Abstract
Purpose
To identify the genetic locus and mutation responsible for autosomal dominant cone dystrophy (adCOD) in a large Chinese family and to describe the phenotypes of the patients.
Methods
Genomic DNA and clinical data were collected from the family. Genome-wide linkage analysis was performed to map the disease locus, and Sanger dideoxy sequencing was used to detect the mutation in a candidate gene.
Results
Initially, genome-wide linkage analysis mapped the disease to 17p13.1 between D17S831 and D17S799, with a maximum lod score of 2.71 for D17S938 and D17S1852 at theta=0. Sequence analysis of the guanylate cyclase 2D gene (GUCY2D) in the linkage interval detected a recurrent heterozygous mutation, c.2513G>A (p.Arg838His). This mutation was present in all eight patients with adCOD, but neither in any of the six unaffected family members nor in 192 control chromosomes.
Conclusions
adCOD in this family is caused by a recurrent mutation in GUCY2D. adCOD can be detected in the first few years after birth in the family by fundus observation and electroretinogram recordings.
Introduction
Cone dystrophy (COD) is a retinal disease characterized by the dysfunction or degeneration of cone photoreceptors that are responsible for central and color vision. COD can be classified into two major types: stationary and progressive. Progressive COD might be difficult to differentiate from cone-rod dystrophy (CORD), since some degree of rod dysfunction develops in the advanced stage of COD [1,2]. It is suggested that COD represents retinal diseases with predominant cone dysfunction with late onset and mild rod involvement, while CORD indicates retinal degeneration with early onset cone dysfunction followed shortly thereafter by significant rod dystrophy [1]. Patients with progressive COD may complain of photophobia, decreased visual acuity, and color vision defects. Fundus changes may be nearly normal or subtle in the early stage. In the advanced stage, macular degeneration can be observed under an ophthalmoscope.
COD can be inherited as an autosomal dominant (adCOD) [3], autosomal recessive (arCOD) [4], or X-linked trait (xlCOD) [5], although it occurs sporadically in most cases. At least five loci have been designed for COD, namely COD1 (OMIM 304020) [5,6], COD2 (OMIM 300085) [7], COD3 (OMIM 602093) [3], COD4 (OMIM 613093) [4], and COD5 (OMIM 303700) [8]. Mutations in several genes have been identified to be responsible for COD, including guanylate cyclase activator 1A (GUCA1A) [3], alpha-prime cone cGMP-specific phosphodiesterase subunit (PDE6C) [4], retinitis pigmentosa GTPase regulator (RPGR) [6], red and green visual pigment genes (OPN1LW and OPN1MW) [8], and guanylate cyclase 2D (GUCY2D) [9,10]. However, no mutation in GUCY2D has been reported in Chinese patients with COD or CORD.
In the present study, progressive COD was found in a three-generation Chinese family with eight affected individuals. A genome-wide linkage study mapped the COD locus to 17p13.1. Sequencing the candidate gene in the linkage interval identified a recurrent c.2513G>A (p.Arg838His) mutation in GUCY2D (OMIM 600179).
Methods
Family with cone dystrophy
adCOD was identified in a three-generation family living in a small town in Guandong province, China. Eight patients and six unaffected individuals in the family participated in this study. Written informed consent was obtained from the participating individuals or their guardians before the collection of clinical data and genomic samples. This study was approved by the Internal Review Board of the Zhongshan Ophthalmic Center, Sun Yat-sen University, Guangzhou, China and followed the tenets of the Declaration of Helsinki and the Guidance of Sample Collection of Human Genetic Diseases (863-Plan) of the Ministry of Public Health of China. Genomic DNA was prepared from venous leukocytes.
Genotyping and linkage analysis
Genotyping for 14 family members was performed using 5′-fluorescently labeled microsatellite markers, as previous described [11]. Briefly, a genome-wide scan was performed using panels 1 to 27 of the ABI PRISM linkage Mapping Set Version 2 (Applied Biosystems, Foster City, CA). PCR was conducted at 94 °C for 8 min, followed by 10 cycles of amplification at 94 °C 15 s, 55 °C 15 s, and 72 °C 30 s; then 20 cycles at 89 °C 15 s, 55 °C 15 s, 72 °C 30 s; and finally at 72 °C for 10 min. After mixing with GENESCANTM 400HD [ROXTM] standard (Applied Biosystems) and deionized formamide, the amplicons were denatured at 95 °C for 5 min and then immediately placed on ice for 5 min. The amplicons were separated on an ABI 3100 Genetic Analyzer (Applied Biosystems). Genotyping data were analyzed using the Gene Mapper version 3.5 software package (Applied Biosystems). Two-point linkage analysis was performed by using the MLINK program of the FASTLINK implementation of the LINKAGE program package [12,13]. COD in the family was analyzed as an autosomal dominant trait with full penetrance and with a disease-gene allele frequency of 0.0001. Haplotypes were generated using the Cyrillic 2.1 program (Cyrillic Software, Wallingford, UK) and confirmed by inspection.
Mutation identification in GUCY2D
PCR was used to amplify the genomic fragments of GUCY2D. Primers employed to amplify the 19 coding exons and their adjacent intronic region of GUCY2D were the same as those previously reported [14]. PCR amplifications were performed in 20 µl reactions containing 80 ng genomic DNA. Touchdown PCR amplification consisted of a denaturizing step at 95 °C for 5 min, followed by 35 cycles of amplification (at 95 °C for 30 s; at 64–57 °C for 30 s, starting from 64 °C and decreasing by 0.5 °C with every cycle for 14 cycles until remaining at 57 °C for 21 cycles; and at 72 °C for 40 s) and a final extension at 72 °C for 10 min. The nucleotide sequences of the amplicons were determined with the ABI BigDye Terminator cycle sequencing kit v3.1 (Applied Biosystems), electrophoresed on an ABI3100 Genetic Analyzer (Applied Biosystems), and analyzed with Seqman software (Lasergene 8.0; DNASTAR, Madison, WI). Any variant detected was initially confirmed by bidirectional sequencing and then evaluated in 192 control chromosomes of 96 normal individuals. Mutation description followed the recommendation of the Human Genomic Variation Society (HGVS).
Results
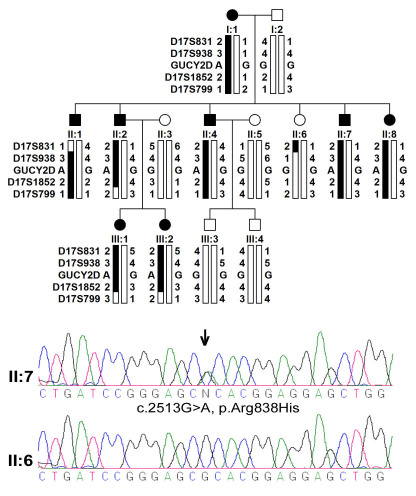
Fourteen individuals in the family participated in this study (Figure 1). The disease in the family had been passed in three generations. Eight individuals in the family were considered to be affected with adCOD based on clinical information (Table 1; Figure 2, and Figure 3).
Figure 1.
Pedigree, genotyping haplotypes on 17p13.1, and GUCY2D mutation. Pedigree and haplotypes are shown at the top. Filled squares (male) or circles (female) represent individuals affected with cone dystrophy. Bars filled with black indicate a disease-associated allele. The sequence chromatograph of GUCY2D is shown at bottom, in which the double peaks (arrow indicated) demonstrated the heterozygous recurrent mutation in GUCY2D.
Table 1. Clinical data of the patients in the Chinese family with an Arg838His mutation in GUCY2D.
| ID# | Gender | Age (years) at | First symptom | Visual acuity | Refraction (diopters) | Fundus changes | ERG responses | Color vision | ||
|---|---|---|---|---|---|---|---|---|---|---|
| |
|
exam |
onset |
|
|
|
|
Cones |
Rods |
|
| I:1 |
F |
54 |
Early childhood |
photophobia |
CF; CF* |
−10.0; −10.0 |
macular atrophy |
N/A |
N/A |
N/A |
| II:1 |
M |
31 |
Early childhood |
photophobia |
0.04; 0.04 |
−11.0; −10.0 |
macular atrophy |
N/A |
N/A |
abnormal |
| II:2 |
M |
29 |
8 years |
photophobia |
0.05; 0.1 |
−8.75; −7.00 |
macular atrophy |
reduced |
normal |
abnormal |
| II:4 |
M |
28 |
7 years |
photophobia |
0.03; 0.03 |
−7.00; −4.75 |
macular atrophy |
reduced |
normal |
abnormal |
| II:7 |
M |
26 |
Early childhood |
photophobia |
0.05; 0.04 |
−2.75; −0.25 |
macular atrophy |
reduced |
normal |
abnormal |
| II:8 |
F |
19 |
6yrs |
photophobia |
0.1; 0.1 |
−6.00; −7.00 |
macular atrophy |
N/A |
N/A |
abnormal |
| III:1 |
F |
2.5 |
N/A |
no symptom |
N/A |
+0.25; plano |
macular white spots |
reduced |
normal |
N/A |
| III:2 | F | 0.75 | N/A | no symptom | N/A | −2.25; −1.00 | macular white spots | reduced | normal | N/A |
Note: *This patient had horizontal nystagmus and mild cortical opacity of both lenses.
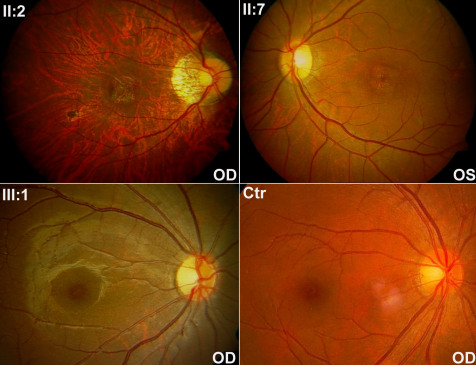
Figure 2.
Fundus photos of three patients and a normal control. II:2 and II:7 demonstrated the typical macular atrophy observed in the six adult patients. III:1 at two and half years old showed mild granular retinal pigment epithelial changes, with the tiny yellowish-white deposits in the macula observed in the two youngest patients. Temporal pallor of the optic disc was observed in patients II:2, II:7, and III:1. Mild artery attenuation was noticed in II:2 and II:7. Ctr: fundus photo of a normal control.
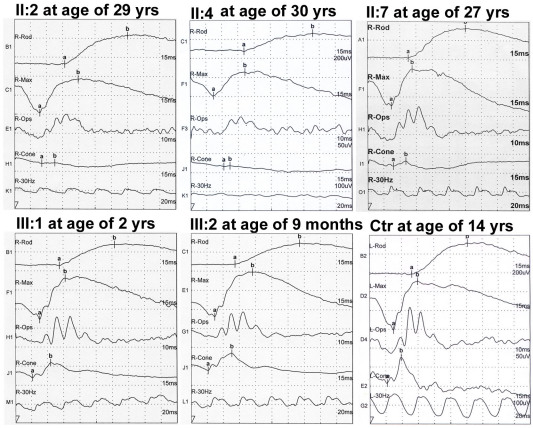
Figure 3.
Electroretinography of five patients and a normal control. Reduced photopic responses, abnormal 30 Hz ERG, and normal scotopic Electroretinography (ERG) were shown in all five patients with ERG recordings (II:2, II:4, II:7, III:1, and III:2). Ctr: A normal control.
Two young affected girls (III:1 and III:2) in the family, aged at two and a half years and nine months, respectively, had no visual symptom at the time of examination (Figure 1 and Table 1). However, fundus observation revealed carpet-like changes with multiple fine yellowish-white spots in the macular region, as well as pallor in the temporal optic discs of these two girls (Figure 2). Electroretinography (ERG) recordings demonstrated reduced cone responses that confirmed the clinical findings (Figure 3).
For the other six adult patients, all had photophobia and blurred vision noticed at about 6 to 8 years old, with progressively decreased visual acuity thereafter. Night vision in these patients was well preserved. Examination of Ishihara color plates on 5 patients suggested color vision defects. Macular atrophy and temporal pallor of the optic disc were present in all six adult patients (Figure 2). Cone responses were significantly reduced in three adult patients who received ERG examination (Figure 3). Rod responses were normal in all five patients (three adults and two young girls), as measured by ERG recordings (Figure 3).
The genome-wide linkage scan excluded linkage to other regions and mapped the adCOD to 17p13.1 between D17S831 and D17S799, with a maximum lod score of 2.71 at theta=0 (Figure 1, Table 2). One gene known to cause CORD, GUCY2D, was present in the linkage interval. Subsequently, sequencing analysis of GUCY2D detected a recurrent heterozygous mutation, c.2513G>A (p.Arg838His). This mutation cosegregated with adCOD in the family with a maximum lod score of 2.71, but was not present in six unaffected family members or in 192 control chromosomes.
Table 2. Two-point lod scores of family for markers around GUCY2D.
| Position | Lod score at theta= | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Markers |
cM* |
Mb# |
0 |
0.01 |
0.05 |
0.1 |
0.2 |
0.3 |
0.4 |
| D17S849 |
0.6 |
0.43 |
-inf |
−3.61 |
−1.61 |
−0.82 |
−0.17 |
0.07 |
0.11 |
| D17S831 |
6.6 |
1.91 |
-inf |
−1.33 |
−0.07 |
0.35 |
0.56 |
0.50 |
0.30 |
| D17S938 |
14.8 |
6.25 |
2.71 |
2.67 |
2.49 |
2.25 |
1.74 |
1.16 |
0.53 |
|
GUCY2D |
|
7.91 |
2.71 |
2.67 |
2.49 |
2.25 |
1.74 |
1.16 |
0.53 |
| D17S1852 |
23.2 |
10.52 |
2.71 |
2.67 |
2.49 |
2.25 |
1.74 |
1.16 |
0.53 |
| D17S799 |
32.8 |
13.17 |
-inf |
−1.33 |
−0.07 |
0.34 |
0.54 |
0.44 |
0.22 |
| D17S921 | 37.3 | 14.26 | -inf | −1.62 | −0.35 | 0.09 | 0.33 | 0.29 | 0.14 |
Genethon, # Homo genome (Build 37.2) Chr1 Primary_Assembly
Discussion
We identified a Chinese family with eight patients showing signs of adCOD that was transmitted as autosomal dominant trait in the family. A genome-wide linkage analysis mapped the disease to 17p13.1 between D17S831 and D17S799. Subsequent mutational screening of a candidate gene in the linkage interval identified a recurrent heterozygous c.2513G>A (p.Arg838His) mutation in GUCY2D. This mutation was present in all eight patients in the family, but was absent in unaffected family members and normal controls. All of these lines of evidence support the view that a mutation in GUCY2D is responsible for the adCOD in the family.
To date, at least 125 mutations in GUCY2D have been identified to be responsible for retinal degeneration, including Leber congenital amaurosis, CORD, COD, and retinitis pigmentosa, based on HGMD® Professional 2011 accessed as of June 24, 2011. A review of the original reports revealed that 10 of the 125 mutations were associated with COD or CORD in 33 families (Table 3). For the 10 mutations in the 33 families, codon 838 accounted for six mutations (66.7%) in 29 families (90.6%). Up to the present, all patients with GUCY2D mutations at codon 838 exhibited COD or CORD except for one patient, who had c.2513G>C (p.Arg838Pro) and Leber congenital amaurosis, but no clinical details were present [15]. It is unclear if there is any functional difference among these different mutations involving codon 838.
Table 3. Reported GUCY2D mutations associated with cone or cone-rod dystrophy.
| No. | Nucleotide change | Residue change | Families | Phenotypes | References |
|---|---|---|---|---|---|
| 1 |
Unclear |
P575L |
1 |
adCOD* |
[17] |
| 2 |
2511_2512delGCinsCA |
[Glu837Asp,Arg838Ser] |
1 |
adCORD |
[10,21] |
| 3 |
2511_2516delGCGCACinsCTGCAT |
[Glu837Asp,Arg838Cys,Thr839Met] |
1 |
adCORD |
[22] |
| 4 |
2512C>T |
Arg838Cys |
16 |
11/adCORD |
[9,10,18,21,23–25] |
| |
|
|
|
5/adCOD |
[9] |
| 5 |
2512C>G |
Arg838Gly |
1 |
adCORD |
[9] |
| 6 |
2513G>A |
Arg838His |
9 |
5/adCORD |
[21,24–27] |
| |
|
|
|
4/adCOD |
[9,16] |
| 7 |
2513G>C |
Arg838Pro |
1 |
1/adCORD |
[27] |
| 8 |
2846T>C |
Ile949Thr |
1 |
arCORD |
[28] |
| 9 |
2540_2542delAGAinsTCC |
[Gln847Leu,Lys848Gln] |
1 |
adCORD |
[29] |
| 10 | 2744_2749delTCATTGinsCCATTC | [I915T,G917R] | 1 | adCORD | [24] |
Of the 33 families with GUCY2D mutations, 22 had adCORD, 1 had arCORD, and 10 had adCOD. For the 10 adCOD families, 5 had the c.2512C>T (p.Arg838Cys) mutation [9], 4 had the c.2513G>A (p.Arg838His) mutation [9,16], and 1 had p.P575L mutation [17]. The phenotypes of the patients were essentially similar but the ages at onset varied significantly. In the Chinese family, photophobia and poor vision was the initial symptom presented at age around 6 to 8 years old, which appeared earlier than those patients with the p.Arg838His mutations in previously reported families. In addition, the visual acuity for the Chinese patients was generally worse than that for patients in previous reports. The rod function in the Chinese patients was well preserved. Our study provided clinical phenotypes for the two youngest patients at ages less than two and half years old, while previous studies only provided phenotypes for patients over 10 years old.
Except for the common signs and symptoms for COD or CORD, myopia has been reported as a common sign in several families, such as myopia greater than 5D in at least one eye in 18 of 29 patients (62%) in a family with the p.Arg838Cys mutation [18], myopia greater than 6D in all 10 patients of two families with the p.Arg838His or Arg838Cys mutation [19], and moderate myopia in patients of a family with the p.[Glu837Asp, Arg838Ser] mutation [20]. In this study, high myopia was present in five of the eight patients.
In summary, adCOD was identified in a Chinese family and was caused by a recurrent mutation in GUCY2D. This is the first report of GUCY2D mutation–associated adCOD in the Chinese population. Fundus and ERG changes in the two patients less than three years old provide the earliest signs for adCOD, which would be valuable in clinical diagnosis.
Acknowledgments
The authors thank all patients and family members for their participation. This study was supported by the National Natural Science Foundation of China (30800615 to X.X.), the National Science Fund for Distinguished Young Scholars (30725044 to Q.Z.), and the Fundamental Research Funds of State Key Lab of Ophthalmology, Sun Yat-sen University.
References
- 1.Traboulsi EI. Genetic diseases of the eye. New York, Oxford, 1998. [Google Scholar]
- 2.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne AM, Downes SM, Bessant DA, Taylor R, Holder GE, Warren MJ, Bird AC, Bhattacharya SS. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum Mol Genet. 1998;7:273–7. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]
- 4.Thiadens AA, den Hollander AI, Roosing S, Nabuurs SB, Zekveld-Vroon RC, Collin RW, De Baere E, Koenekoop RK, van Schooneveld MJ, Strom TM, van Lith-Verhoeven JJ, Lotery AJ, van Moll-Ramirez N, Leroy BP, van den Born LI, Hoyng CB, Cremers FP, Klaver CC. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet. 2009;85:240–7. doi: 10.1016/j.ajhg.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Peachey NS, Moshfeghi DM, Thirumalaichary S, Chorich L, Shugart YY, Fan K, Zhang K. Mutations in the RPGR gene cause X-linked cone dystrophy. Hum Mol Genet. 2002;11:605–11. doi: 10.1093/hmg/11.5.605. [DOI] [PubMed] [Google Scholar]
- 6.Demirci FY, Rigatti BW, Wen G, Radak AL, Mah TS, Baic CL, Traboulsi EI, Alitalo T, Ramser J, Gorin MB. X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am J Hum Genet. 2002;70:1049–53. doi: 10.1086/339620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergen AA, Pinckers AJ. Localization of a novel X-linked progressive cone dystrophy gene to Xq27: evidence for genetic heterogeneity. Am J Hum Genet. 1997;60:1468–73. doi: 10.1086/515458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nathans J, Davenport CM, Maumenee IH, Lewis RA, Hejtmancik JF, Litt M, Lovrien E, Weleber R, Bachynski B, Zwas F, Klingaman R, Fishman G. Molecular genetics of human blue cone monochromacy. Science. 1989;245:831–8. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- 9.Kitiratschky VB, Wilke R, Renner AB, Kellner U, Vadala M, Birch DG, Wissinger B, Zrenner E, Kohl S. Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Invest Ophthalmol Vis Sci. 2008;49:5015–23. doi: 10.1167/iovs.08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelsell RE, Gregory-Evans K, Payne AM, Perrault I, Kaplan J, Yang RB, Garbers DL, Bird AC, Moore AT, Hunt DM. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet. 1998;7:1179–84. doi: 10.1093/hmg/7.7.1179. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Zulfiqar F, Xiao X, Amer Riazuddin S, Ayyagari R, Sabar F, Caruso R, Sieving PA, Riazuddin S, Fielding Hejtmancik J. Severe autosomal recessive retinitis pigmentosa maps to chromosome 1p13.3-p21.2 between D1S2896 and D1S457 but outside ABCA4. Hum Genet. 2005;118:356–65. doi: 10.1007/s00439-005-0054-4. [DOI] [PubMed] [Google Scholar]
- 12.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–5. [PMC free article] [PubMed] [Google Scholar]
- 13.Schäffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44:225–37. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Xiao X, Li S, Jia X, Wang P, Guo X, Jiao X, Zhang Q, Hejtmancik JF. Detection of variants in 15 genes in 87 unrelated Chinese patients with Leber congenital amaurosis. PLoS ONE. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auz-Alexandre CL, Vallespin E, Aguirre-Lamban J, Cantalapiedra D, Avila-Fernandez A, Villaverde-Montero C, Ainse E, Trujillo-Tiebas MJ, Ayuso C. Novel human pathological mutations. Gene symbol: GUCY2D. Disease: Leber congenital amaurosis. Hum Genet. 2009;125:349. [PubMed] [Google Scholar]
- 16.Kim BJ, Ibrahim MA, Goldberg MF. Use of Spectral Domain OCT to Visualize Photoreceptor Abnormalities in Cone/Rod Dystrophy-6. Retin Cases Brief Rep. 2011;5:56–61. doi: 10.1097/ICB.0b013e3181cd1d8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Small KW, Silva-Garcia R, Udar N, Nguyen EV, Heckenlively JR. New mutation, P575L, in the GUCY2D gene in a family with autosomal dominant progressive cone degeneration. Arch Ophthalmol. 2008;126:397–403. doi: 10.1001/archopht.126.3.397. [DOI] [PubMed] [Google Scholar]
- 18.Smith M, Whittock N, Searle A, Croft M, Brewer C, Cole M. Phenotype of autosomal dominant cone-rod dystrophy due to the R838C mutation of the GUCY2D gene encoding retinal guanylate cyclase-1. Eye (Lond) 2007;21:1220–5. doi: 10.1038/sj.eye.6702612. [DOI] [PubMed] [Google Scholar]
- 19.Ito S, Nakamura M, Ohnishi Y, Miyake Y. Autosomal dominant cone-rod dystrophy with R838H and R838C mutations in the GUCY2D gene in Japanese patients. Jpn J Ophthalmol. 2004;48:228–35. doi: 10.1007/s10384-003-0050-y. [DOI] [PubMed] [Google Scholar]
- 20.Gregory-Evans K, Kelsell RE, Gregory-Evans CY, Downes SM, Fitzke FW, Holder GE, Simunovic M, Mollon JD, Taylor R, Hunt DM, Bird AC, Moore AT. Autosomal dominant cone-rod retinal dystrophy (CORD6) from heterozygous mutation of GUCY2D, which encodes retinal guanylate cyclase. Ophthalmology. 2000;107:55–61. doi: 10.1016/s0161-6420(99)00038-x. [DOI] [PubMed] [Google Scholar]
- 21.Payne AM, Morris AG, Downes SM, Johnson S, Bird AC, Moore AT, Bhattacharya SS, Hunt DM. Clustering and frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J Med Genet. 2001;38:611–4. doi: 10.1136/jmg.38.9.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrault I, Rozet JM, Gerber S, Kelsell RE, Souied E, Cabot A, Hunt DM, Munnich A, Kaplan J. A retGC-1 mutation in autosomal dominant cone-rod dystrophy. Am J Hum Genet. 1998;63:651–4. doi: 10.1086/301985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Ghelue M, Eriksen HL, Ponjavic V, Fagerheim T, Andreasson S, Forsman-Semb K, Sandgren O, Holmgren G, Tranebjaerg L. Autosomal dominant cone-rod dystrophy due to a missense mutation (R838C) in the guanylate cyclase 2D gene (GUCY2D) with preserved rod function in one branch of the family. Ophthalmic Genet. 2000;21:197–209. [PubMed] [Google Scholar]
- 24.Ito S, Nakamura M, Nuno Y, Ohnishi Y, Nishida T, Miyake Y. Novel complex GUCY2D mutation in Japanese family with cone-rod dystrophy. Invest Ophthalmol Vis Sci. 2004;45:1480–5. doi: 10.1167/iovs.03-0315. [DOI] [PubMed] [Google Scholar]
- 25.Udar N, Yelchits S, Chalukya M, Yellore V, Nusinowitz S, Silva-Garcia R, Vrabec T, Hussles Maumenee I, Donoso L, Small KW. Identification of GUCY2D gene mutations in CORD5 families and evidence of incomplete penetrance. Hum Mutat. 2003;21:170–1. doi: 10.1002/humu.9109. [DOI] [PubMed] [Google Scholar]
- 26.Weigell-Weber M, Fokstuen S, Torok B, Niemeyer G, Schinzel A, Hergersberg M. Codons 837 and 838 in the retinal guanylate cyclase gene on chromosome 17p: hot spots for mutations in autosomal dominant cone-rod dystrophy? Arch Ophthalmol. 2000;118:300. doi: 10.1001/archopht.118.2.300. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Hoyos M, Auz-Alexandre CL, Almoguera B, Cantalapiedra D, Riveiro-Alvarez R, Lopez-Martinez MA, Gimenez A, Blanco-Kelly F, Avila-Fernandez A, Trujillo-Tiebas MJ, Garcia-Sandoval B, Ramos C, Ayuso C. Mutation analysis at codon 838 of the Guanylate Cyclase 2D gene in Spanish families with autosomal dominant cone, cone-rod, and macular dystrophies. Mol Vis. 2011;17:1103–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Ugur Iseri SA, Durlu YK, Tolun A. A novel recessive GUCY2D mutation causing cone-rod dystrophy and not Leber's congenital amaurosis. Eur J Hum Genet. 2010;18:1121–6. doi: 10.1038/ejhg.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida S, Yamaji Y, Yoshida A, Kuwahara R, Yamamoto K, Kubata T, Ishibashi T. Novel triple missense mutations of GUCY2D gene in Japanese family with cone-rod dystrophy: possible use of genotyping microarray. Mol Vis. 2006;12:1558–64. [PubMed] [Google Scholar]