acta Paediatr Scand. 1991;80:1220–1228. doi: 10.1111/j.1651-2227.1991.tb11812.x. [DOI] [PubMed] [Google Scholar]
- 22.Vancura RW, Kepes JJ, Newell KL, Ha TM, Arnold PM. Secondary intracranial neoplasms exhibiting features of astrocytoma and neuroblastoma in 2 children treated for acute lymphoblastic leukemia: report of 2 cases. Surg Neurol. 2006;65:490–494. doi: 10.1016/j.surneu.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 23.Bo J, Schroder H, Kristinsson J, Madsen B, Szumlanski C, et al. Possible carcinogenic effect of 6-mercaptopurine on bone marrow stem cells: relation to thiopurine metabolism. Cancer. 1999;86:1080–1086. doi: 10.1002/(sici)1097-0142(19990915)86:6<1080::aid-cncr26>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Relling MV, Rubnitz JE, Rivera GK, Boyett JM, Hancock ML, et al. High incidence of secondary brain tumours after radiotherapy and antimetabolites. Lancet. 1999;354:34–39. doi: 10.1016/S0140-6736(98)11079-6. [DOI] [PubMed] [Google Scholar]
- 25.Overall RW, Kempermann G, Peirce J, Lu L, Goldowitz D, et al. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front Neurosci. 2009;3:55. doi: 10.3389/neuro.15.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szumlanski CL, Honchel R, Scott MC, Weinshilboum RM. Human liver thiopurine methyltransferase pharmacogenetics: biochemical properties, liver-erythrocyte correlation and presence of isozymes. Pharmacogenetics. 1992;2:148–159. [PubMed] [Google Scholar]
- 27.Van Loon JA, Weinshilboum RM. Thiopurine methyltransferase biochemical genetics: human lymphocyte activity. Biochemical genetics. 1982;20:637–658. doi: 10.1007/BF00483962. [DOI] [PubMed] [Google Scholar]
- 28.Van Loon JA, Szumlanski CL, Weinshilboum RM. Human kidney thiopurine methyltransferase. Photoaffinity labeling with S-adenosyl-L-methionine. Biochem Pharmacol. 1992;44:775–785. doi: 10.1016/0006-2952(92)90416-g. [DOI] [PubMed] [Google Scholar]
- 29.McLeod HL, Relling MV, Liu Q, Pui CH, Evans WE. Polymorphic thiopurine methyltransferase in erythrocytes is indicative of activity in leukemic blasts from children with acute lymphoblastic leukemia. Blood. 1995;85:1897–1902. [PubMed] [Google Scholar]
- 30.Yuan B, O'Connor TR, Wang Y. 6-Thioguanine and S-methylthioguanine are mutagenic in human cells. ACS chemical biology. 2010;5:1021–1027. doi: 10.1021/cb100214b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 32.Yuan B, Wang Y. Mutagenic and cytotoxic properties of 6-thioguanine, S6-methylthioguanine, and guanine-S6-sulfonic acid. J Biol Chem. 2008;283:23665–23670. doi: 10.1074/jbc.M804047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan T, Berry SE, Desai AB, Kinsella TJ. DNA mismatch repair (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clin Cancer Res. 2003;9:2327–2334. [PubMed] [Google Scholar]
- 34.Weinshilboum RM, Raymond FA, Pazmino PA. Human erythrocyte thiopurine methyltransferase: radiochemical microassay and biochemical properties. Clin Chim Acta. 1978;85:323–333. doi: 10.1016/0009-8981(78)90311-x. [DOI] [PubMed] [Google Scholar]
- 35.Hartford C, Vasquez E, Schwab M, Edick MJ, Rehg JE, et al. Differential effects of targeted disruption of thiopurine methyltransferase on mercaptopurine and thioguanine pharmacodynamics. Cancer Res. 2007;67:4965–4972. doi: 10.1158/0008-5472.CAN-06-3508. [DOI] [PubMed] [Google Scholar]
- 36.Christie NT, Drake S, Meyn RE, Nelson JA. 6-Thioguanine-induced DNA damage as a determinant of cytotoxicity in cultured Chinese hamster ovary cells. Cancer Res. 1984;44:3665–3671. [PubMed] [Google Scholar]
- 37.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 38.Kinsella TJ. Coordination of DNA mismatch repair and base excision repair processing of chemotherapy and radiation damage for targeting resistant cancers. Clin Cancer Res. 2009;15:1853–1859. doi: 10.1158/1078-0432.CCR-08-1307. [DOI] [PubMed] [Google Scholar]
- 39.Chalastanis A, Penard-Lacronique V, Svrcek M, Defaweux V, Antoine N, et al. Azathioprine-induced carcinogenesis in mice according to Msh2 genotype. J Natl Cancer Inst. 2010;102:1731–1740. doi: 10.1093/jnci/djq389. [DOI] [PubMed] [Google Scholar]
- 40.Cooley N, Elder RH, Povey AC. The effect of Msh2 knockdown on methylating agent induced toxicity in DNA glycosylase deficient cells. Toxicology. 2010;268:111–117. doi: 10.1016/j.tox.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 42.Stanulla M, Schaeffeler E, Moricke A, Coulthard SA, Cario G, et al. Thiopurine methyltransferase genetics is not a major risk factor for secondary malignant neoplasms after treatment of childhood acute lymphoblastic leukemia on Berlin-Frankfurt-Munster protocols. Blood. 2009;114:1314–1318. doi: 10.1182/blood-2008-12-193250. [DOI] [PubMed] [Google Scholar]
- 43.Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, et al. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14:407–417. doi: 10.1097/01.fpc.0000114745.08559.db. [DOI] [PubMed] [Google Scholar]
- 44.Yue J, Sheng Y, Ren A, Penmatsa S. A miR-21 hairpin structure-based gene knockdown vector. Biochem Biophys Res Commun. 2010;394:667–672. doi: 10.1016/j.bbrc.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]