Abstract
Fungus-growing ants employ several defenses against diseases, including disease-suppressing microbial biofilms on their integument and in fungal gardens. Here, we compare the phenology of microbiomes in natural nests of the temperate fungus-growing ant Trachymyrmex septentrionalis using culture-dependent isolations and culture-independent 16S-amplicon 454-sequencing. 454-sequencing revealed diverse actinobacteria associated with ants, including most prominently Solirubrobacter (12.2–30.9% of sequence reads), Pseudonocardia (3.5–42.0%), and Microlunatus (0.4–10.8%). Bacterial abundances remained relatively constant in monthly surveys throughout the annual active period (late winter to late summer), except Pseudonocardia abundance declined in females during the reproductive phase. Pseudonocardia species found on ants are phylogenetically different from those in gardens and soil, indicating ecological separation of these Pseudonocardia types. Because the pathogen Escovopsis is not known to infect gardens of T. septentrionalis, the ant-associated microbes do not seem to function in Escovopsis suppression, but could protect against ant diseases, help in nest sanitation, or serve unknown functions.
Fungus-growing ants (Attini, Formicidae) depend on fungal gardens cultivated for food. The cultivated fungi (Agaricales, Basidiomycotina) grow in the gardens alongside an assortment of secondary microbes that interact mutualistically, commensally, or antagonistically with the cultivated fungi and the ants1,2,3,4,5,6. Attine gardens are susceptible to diseases, the best studied of which are filamentous fungi in the genus Escovopsis7,8. Attine workers employ a number of defenses against their diseases, including management of auxiliary microbes (bacteria, yeasts) with potential disease-suppressing functions2,3,4.
Actinomycete bacteria such as Pseudonocardia, Streptomyces, Amycolatopsis, Kribbella, Tsukamurella, and Nocardioides have been isolated from lab-maintained attine ants and their gardens using culture-dependent methods5,9,10,11. Of the known auxiliary microbes, Pseudonocardia bacteria have attracted the greatest attention for their possible mutualistic roles in suppression of diseases of gardens or of the ants. Pseudonocardia can be isolated readily from microbial biofilms on the ant's integument, and several ant-associated Pseudonocardia species secrete chemicals in vitro that inhibit the growth of Escovopsis, as well as the growth of a great diversity of other fungi and bacteria5,11,12,13,14,15,16. Typical for actinomycete bacteria, Pseudonocardia bacteria possess polyene gene clusters regulating the synthesis of antifungal polyketide compounds17, and whole-genome sequencing has confirmed the presence of such polyene genes in Pseudonocardia strains isolated from a leafcutter ant14,18.
Ant-associated Pseudonocardia strains were initially thought to be antibiotically highly specialized and primarily vertically transmitted between ant generations, giving rise to the view of long-term co-evolutionary interactions between ants and their Pseudonocardia associates8,19. Under the original co-evolutionary interpretation of the ant-Escovopsis-Pseudonocardia association, attine ants were believed to manage integumental Pseudonocardia as a specific co-evolved antibiotic defense targeted against Escovopsis fungi, which were thought not to evolve resistance to Pseudonocardia antibiotics due to some unknown disadvantages in a co-evolutionary arms race10,13,19. However, more comprehensive recent studies have begun to question the postulated tight ant-Pseudonocardia co-evolution, suggesting instead frequent recruitment of Pseudonocardia from environmental sources (plant material, soil) into the integumental biofilms and possible horizontal transmission of microbes by males during mating5,9,15,18,20,21,22.
We study here the fungus-growing ant Trachymyrmex septentrionalis to characterize seasonal changes in ant-bacteria associations in undisturbed field colonies. T. septentrionalis provides unique opportunities to explore the potential roles of auxiliary, secondary microbes in the attine ant-microbe symbiosis, for two main reasons. First, despite the frequent presence of whitish accretions (presumed actinomycete growth) on the integument of T. septentrionalis, Escovopsis, a prevalent pathogen infecting gardens of tropical attine ants, does not appear to infect gardens of the temperate T. septentrionalis21,23. Our study population of T. septentrionalis was previously surveyed for non-cultivar filamentous fungi and yeasts growing in gardens alongside the cultivated fungus23. No Escovopsis could be isolated from any of 16 surveyed nests (4 nests surveyed every three months over an entire year), nor from dozens of gardens of two other sympatric attine species (Atta texana, Cyphomyrmex wheeleri23). Moreover, another survey covering the full range of T. septentrionalis (Texas, Illinois, Florida, New York) had failed to find any Escovopsis infection in gardens21. The presence of biofilms on the integument of T. septentrionalis in the apparent absence of Escovopsis therefore suggested possible novel functions of the microbial accretions on the integument of T. septentrionalis (e.g., protection of the ants against predators, entomopathogenic diseases, or parasites such as mites; protection against other garden diseases).
A second reason T. septentrionalis represents a unique study system is that this temperate ant exhibits life-history features that permit testing for seasonal patterns of microbial association and abundance. Gardens are collapsed in winter (sometimes to small fragments), then are reactivated in spring by workers, and gardens reach the largest sizes throughout early summer when alates emerge and feed on gardens until rains stimulate mating flights24,25. Seasonal correlations between the abundance of microbial associates, garden size, worker activity, and production of reproductives can therefore provide clues for the possible roles of the microbial associates in the temperate T. septentrionalis, whereas such correlations are less obvious in the comparatively stable, tropical ant-microbe symbioses.
To generate insights into such phenological changes in ant-associated bacterial microbiomes, we performed culture-dependent isolation and culture-independent 16S-amplicon 454-sequencing of T. septentrionalis nests collected between January and September 2009. No study to date has quantified the relative abundance of bacteria in natural, field-collected attine nests.
Results
Culture-dependent screens
Culture-dependent isolations identified eleven genera from the order Actinomycetales among 914 bacterial colonies isolated from garden workers, outside workers, reproductives, garden, and soil samples (Table 1). Isolates were morphotyped (by color and growth morphology on PDA medium), and 264 representative isolates were identified through partial 16S-sequencing. Pseudonocardia, Kribbella, Amycolatopsis, and Streptomyces accounted for 98% (n = 890) of the isolated colonies (Table 1). Rare isolates included the genera Actinomadura, Actinomycetospora, Nocardioides, Nocardia, Nonomuraea, and Sphaerisporangium. To test whether bacterial growth was specific to ant body segments (head, mesosoma, metasoma), we also streaked these segments individually on chitin medium. We found P. cf. carboxydivorans and Streptomyces on all body segments of workers, males, and females (Table 1). Additionally, Amycolatopsis was found on all body segments of males.
Table 1. Bacteria from eleven actinomycete genera isolated from the fungus-growing ant Trachymyrmex septentrionalis with culture-dependent methods. Whole ants, garden, chamber soil, and excavated soil were plated as macerated buffer suspensions on minimum-carbon chitin medium favoring growth of autotrophic bacteria. Ants were screened whole, but the three main body segments of ants (head, mesosoma, and metasoma) were also streaked separately on chitin medium. Bacterial counts per sample type need to be interpreted with caution because sampling effort could not be completely standardized across all sample types. Pseudonocardia and Streptomyces were isolated in all except one sample type and from all three body segments. Empty cells indicate an isolation count of zero. See Supplementary Methods for details on the combined 16S-sequencing and morphotyping approach to identify bacteria to genus.
| Actinomycete genera isolated from Trachymyrmex septentrionalis samples | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Type | # Samples | Pseudonocardia | Streptomyces | Kribbella | Amycolatopsis | Nocardia | Nocardioides | Actinomadura | Actinomycetospora | Nonomuraea | Sphaerisporangium | Unclassified actinobacterium | |
| Garden worker | whole ant | 31 | 113 | 48 | 29 | 14 | 5 | 2 | |||||
| head | 16 | 16 | 3 | ||||||||||
| mesosoma | 16 | 10 | 3 | 1 | |||||||||
| metasoma | 16 | 8 | 1 | ||||||||||
| Outside worker | whole ant | 29 | 124 | 7 | 14 | 6 | 2 | 3 | 1 | ||||
| head | 15 | 30 | 7 | ||||||||||
| mesosoma | 15 | 56 | 11 | 1 | |||||||||
| metasoma | 15 | 25 | 2 | ||||||||||
| Male | whole ant | 2 | 12 | 1 | 1 | ||||||||
| head | 2 | 13 | 2 | 1 | |||||||||
| mesosoma | 2 | 9 | 6 | 3 | |||||||||
| metasoma | 2 | 11 | 4 | 2 | |||||||||
| Reproductive female | whole ant | 6 | 106 | 3 | 3 | 2 | |||||||
| head | 5 | 13 | 6 | ||||||||||
| mesosoma | 5 | 11 | 1 | ||||||||||
| metasoma | 5 | 20 | 5 | ||||||||||
| Garden | 28 | 29 | 28 | 1 | 2 | 3 | 2 | ||||||
| Excavated soil | 30 | 8 | 1 | 2 | 1 | ||||||||
| Chamber soil | 30 | 3 | 34 | 19 | 2 | 2 | |||||||
| Total number of isolates identified to this genus | 609 | 179 | 69 | 33 | 8 | 8 | 2 | 2 | 2 | 1 | 1 | ||
| Fraction of this genus among all bacteria identified | 66.6 | 19.6 | 7.5 | 3.6 | 0.9 | 0.9 | 0.2 | 0.2 | 0.2 | 0.1 | 0.1 | ||
| % of samples identified by 16S sequencing | 20.5 | 41.9 | 31.9 | 66.7 | 87.5 | 37.5 | 50 | 50 | 100 | 100 | 100 | ||
| % of samples identified by morphotyping only | 79.5 | 58.1 | 68.1 | 33.3 | 12.5 | 62.5 | 50 | 50 | 0 | 0 | 0 | ||
Sequenced Pseudonocardia from the culture-dependent isolations had identical partial 16S-sequences, which were identified as Pseudonocardia cf. carboxydivorans via BLASTn (99% identity to GenBank accession FJ532384). The only exceptions were three isolates from garden samples in June, which were closely related to Pseudonocardia spinosispora (696 bp at 98% identity to GenBank accession NR_025367). Although all the isolates identified as P. cf. carboxydivorans had identical 16S-sequences, we found four distinct morphotypes among these isolates (Supplementary Fig. S1). Interestingly, more than one P. cf. carboxydivorans morphotype could be found within a single nest. Four nests had two morphotypes and one nest had three morphotypes found on the workers of the same nest (Table 2).
Table 2. Number of Pseudonocardia strains found per nest of Trachymyrmex septentrionalis. The culture-dependent counts are presented as the number of P. cf. carboxydivorans isolates of morphotype A–D found in a particular nest. The culture-independent counts list the total number of Pseudonocardia 454-sequences found per ant caste and the number of different Pseudonocardia strains among these sequences. Empty cells indicate absence of Pseudonocardia, and “na” indicates that a particular sample type (e.g., reproductive ant) was not found in a nest at the time of collection. Because of an oversight, nests JLM090322-04 and JLM090630-02B were only screened in the culture-independent screen. In our custom Pseudonocardia BLAST, the majority of the sequences were identified as P. cf. carboxydivorans (i.e., sequence accession FJ490549 was hit in the BLAST of every ant sample; see also BLAST-results identifying clade-3 Pseudonocardia in Supplementary Table S5).
| Culture-Dependent Screen | Culture-Independent Screen | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nest ID (Abbreviated nest ID) | P. cf. carboxydivorans Cultured Morphotypes | |||||||||||||||
| Month | A | B | C | D | # of Pseudonocardia 454-sequences | Distinct # of Pseudonocardia strains per nest | ||||||||||
| garden worker | outside worker | male | female | outside worker | garden worker | outside worker | female | garden worker | outside worker | garden worker | outside worker | female | male | |||
| JLM090131-01 | Jan | 2 | na | na | na | na | na | na | na | na | ||||||
| JLM090131-03 | Jan | 1 | na | na | na | na | na | na | na | na | ||||||
| JLM090207-01 (J0201) | Feb | na | na | na | 30 | 29 | na | na | 1 | |||||||
| JLM090322-01 (J8-12) | Mar | na | na | na | 74 | 16 | na | na | 2 | |||||||
| JLM090322-2A (J0302A) | Mar | na | na | na | 1831 | 47 | na | na | 6 | |||||||
| JLM090322-2B (J0302B) | Mar | na | na | na | 124 | 67 | na | na | 4 | |||||||
| JLM090322-03 (J0303) | Mar | na | na | na | 157 | 36 | na | na | 4 | |||||||
| JLM090322-04 (J0304) | Mar | na | na | na | na | na | na | na | na | na | na | 20 | 11 | na | na | <1?h=-12pt?>2 |
| JLM090405-01 (J0401) | Apr | 11 | 6 | na | na | na | 59 | 100 | na | na | <1?h=-12pt?>2 | |||||
| JLM090405-02 (J0402) | Apr | 3 | na | na | na | 66 | 108 | na | na | 1 | ||||||
| JLM090405-03 (J0403) | Apr | 7 | 3 | na | na | na | 132 | 182 | na | na | 1 | |||||
| JLM090405-04 (J0404) | Apr | 3 | 1 | na | na | na | 1 | 55 | 128 | na | na | 3 | ||||
| JLM090528-01 (J0501) | May | 1 | 1 | na | na | 2 | 3 | na | 1 | 46 | 96 | na | na | 2 | ||
| JLM090528-02 (J0502) | May | 7 | 7 | na | na | na | 46 | 59 | na | na | <1?h=-12pt?>1 | |||||
| JLM090528-03 (J0503) | May | na | na | na | 233 | 204 | na | na | 3 | |||||||
| JLM090528-04 (J0504) | May | 3 | 9 | na | na | na | 2 | 218 | 144 | na | na | 5 | ||||
| JLM090627-01 (J0601) | Jun | 10 | na | 436 | 357 | na | na | <1?h=-12pt?>3 | ||||||||
| JLM090627-02 (J0602) | Jun | 55 | na | na | 52 | 299 | na | na | 1 | |||||||
| JLM090627-03 (J0603) | Jun | 13 | na | na | 124 | 56 | na | na | 2 | |||||||
| JLM090630-02 (J1-J7) | Jun | 6 | 25 | 35 | 24 | 273 | 125 | 3081 | 1806 | 7 | ||||||
| JLM090630-02B (J0630-02) | Jun | na | na | na | na | na | na | na | na | na | na | na | na | 5440 | 286 | 3 |
| JLM090630-03 (J0630-03) | Jun | 10 | 9 | na | 17 | 72 | 93 | 521 | na | 2 | ||||||
| RS090713-01 | Jul | 3 | 3 | na | na | na | na | na | na | na | ||||||
| JLM090713-01 (J0701) | Jul | 25 | 28 | na | 27 | 83 | 84 | 729 | na | 2 | ||||||
| JLM090713-02 (J0702) | Jul | 8 | 33 | na | na | 152 | 50 | na | na | <1?h=-12pt?>2 | ||||||
| JLM090713-04 (J0704) | Jul | na | 4 | 56 | 233 | 153 | 1513 | na | <1?h=-12pt?>7 | |||||||
| JLM090830-01 (J0801) | Aug | 29 | 47 | na | 22 | 184 | 171 | 110 | na | <1?h=-12pt?>2 | ||||||
| JLM090830-02 (J0802) | Aug | na | na | 5 | 2476 | 167 | na | na | 2 | |||||||
| JLM090830-03 (J0803) | Aug | 6 | na | 82 | 181 | 681 | na | 6 | ||||||||
| Average | 290.32 | 118.52 | 1725 | 1046 | 2.92 | |||||||||||
The P. cf. carboxydivorans morphotypes fell into four categories (morphotype A–D; Supplementary Fig. S1). Morphotype A was the most predominant type isolated from garden workers, outside workers, reproductives, and gardens, although a large number of morphotype B was found associated with reproductive females in nests collected in July 2009 (Table 2). We were unable to culture Pseudonocardia bacteria from any soil samples due to rapid fungal contamination of chitin isolation plates.
In the culture-dependent screens, Pseudonocardia, Amycolatopsis, Kribbella, and Streptomyces were isolated in greater numbers from samples collected in June and July than in any other month (Table 3), with Amycolatopsis and Kribbella isolations almost completely restricted to June and July. Streptomyces was isolated in all months (except January) and in all sample types (Table 3). The culture-dependent screen yielded different results than the parallel culture-independent screen (Table 3), confirming expected isolation biases of the minimum-carbon chitin medium5,22,26.
Table 3. Pseudonocardia, Amycolatopsis, Kribbella, Streptomyces, Solirubrobacter, and Microlunatus bacteria found in culture-dependent and culture-independent monthly surveys in the fungus-growing ant Trachymyrmex septentrionalis. The + sign indicates presence; an empty cell indicates absence; cells with “na” indicate that a sample was not available for screening. The presence of the specific bacterial genera listed was consistent across the monthly samples in the culture-independent screens. Pseudonocardia, Amycolatopsis, Kribbella, and Streptomyces were most frequently isolated in the months of June and July in the culture-dependent screens. Culture-independent screens failed to detect Kribbella in ant samples, whereas culture-dependent screens readily detected Kribbella. It is possible that Kribbella was not detected in the culture-independent screens because it is actually rare, but it grows well on the minimum-carbon chitin medium favoring autotrophic bacteria. In contrast to Kribbella, both Solirubrobacter and Microlunatus were found abundantly in the culture-independent screens, but were never isolated in the culture-dependent screens.
| Culture-Dependent Screen (chitin-medium isolation) | Culture-Independent Screen (454 sequencing) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterium Culture | Sample Type | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | Sep | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug |
| # nests screened | 2 | 1 | 4 | 4 | 4 | 5 | 4 | 3 | 3 | 0 | 1 | 5 | 4 | 4 | 5 | 3 | 3 | |
| Pseudonocardia | Excavated Soil | + | + | + | + | + | + | + | ||||||||||
| Chamber Soil | + | + | + | + | + | + | ||||||||||||
| Garden | + | + | + | + | + | + | + | |||||||||||
| Garden Worker | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| Outside Worker | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| Reproductive Female | na | na | na | na | na | + | + | + | na | na | na | na | na | + | + | + | ||
| Male | na | na | na | na | na | + | na | na | na | na | na | na | na | + | na | na | ||
| Amycolatopsis | Excavated Soil | + | + | + | + | |||||||||||||
| Chamber Soil | + | + | + | + | + | + | + | |||||||||||
| Garden | + | + | ||||||||||||||||
| Garden Worker | + | + | + | + | + | |||||||||||||
| Outside Worker | + | + | + | + | ||||||||||||||
| Reproductive Female | na | na | na | na | na | + | na | na | na | na | na | + | + | |||||
| Male | na | na | na | na | na | + | na | na | na | na | na | na | na | + | na | na | ||
| Kribbella | Excavated Soil | + | + | + | + | |||||||||||||
| Chamber Soil | + | + | + | + | ||||||||||||||
| Garden | + | + | ||||||||||||||||
| Garden Worker | + | + | + | |||||||||||||||
| Outside Worker | + | + | ||||||||||||||||
| Reproductive Female | na | na | na | na | na | + | na | na | na | na | na | |||||||
| Male | na | na | na | na | na | na | na | na | na | na | na | na | na | na | ||||
| Streptomyces | Excavated Soil | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| Chamber Soil | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| Garden | + | + | + | + | + | + | + | + | ||||||||||
| Garden Worker | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| Outside Worker | + | + | + | + | + | + | + | + | + | + | + | |||||||
| Reproductive Female | na | na | na | na | na | + | + | + | na | na | na | na | na | + | + | + | ||
| Male | na | na | na | na | na | + | na | na | na | na | na | na | na | + | na | na | ||
| Solirubrobacter | Excavated Soil | + | + | + | + | + | + | + | ||||||||||
| Chamber Soil | + | + | + | + | + | + | + | |||||||||||
| Garden | + | + | + | + | + | + | ||||||||||||
| Garden Worker | + | + | + | + | + | + | + | |||||||||||
| Outside Worker | + | + | + | + | + | + | + | |||||||||||
| Reproductive Female | na | na | na | na | na | na | na | na | na | na | + | + | + | |||||
| Male | na | na | na | na | na | na | na | na | na | na | na | na | + | na | na | |||
| Microlunatus | Excavated Soil | + | + | + | + | + | + | |||||||||||
| Chamber Soil | + | + | + | + | ||||||||||||||
| Garden | + | |||||||||||||||||
| Garden Worker | + | + | + | + | + | + | + | |||||||||||
| Outside Worker | + | + | + | + | + | + | + | |||||||||||
| Reproductive Female | na | na | na | na | na | na | na | na | na | na | + | + | + | |||||
| Male | na | na | na | na | na | na | na | na | na | na | na | na | + | na | na | |||
Culture-independent screens
454-sequencing BLAST results were analyzed by the closest operational taxonomic level (Supplementary Table S1) and by genus in a forced-genus BLAST match (Supplementary Table S2) to estimate average relative sequence-abundances by sample type. Rarefaction analyses at a 97% sequence-similarity criterion revealed that most of the garden workers, outside workers, and reproductives (males, females) were adequately sampled (species-accumulation curves reached an asymptote with 2000–5000 sequences sampled; Supplementary Fig. S2). However, chamber soil, excavated soil, and most garden samples were undersampled (Supplementary Fig. S2). As expected, soil samples require a far greater sequencing depth to evaluate bacterial diversities. Garden samples proved more difficult to sequence, resulting in lower sequence yields, with an average of 2,661 sequences per garden as compared to an average of 4,007 sequences from all other sample types. Additionally, sequences obtained from garden samples also had fewer reliable BLAST hits, and many of these sequences therefore remained unidentified (Supplementary Tables S1 and S2).
We used an unweighted principal coordinate analysis (PCoA) in Fast Unifrac to visualize the differences between microbial communities for each of our sample types (Fig. 1). Bacterial communities associated with soil samples were significantly different than the ant-associated bacterial communities (R2 = 0.21, adonis test P<0.01). Chamber soil samples clustered more tightly together than the excavated soil (Fig. 1), but both soils had overlapping microbial community compositions. Some garden samples clustered with soil samples, whereas other garden samples separated out distinctly (Fig. 1).
Figure 1. Fast UniFrac unweighted principal coordinate analysis (PCoA) for Trachymyrmex septentrionalis ants, gardens, soil samples, and control ants (Pheidole).
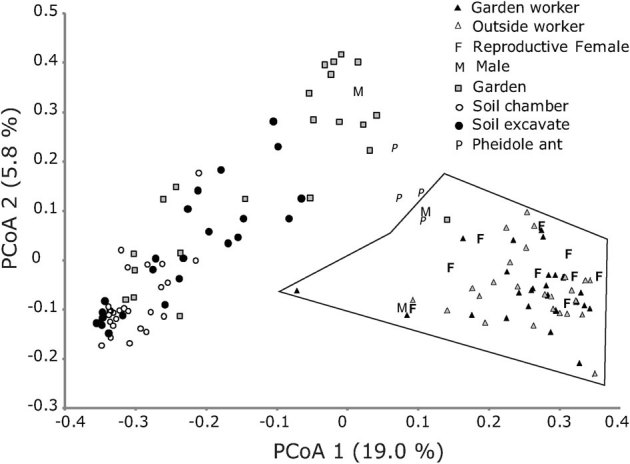
The PCoA uses a phylogenetic approach to compare the whole microbial communities between samples. The polygon contains all ant samples except for one male ant; the latter grouped near the garden and Pheidole samples. The chamber-soil and excavated-soil samples cluster loosely together, but the garden samples range widely, with several samples grouping mostly among the soils; despite careful excavation, gardens may have become contaminated with soil dust that spread to gardens accidentally by agitated workers.
The combined ant samples had an average of 88 (+/− 26) bacterial genera identified per nest (Supplementary Table S3). Soil samples contained significantly more bacterial genera (average of 259 (+/− 50) genera per nest; t-test t = 15.641, df = 24, P<0.001). Ants shared an average of 65% of their bacterial genera with those found in soil; however, these shared bacterial genera had typically low abundance (i.e., abundant genera were typically not shared). Differences between bacterial compositions of ants and soil are clearly evident when compared at the order taxonomic level (Supplementary Fig. S3).
454-sequencing revealed 19 bacterial genera frequently associated with T. septentrionalis ants (each of these genera had a minimum of 1% sequence reads averaged across all samples from all ant castes; Table 4). Eleven of these 19 genera belong to the class Actinobacteria, including most prominently Solirubrobacter (order Solirubrobacterales), Microlunatus, Pseudonococardia, Aeromicrobium, Phycicoccus, and Agrococcus (all in the order Actinomycetales; Table 4). The two actinobacterial orders Actinomycetales and Solirubrobacterales comprise almost 70% of the total bacterial sequence reads identified from T. septentrionalis ants (Table 4, Supplementary Fig. S3). Other bacterial genera (e.g., Bacillus, Burkholderia, Corynebacterium, Mesoplasma, Spiroplasma) were detected occasionally as abundant sequence reads, but this was the case in only a few individual ants. Overall, the 19 common genera comprise more than 90% of all the bacterial sequence reads identified for workers and female reproductives (Table 4). Reproductive males also had abundant sequence reads of Comamonas, Escherichia, and Propionibacterium bacteria (Supplemental Table S2), but not consistently so in all males. The significance of the male-associated bacteria is unclear because only three males were screened.
Table 4. Average percent sequence-reads of common bacterial genera estimated by culture-independent 16S-amplicon 454-sequencing in T. septentrionalis ant samples (outside workers, garden workers, female reproductives, males). The information presents the average percent sequence-reads per ant sample type (+/− standard deviation) according to the assigned reference genus in the forced-genus BLAST (see Supplementary Table S2 for assignments in the forced-genus BLAST-identification). As a measure of consistency of association between samples, numbers in parentheses give the number of samples containing the respective bacterial genus per total number of samples screened.
| Genus | Order | Class | Garden Worker | Outside Worker | Reproductive Female | Male |
|---|---|---|---|---|---|---|
| Aeromicrobium | Actinomycetales | Actinobacteria | 6.37% +/− 6.05 (24/25) | 8.24% +/− 5.01 (25/25) | 3.94% +/− 5.03 (8/9) | .05% +/− .06 (2/3) |
| Agrococcus | Actinomycetales | Actinobacteria | 7.42% +/− 4.06 (25/25) | 7.15% +/− 3.15 (25/25) | 10.75% +/− 8.93 (8/9) | .02% +/− .04 (1/3) |
| Corynebacterium | Actinomycetales | Actinobacteria | 1.90% +/− 7.00 (12/25) | 0.17% +/− .35 (9/25) | 0% | 0.83% +/− 1.44 (1/3) |
| Dermacoccus | Actinomycetales | Actinobacteria | 1.05% +/− 3.38 (13/25) | 0.62% +/− 1.45 (13/25) | 0.81% +/− 0.82 (6/9) | 1.16% +/− 1.89 (2/3) |
| Microlunatus | Actinomycetales | Actinobacteria | 10.76% +/− 4.62 (25/25) | 10.10% +/− 4.23 (25/25) | 4.00% +/− 4.07 (9/9) | 0.36% +/− .62 (1/3) |
| Nocardiodes | Actinomycetales | Actinobacteria | 1.23% +/− 2.49 (21/25) | 1.46% +/− 2.65 (22/25) | 0.12% +/− 0.21 (4/9) | .25% +/− .44 (1/3) |
| Phycicoccus | Actinomycetales | Actinobacteria | 6.82% +/− 3.43 (24/25) | 8.14% +/− 4.09 (25/25) | 4.58% +/− 5.55 (7/9) | 0% |
| Ponticoccus | Actinomycetales | Actinobacteria | 1.72% +/− 1.44 (25/25) | 1.29% +/− 0.85 (25/25) | 0.93% +/− .96 (6/9) | 0.05% +/− 0.10 (1/3) |
| Pseudonocardia | Actinomycetales | Actinobacteria | 7.05% +/− 17.15 (25/25) | 3.52% +/− 2.55 (25/25) | 41.99% +/− 29.00 (9/9) | 17.35% +/− 21.73 (3/3) |
| Solirubrobacter | Solirubrobacterales | Actinobacteria | 30.93% +/− 11.08 (25/25) | 29.21% +/− 10.17 (25/25) | 15.47% +/− 11.92 (9/9) | 12.19% +/− 19.26 (3/3) |
| unknown genus | Actinomycetales | Actinobacteria | 2.66% +/− 1.8 (25/25) | 2.95% +/− 2.01 (25/25) | 1.06% +/− 1.28 (8/9) | 0.03% +/− 0.06 (1/3) |
| Bacillus | Bacillales | Bacilli | 0.11% +/− 0.27 (10/25) | 1.46% +/− 7.11 (10/25) | 0.04% +/− 0.08 (2/9) | 0.08% +/− 0.09 (2/3) |
| Bacteriodes | Bacteroidales | Bacteroidia | 0.24% +/− 0.44 (14/25) | 0.14% +/− 0.40 (9/25) | 2.54% +/− 5.30 (4/9) | 4.80% +/− 8.31 (1/3) |
| Burkholderia | Burkholderiales | Betaproteobacteria | 0.08% +/− 0.17 (11/25) | 0.08% +/− .14% (12/25) | 3.33% +/− 6.62 (2/9) | 0.08% +/− 0.14 (1/3) |
| Derxia | Burkholderiales | Betaproteobacteria | 8.69% +/− 5.70 (25/25) | 8.32% +/− 5.35 (25/25) | 2.05% +/− 2.35 (7/9) | 0.68% +/− 1.18 (1/3) |
| Pseudomonas | Pseudomonadales | Gammaproteobacteria | 0.18% +/− 0.35 (15/25) | 0.12% +/− 0.16 (20/25) | 0.52% +/− 0.92 (6/9) | 9.95% +/− 14.25 (3/3) |
| Xanthomonas | Xanthomonadales | Gammaproteobacteria | 1.35% +/− 1.91 (19/25) | 1.49% +/− 1.91 (17/25) | 0% | 0% |
| Mesoplasma | Entomoplasmatales | Mollicutes | 1.80% +/− 6.32 (2/25) | 1.54% +/− 6.96 (3/25) | 0% | 0% |
| Spiroplasma | Entomoplasmatales | Mollicutes | 2.59% +/− 9.75 (4/25) | 7.28% +/− 19.83 (6/25) | 0.01% +/− 0.03 (2/9) | 0% |
As a quality check of the 454-BLAST reference assignments generated from the Medical Biofilm's reference database, we also used the Ribosomal Database Classifier (RDC) to assign identities to observed 454-sequences27. The comparison between the bacterial assignments derived from the two databases revealed comparable results for the common bacteria found in ant samples. Specifically, Solirubrobacter and Pseudonocardia had 98% and 93% identical hits from both reference databases. The soil and garden samples showed less similarity between the two BLAST databases, suggesting caution in the interpretation of the relative bacterial abundances in these samples. A sub-set of the RDC results is presented in Supplementary Table S4.
Phenology of ant-associated bacterial microbiomes
The culture-independent screen captured the bacterial microbiomes of T. septentrionalis nests from late winter to late summer, covering reactivation of the fungal gardens in spring, emergence of reproductives in early summer, and the period of post-mating flights in late summer. Outside and garden workers showed few changes in their bacterial profiles from winter to summer, with the exception of Spiroplasma bacteria, which were observed only in March and June (Fig. 2). However, in late summer (30-Aug) the percent sequence reads of Pseudonocardia increased in both worker types, but especially in garden workers (from 5% in July to 34% in August). Average percent of sequence reads of Pseudonocardia on reproductive females collected in June was initially high (68%), but declined by August (10%). Dealate females collected on 30-Aug (i.e., females that lost their wings and had failed to leave the nest for a mating flight) showed a low percentage of Pseudonocardia sequence reads, but showed increased percentage sequence-reads of other bacteria commonly found on workers (Solirubrobacter, Microlunatus, Phycicoccus, and Aeromicrobium).
Figure 2. Phenology of common bacterial genera associated with T. septentrionalis garden workers, outside workers, and reproductive ants as determined by 16S-amplicon 454-sequencing.
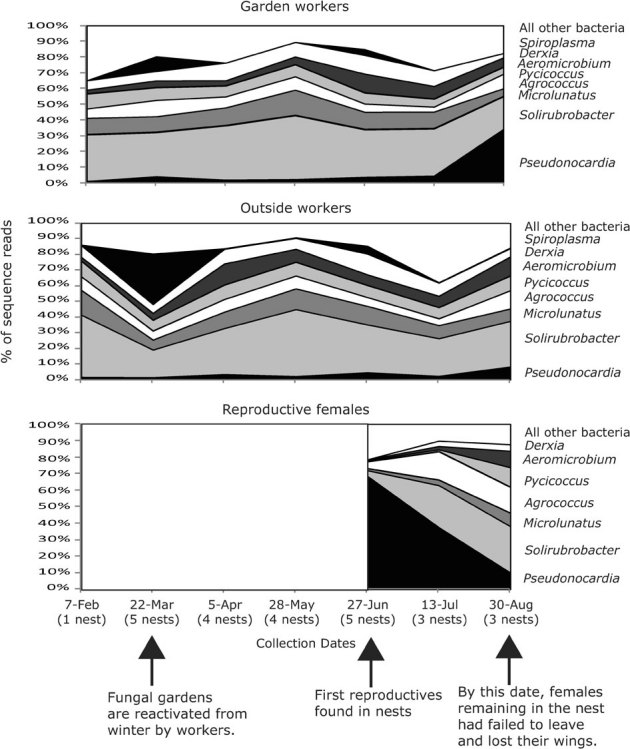
Percent sequence-reads for the common bacteria for the ant castes were averaged by month. Reproductive males are not included in this figure because males were only collected in June. Arrows indicate the month of important life history stages in the annual reproductive cycle of T. septentrionalis. Garden and outside workers showed relatively constant bacterial-community profiles, except for increases of Spiroplasma in March and Pseudonocardia in August. Reproductive females showed initially (June) a high percentage of sequence-reads for Pseudonocardia, but percentage of Pseudonocardia sequence-read decreased in females that had not dispersed from the nest by late August.
Pseudonocardia
We found Pseudonocardia sequences in each of the ant samples of garden workers (n = 25), outside workers (n = 25), and male and female reproductives (n = 12). In the custom BLAST of 26,965 Pseudonocardia 454-sequences, we found 14 distinct Pseudonocardia strains among the garden workers, 14 distinct strains among outside workers, and 6 distinct strains among reproductive ants (Supplementary Table S5). These strains covered much of the known Pseudonocardia diversity, including representatives from 7 of the 10 known Pseudonocardia clades (Supplementary Table S5; Fig. 2 from22). Eighty percent of the nests (n = 25) had more than one Pseudonocardia strain on workers per nest, with an average of 2.9 strains on workers from the same nest (here a strain refers to a unique Pseudonocardia sequence; Table 2; see Methods). However, most of the strains were rare with the exception of one predominant Pseudonocardia strain from clade 3 (the so-called P. nitrificans/alni/carboxydivorans clade22), which received 99.7% of the Pseudonocardia BLAST hits (Supplementary Table S5). Soils contained a greater diversity of Pseudonocardia strains (chamber soil and excavated soil revealed, respectively, 45 and 44 strains, which were primarily from clades 6–10; Supplementary Table S5). Compared to all BLAST results, Pseudonocardia was found at low percent sequence reads in the soils, comprising only 1.33% (chamber soil) and 1.05% (excavated soil) of the total sequences obtained.
Discussion
This study is the first to compare natural microbiomes of field-collected fungus-growing ants using both culture-dependent and culture-independent methods. We find that the ant- and garden-associated microbiomes of T. septentrionalis ants are complex communities of bacteria, including most prominently the actinobacterial genera Solirubrobacter, Microlunatus, Pseudonococardia, Aeromicrobium, Phycicoccus, and Agrococcus (Table 4; Supplementary Table S2). The consistent association of diverse actinobacterial bacteria with T. septentrionalis, as well as the consistent association of multiple Pseudonocardia species on workers of a single ant nest, corroborate for field colonies the finding of complex integumental biofilm communities that were first reported for laboratory colonies of T. septentrionalis5 and that have recently been documented for other attine ants14,18. The presence of multiple actinobacterial lineages suggests that either the ants are readily colonized by a far greater diversity of transient, commensal actinobacteria than previously realized12,15,28, or that the ants accumulate complex biofilms that may include actinobacteria with beneficial complementary or synergistic properties2,5,11,14,16,18,20,29,30.
An unweighted Unifrac analysis revealed that different ant castes carry similar bacterial communities (Figure 1), except that the percentage of Pseudonocardia sequence reads on reproductive females early in the mating season appears to exceed the corresponding Pseudonocardia percentages on nestmate workers collected at the same time (Figure 2). However, this difference in percent sequence reads may be confounded by the difference in body size between castes and by the different number of individuals pooled in each caste sample. Moreover, 454-sequencing-read abundance patterns need to be interpreted with caution because of possible amplification biases31.
While between-caste comparisons are complicated by size differences and possible amplification biases, bacterial comparisons within a caste at different time points could be less subject to such complications. For example, female reproductives collected in June showed a high percentage of Pseudonocardia sequence reads, but sequence percentages declined during the next two months (Figure 2). By late summer, female reproductives that failed to leave the nest (likely because of a severe drought in 2009 that reduced opportunities for mating flights) had shed their wings. These dealate females had bacterial read-abundances similar to outside and garden workers. The seasonal change in percent sequence reads suggests either a decrease of Pseudonocardia on the integument of dealate females, or increased prevalence of other microbial types on the integument, or both.
Throughout the seasons, both garden and outside workers showed constant proportions of sequence reads for the common bacterial genera, with the exception of an increase of Pseudonocardia sequence reads in late August. This late-season increase was more pronounced in garden workers than outside workers (Figure 2). Future studies could test whether a caste-specific late-season Pseudonocardia increase could be related to the accumulation of specific diseases or enemies (e.g., mites, predators) that may increasingly impact the ants or their gardens in late summer.
If components of the microbiomes are mutualists of the ant host, as is likely true for tropical attine ants3,5,18,29,32,33, these putative, beneficial microbes could serve distinct functions in different ant castes of T. septentrionalis. We observed high sequence reads of Pseudonocardia in male and female reproductives in June, whereas workers had several common actinobacteria in addition to Pseudonocardia; this could implicate functional differences of bacterial mutualists between castes, or reflect differential bacterial acquisition resulting from behavioral differences. Bacterial associates could potentially be used by male and female reproductives for antibiotic protection against entomopathogenic diseases, or in the case of females, protection of the nest environment or the garden after nest establishment. In contrast, garden and outside workers may accumulate a more diversified array of bacteria for protection against a diverse set of harmful microbes encountered during foraging and gardening. Overall, the roles of any beneficial microbes within the nest could be diverse, including suppression of ant diseases34,35,36,37; general sanitation of the nest environment (e.g., chamber walls); suppression of mites, nematodes, or thieving fungivores in a nest38; or detoxification39.
Because gardens of the temperate attine ant T. septentrionalis are rarely or never infected with the pathogen Escovopsis, the actinobacteria (e.g., Pseudonocardia) carried by T. septentrionalis are unlikely to represent a specific defense against Escovopsis. This suggests that either ant-associated Pseudonocardia can occupy roles different than specific defense against Escovopsis, or that the integumental Pseudonocardia may serve no beneficial function (at least in some attine ant populations). High Pseudonocardia loads on T. septentrionalis may be accidental (e.g., inert integumental accretions may accidentally accumulate autotrophic microbes) or may serve one or several of the functions mentioned above. Nevertheless, the documented prevalence of P. cf. carboxydivorans in both the culture-dependent and culture-independent screens suggest that P. cf. carboxydivorans likely occupies an important ecological role in the microbial ecology of T. septentrionalis.
Pseudonocardia can be readily isolated from the head and metasoma of the ants in addition to the mesosoma (propleural thoracic plates; Table 1). This finding contradicts the current belief that Pseudonocardia occurs primarily on the mesosoma of attine ants and that Pseudonocardia biomass on ants correlates with the amount of whitish integumental accretion that is visible on the propleural thoracic plates in tropical Trachymyrmex13. The occurrence of Pseudonocardia on multiple body segments (Table 1) could imply that the extent of visible, whitish accretions on the integument of attine ants therefore may not be a reliable indicator of Pseudonocardia abundance.
Of all Pseudonocardia species identified, P. cf. carboxydivorans was the dominant species found in culture-dependent isolations (99.5%) and culture-independent screens (99.7%), consistent with recent reports15,22 that P. cf. carboxydivorans and closely relatives are most frequently isolated from Trachymyrmex and Acromyrmex ants in the tropics. Sequence reads assigned to P. cf. carboxydivorans with 100% sequence identity could be further subcategorized into four distinct morphotypes (Supplementary Fig. S1). All four of these P. cf. carboxydivorans morphotypes occurred on T. septentrionalis workers. In some cases, we observed the presence of two distinct P. cf. carboxydivorans morphotypes on ants from the same nest (Table 2), suggesting cryptic diversity on the ant integument that is not distinguished by 16S sequencing. Because this cryptic diversity may represent biochemical variants arising through mutation within the nest, it will be important in future screens to isolate multiple strains (perhaps dozens or hundreds strains) to characterize the true biochemical and metabolic diversity of Pseudonocardia associates, contrary to the prevailing belief that identification of one or a few isolates adequately characterizes the Pseudonocardia associates of an ant nest10,15. Moreover, it appears that the diversity of Pseudonocardia coexisting on a single ant worker does not become apparent until microbes are subcultured and allowed to grow for sufficient time. Specifically, the morphologically homogenous growth on chitin plates [e.g., Electronic supplementary material S1b-d in15] appears to be insufficient evidence that only one Pseudonocardia type grows on the ants. In fact, we found in our isolations that bacteria from several actinomycete genera can be intermixed in deceptively “homogenous” cultures.
The culture-independent 16S-sequencing found other Pseudonocardia strains besides P. cf. carboxydivorans associated with T. septentrionalis ants. On average, garden workers and outside workers from the same nest carried one P. cf. carboxydivorans strain and two additional Pseudonocardia strains per nest (Table 2). Although these additional Pseudonocardia strains were rare (as judged by sequence abundance in the 454-screens), these Pseudonocardia strains spanned essentially the full diversity within the genus Pseudonocardia [Supplementary Table S522]. This suggests that different Pseudonocardia strains could compete with each other for resources, perhaps evolving features that confer competitive advantages but that are simultaneously detrimental to the ant hosts5,40.
Finally, we found a great diversity of Pseudonocardia strains in the soil samples, but as a low percentage of overall 454-sequence reads (45 different Pseudonocardia strains among 2,067 Pseudonocardia sequences from 202,908 total sequences characterized for soil). Detection of Pseudonocardia in soil samples could be more difficult than in ant samples because soil samples may be more difficult to extract, contain more PCR inhibitors, or facilitate primer competition41. Pseudonocardia were more difficult to detect in culture-dependent screens of soil because, unlike the plates for ant and garden isolation, soil-isolation plates were more readily overgrown with fungi. Thus, estimates of Pseudonocardia prevalence in soil and ant nests cannot be directly compared in our culture-dependent screens. The 45 strains from the culture-independent analysis covered the full known Pseudonocardia diversity, but the majority of Pseudonocardia from soil belonged to clades 6–10 as defined by22 (Supplementary Table S5). Only 16 sequences from the soil samples belonged to clade 3 (the P. cf. carboxydivorans clade), suggesting that either colonization of the ant integument by Pseudonocardia from soil is highly selective [i.e., symbiont-acquisition is highly selective; sensu18,42,43], or soil is an unlikely source for environmental acquisition of Pseudonocardia. Instead, plant material, which frequently contains endophytic Pseudonocardia closely related to P. cf. carboxydivorans22, may be a more likely external source of Pseudonocardia colonizing the ant integument.
Overall, bacterial communities of ants and soil are easily differentiated from each other (Fig. 1, Supplementary Fig. S3). Bacterial communities of gardens showed some similarity with soil communities (Fig. 1), possibly reflecting the fact that ants forage for garden substrate on the ground. During our sample collections, gardens were more difficult to collect than the ants, and therefore we cannot completely rule out occasional contamination of gardens by accidental soil fallout during excavation (such contamination may also apply to the single male whose bacterial community clustered among gardens and soil; Fig. 1).
454-sequencing revealed some previously unknown and potentially important bacterial associates such as Solirubrobacter (order Solirubrobacterales), which comprised approximately a third of the total sequence reads from the ant samples. Solirubrobacter has previously been reported in agricultural soils, soil crust, and earthworm burrows44,45,46, but the biology of these bacteria is otherwise unknown. A prior study also found Solirubrobacter in one lab colony of T. septentrionalis5. Independent screening of the internal (e.g., gut) and external microbiomes will be needed to identify the exact location of Solirubrobacter associated with T. septentrionalis ants. In addition, future culture-dependent surveys of attine integumental microbiomes need to improve isolation methods, which failed so far to reveal the abundant presence of Solirubrobacter and Microlunatus on any attine ant using a minimum-carbon chitin medium for screening (Table 3).
Additional bacterial associates of the ants were Bacillus, Burkholderia, Corynebacterium, Mesoplasma, and Spiroplasma, which were detected as a high percentage of sequence reads only in a few ant samples. This suggests that these bacteria may be pathogens that infect only the odd ant, or these bacteria are acquired by ants only under unique circumstances, such as a unique foraging history. Both Spiroplasma and the closely related Mesoplasma are known insect pathogens, but the frequency of Spiroplasma in ants is relatively unknown47,48,49,50, whereas Mesoplasma and other Entoplasmatales bacteria have been found in diverse ant lineages51. Future research should elucidate the roles of these bacterial associates in the biology of T. septentrionalis.
While culture-dependent methods capture some of the overall actinobacterial diversity associated with T. septentrionalis, our study shows that the traditional minimum-carbon chitin medium appears insufficient to adequately characterize dominant actinobacterial associates (e.g., Solirubrobacter, Microlunatus) and favor growth of specific bacteria (e.g., Pseudonocardia, Amycolatopsis, Kribbella; Table 3). The observed differences between culture-dependent and culture-independent screens are undoubtedly due to isolation biases known for culture-dependent methods5,22,26. While 16S-amplicon 454-sequencing reflects more accurately the total bacterial community composition, greater sequencing depths are needed to characterize the diversity of rare bacterial associates. Moreover, the finding of several Pseudonocardia species on workers of the same ant nest, and the finding of cryptic Pseudonocardia diversity (i.e., several P. cf. carboxydivorans morphotypes coexisting on workers of the same nest) calls for more careful characterization of the ant- and garden-associated microbiomes in future studies, and for continued rethinking of the interaction complexity inherent in such diversity [e.g., possible competition between bacteria occupying the same niche on the ant integument5,40,43].
Complex interactions within host-associated microbiomes do not necessarily rule out overall beneficial contributions of microbes to host fitness18,43, but research into such complex associations faces a series of challenges: the roles of symbiotic microbes are likely diverse and context-dependent; the roles can change rapidly over evolutionary time; strongly interacting host-microbe associations may be difficult to distinguish from unimportant transient associations; significant contribution of a particular microbe to host fitness can not readily be inferred from microbe abundance; and strongly-interacting hosts and microbes often lack the footprints of evolutionary modification that document co-evolutionary interplay. A pluralistic conceptual approach acknowledging diverse functions of microbiomes is therefore most likely to advance understanding of the attine-associated microbes37.
Methods
Sample Collection
Every month between January and September 2009, we searched for T. septentrionalis nests along a 200 meter trail segment at Stengl “Lost Pines” Biological Station, Smithville, Bastrop County, Texas (GPS N30.086, W97.168, elevation: 145 m; see Supplementary Methods). We collected ants, fungal gardens, soil from the nest chamber, and worker-excavated soil from four T. septentrionalis nests every month, except in January (2 nests), February (1 nest), and September (3 nests). All samples were collected into two sterile vials, one vial with 1 mL of 100% ethanol for culture-independent 454-pyrosequencing analyses of bacterial community composition, and a second vial with saline buffer solution for culture-dependent isolation of actinomycete bacteria.
We collected from all nests so-called “outside workers” (exiting or incoming extranidal ants collected from the nest mound before nest excavation), “garden workers” (intranidal ants collected from the surface of the garden), and, when present (June – August), reproductive females and males. The great majority of female reproductives were collected as winged individuals in the nest (pre-mating flights), but a few females encountered late in the mating season (August) had shed their wings inside their natal nests. We assumed that such dealate females found late in the season had failed to leave the nest for a mating flight, and were in the process of gradually assuming a worker-like role; such caste transition has been observed in a number of attine species38. When possible, five ants were collected for each worker sample, but in some cases fewer ants had to be collected because colonies were too small, or showed little or no foraging activity at the time of collecting. Samples of reproductive females or males were pooled with one to three individuals per vial. A few live garden workers, outside workers, males, and female reproductives were also collected in empty, sterile vials and analyzed for the body-segment-specific presence of actinomycete bacteria using culture-dependent isolation (i.e., targeting bacteria on the head, mesosoma, or metasoma).
Each nest was excavated by first digging a 40–50 cm deep hole at ∼15 cm distance from the entrance tunnel, and then carefully excavating laterally until a garden chamber was found. Garden chambers were carefully accessed from the side to minimize fallout of soil that would contaminate the garden, but subtle, inadvertent contamination with soil may have occurred in some garden samples. A ∼1 cm3 fragment of uncontaminated garden was collected from the chamber and placed immediately into vials with ethanol or buffer. Additionally, ∼5 mg of soil was collected from the chamber wall by scraping a 1–2 mm thick layer off the wall (chamber soil), and ∼5 mg freshly deposited soil excavate was collected from the edge of the nest mound where such excavate had been deposited by workers (excavated soil). All soil and garden samples were collected with flame-sterilized spatulas and forceps, which were allowed to cool before each collection. After collection, the chambers were carefully closed and the excavated hole was refilled with soil so that the nest would survive. For comparison, we also included four samples of non-attine workers (Pheidole sp.) collected about 50 meters distant from the T. septentrionalis collection site (n = 3 samples of Pheidole including 3–5 workers, July) or next to a T. septentrionalis nest (n = 1 samples of 3 Pheidole workers, August).
The samples collected in January and September 2009 were analyzed only with culture-dependent methods, but all other samples were analyzed with both culture-dependent and culture-independent methods. Samples in saline buffer solution and live ants from the field were processed via culture-dependent methods in the lab on the same collection day. Ethanol-preserved samples were stored at −80°C and shipped at the end of the study as a single batch to the Biofilms Institute in Lubbock, TX for 454-pyrosequencing of 16S rDNA amplicons from bacteria, following the methods of5,47,52.
Culture-dependent isolation and identification
Actinomycete bacteria were isolated on chitin-medium5, a minimum-carbon medium that appears to favor growth of autotrophic bacteria5,22,26. See Supplementary Methods for detailed culturing methods. We identified the actinomycete morphotypes by Sanger-sequencing a portion of the 16S rDNA gene (Supplementary Table S6), and classifying in fall 2009 the sequences to genus or species based on the closest hit via BLASTn at the National Center for Biotechnology Information (NCBI) Core Nucleotide database (Supplementary Methods). Sequences are deposited at Genbank (accessions JN413390-JN413649).
Bacterial Tag-encoded FLX 454-Pyrosequencing (bTEFAP) and BLAST
bTEFAP was performed on a FLX Genome Sequencer using Titanium protocols and reagents (Roche, Indianapolis, IN) at the Medical Biofilm Research Institute (MBRI; Lubbock, TX), following methods described previously47,52,53,54,55. 16S-amplicons from whole bacterial communities associated with garden workers (n = 25 samples), outside workers (n = 25), female reproductives (n = 9), males (n = 3), gardens (n = 22), chamber soils (n = 25), excavated soils (n = 24), and non-attine control ants (Pheidole sp., n = 4) were PCR-amplified with primers designed to span the 16S variable regions V1–V3 using primers Gray28F 5′GAGTTTGATCNTGGCTCAG and Gray519R 5′GTNTTACNGCGGCKGCTG.
Raw sequences generated from bTEFAP were screened and trimmed based upon quality scores, depleted of short reads (<300 bp) and of chimeras using the black box chimera check software (B2C2)56. The 574,008 sequences remaining after these quality checks were assigned to bacterial types via BLAST at a minimum identity match of 75% using a reference database curated at the MBRI. The reference database contained only high-quality sequences derived from the National Center for Biotechnology Information, using selection criteria described at the Ribosomal Database Project version 957. All sequences that did not match the minimum of 75% identity (9.1% of the total sequences) were excluded from the BLAST output and were assumed to be either of poor quality or derived from unclassified bacteria. The BLAST hit from each individual sequence was analyzed separately for each sample, and presented as a percentage of sequence reads for each bacterial genus found in individual samples. These percentages are generally interpreted as estimates of relative bacterial abundances, but biases in 454-sequencing can reduce accuracy of abundance estimates31.
In a first identification by BLAST against the reference database, 454-sequences were identified to their closest taxonomic level (Supplementary Table S1). Specifically, the reference BLAST-assignments were classified by percent sequence-identity to particular taxonomic levels (i.e., sequences with greater than 97% sequence-identity match were resolved to species: between 95–97% to genus, between 90–95% to family, between 85–90% to order, 80–85% to class, and 75–80% to phylum).
A second identification interpreted the above BLAST-assignments differently by forcing assignments to the closest genus match at a minimum stringency of 75% identity (Supplementary Table S2). This forced BLAST table was not intended to generate a strict reference assignment, but was useful to compare the sequence-read variation observed within sample types. Despite a 75% identity minimum, the average BLAST assignment used in this table had a 93.8% identity (standard deviation of 4.2%), suggesting confidence in the majority of the genus reference assignments. However, many of the rare (singleton) genus assignments are likely contentious. The raw sequence reads (or tags), the % identity score, and the genus reference assignments can be found in Supplementary Table S7. To test the validity of the database curated at the MBRI, all the raw sequences were also referenced using the Ribosomal Database Project's classifier (http://rdp.cme.msu.edu/classifier/classifier.jsp)27. Short-nucleotide 454-reads of microbial communities have been deposited at the NCBI Short Read Archive accession SRP008669, with the exception of 12 samples JLM-J1-J12 for which raw sff files were accidentally lost (Supplementary Methods).
From the forced-genus BLAST, we calculated the average number of bacterial genera for ant samples (garden worker, outside worker, and male and female reproductives combined) and for the soils (chamber and excavate combined) from each nest (n = 25) and statistically compared to each other using a 2-tailed t-test. As a rough measure of possible ecological links between ant-associated microbes and microbes in the soil, we counted the bacterial genera shared by the ant and soil samples collected from each nest.
454 Sequencing Fast UniFrac Analyses
For community comparison analyses, we randomly sampled 1000 454-sequences from each sample (Supplementary Methods). The program CD-Hit58 was used to cluster similar sequences at a 97% identity and an alignment was generated using the SILVA database in the Mothur sequencing pipeline59. An approximate maximum-likelihood tree was generated using FastTree60. We used an unweighted principal coordinate analysis (PCoA) in Fast Unifrac61 to assess the differences between the bacterial communities of ant, garden and soil samples. We additionally used the UniFrac distance matrix to test if the ant and soil bacterial communities were statistically different by running the adonis function in the vegan package using R 2.8.1 with 200 permutations with ant and soil as the category variables (www.r-project.org).
Pseudonocardia BLAST to species designation
We performed a custom BLAST to determine the number of different Pseudonocardia strains identified through 454-pyrosequencing in each sample. The initial BLAST of the 454-sequences (above) included only a few reference species of Pseudonocardia, so we explored the observed Pseudonocardia diversity at higher resolution with the help of a custom BLAST against a more complete Pseudonocardia database. This database contained 116 unique Pseudonocardia 16S-sequences (accessions listed in Supplementary Methods) selected from a comprehensive Pseudonocardia phylogeny reported in22. The BLAST results were parsed into a table (Supplementary Table S8) using the BioPerl Bio::SearchIO module [www.bioperl.org/wiki/BioPerl62].
Author Contributions
JLM and RS collected T. septentrionalis field colonies and performed the culture-dependent isolations. SED and the Medical Biofilm Research Institute performed the 454-sequencing and BLAST analyses. EM wrote custom Perl scripts, aided in the analysis of the 454 dataset, and performed statistics in R. HDI sequence-identified the bacterial isolates and analyzed the 454-sequence information. UGM conceived of the study, and JLM and RS participated in its design. HDI, JLM, and UGM wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Dataset
Supplementary Tables S1, S2, S4, S8
Supplementary Table S7
Acknowledgments
We thank Franziska Rall, Keith Miller, Cindy Miller, and Vera Magero for help with ant collection; Thinh Dang for help with isolations and vouchering. Quinn McFrederick, Jon Seal, Katrin Kellner, Sabrina Amador, and two anonymous reviewers for constructive comments. The work was supported by the National Science Foundation (NSF-REU supplement to JLM; DEB-0920138, DEB-0919519, and DEB-0639879 to UGM); an Undergraduate Research Fellowship from the University of Texas at Austin to JLM; and the W.M. Wheeler Lost Pines Endowment.
References
- Carreiro S. C. et al. Yeasts associated with nests of the leaf-cutting ant Atta sexdens rubropilosa Forel, 1908. Anton. Leeuw. 71, 243–248 (1997). [DOI] [PubMed] [Google Scholar]
- Santos A. V., Dillon R. J., Dillon V. M., Reynolds S. E. & Samuels R. I. Occurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol. Lett. 239, 319–323 (2004). [DOI] [PubMed] [Google Scholar]
- Mueller U. G., Gerardo N. M., Aanen D. K., Six D. L. & Schultz T. R. The evolution of agriculture in insects. Annu. Rev. Ecol. Evol. Syst. 36, 563–595 (2005). [Google Scholar]
- Rodrigues A., Cable R. N., Mueller U. G., Bacci M. & Pagnocca F. C. Antagonistic interactions between garden yeasts and microfungal garden pathogens of leaf-cutting ants. Anton. Leeuw. 96, 331–342 (2009). [DOI] [PubMed] [Google Scholar]
- Sen R. et al. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. USA 106, 17805–17810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen G. et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 6, e1001129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C. R., Mueller U. G. & Malloch D. The agricultural pathology of ant fungus gardens. Proc. Natl. Acad. Sci. USA 96, 7998–8002 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C. R., Bot A. N. M. & Boomsma J. J. Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos 101, 91–102 (2003). [Google Scholar]
- Mueller U. G., Dash D., Rabeling C. & Rodrigues A. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution 62, 2894–2912 (2008). [DOI] [PubMed] [Google Scholar]
- Poulsen M., Erhardt D. P., Molinaro D. J., Lin T.-L. & Currie C. R. Antagonistic bacterial interactions help shape host-symbiont dynamics within the fungus-growing ant-microbe mutualism. PLoS ONE 2, e960 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeder S., Wirth R., Herz H. & Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 106, 4742–4746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro M. J. & Currie C. R. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can. J. Microbiol. 51, 441–446 (2005). [DOI] [PubMed] [Google Scholar]
- Currie C. R. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311, 81–83 (2006). [DOI] [PubMed] [Google Scholar]
- Barke J. et al. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 8, 109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro M. J. et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc. Roy. Sci. B-Biol. Sci. (2011).10.1098/rspb.2010.2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenian I. et al. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc. Natl. Acad. Sci. USA 108, 1955–1960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-G. et al. Identification of functionally clustered nystatin-like biosynthetic genes in a rare actinomycetes, Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 36, 1425–1434 (2009). [DOI] [PubMed] [Google Scholar]
- Barke J., Seipke R. F., Yu D. W. & Hutchings M. I. A mutualistic microbiome: How do fungus-growing ants select their antibiotic-producing bacteria? Commun. Integr. Biol. 4, 1–3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C. R., Scott J. A., Summerbell R. C. & Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398, 701–704 (1999). [Google Scholar]
- Kost C. et al. Non-specific association between filamentous bacteria and fungus-growing ants. Naturwissenschaften 94, 821–828 (2007). [DOI] [PubMed] [Google Scholar]
- Mikheyev A. S., Vo T. & Mueller U. G. Phylogeography of post-Pleistocene population expansion in a fungus-gardening ant and its microbial mutualists. Mol. Ecol. 17, 4480–4488 (2008). [DOI] [PubMed] [Google Scholar]
- Mueller U. G., Ishak H. D., Lee J. C., Sen R. & Gutell R. R. Placement of attine ant-associated Pseudonocardia in a global Pseudonocardia phylogeny (Pseudonocardiaceae, Actinomycetales): a test of two symbiont-association models. Anton. Leeuw. 98, 195–212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A., Mueller U. G., Ishak H. D., Bacci M. J. & Pagnocca F. C. Ecology of microfungal communities in gardens of fungus-growing ants (Hymenoptera: Formicidae): A year-long survey of three species of attine ants in Central Texas. FEMS Microbiol. Ecol. 78, 244–255 (2011). [DOI] [PubMed] [Google Scholar]
- Seal J. N. & Tschinkel W. R. Colony productivity of the fungus-gardening ant Trachymyrmex septentrionalis (Hymenoptera: Formicidae) in a Florida pine forest. Ann. Entomol. Soc. Am. 99, 673–682 (2006). [Google Scholar]
- Rabeling C., Cover S. P., Johnson R. A. & Mueller U. G. A review of the North American species of the fungus-gardening ant genus Trachymyrmex (Hymenoptera: Formicidae). Zootaxa 1664, 1–53 (2007). [Google Scholar]
- Hamady M. & Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 19, 1141–1152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M., Cafaro M., Boomsma J. J. & Currie C. R. Specificity of the mutualistic association between actinomycete bacteria and two sympatric species of Acromyrmex leaf-cutting ants. Mol. Ecol. 14, 3597–3604 (2005). [DOI] [PubMed] [Google Scholar]
- Pinto-Tomás A. A. et al. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326, 1120 –1123 (2009). [DOI] [PubMed] [Google Scholar]
- Zucchi T. D., Guidolin A. S. & Cônsoli F. L. Isolation and characterization of actinobacteria ectosymbionts from Acromyrmex subterraneus brunneus (Hymenoptera, Formicidae). Microbiol. Res. 166, 68–76 (2011). [DOI] [PubMed] [Google Scholar]
- Amend A. S., Seifert K. A. & Bruns T. D. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol. Eco. 19, 5555–5565 (2010). [DOI] [PubMed] [Google Scholar]
- Bacci M. J., Ribeiro S. B., Casarotto M. E. F. & Pagnocca F. C. Biopolymer-degrading bacteria from nests of the leaf-cutting ant Atta sexdens rubopilosa. Braz. J. Med. Biol. Res. 28, 79–82 (1995). [Google Scholar]
- Caldera E. J., Poulsen M., Suen G. & Currie C. R. Insect symbioses: a case study of past, present, and future fungus-growing ant research. Environ. Entomol 38, 78–92 (2009). [DOI] [PubMed] [Google Scholar]
- Hughes W. O. H., Pagliarini R., Madsen H. B., Dijkstra M. B. & Boomsma J. J. Antimicrobial defense shows an abrupt evolutionary transition in the fungus-growing ants. Evolution 62, 1252–1257 (2008). [DOI] [PubMed] [Google Scholar]
- Hughes D. P., Evans H. C., Hywel-Jones N., Boomsma J. J. & Armitage S. A. O. Novel fungal disease in complex leaf-cutting ant societies. Ecol. Entomol. 34, 214–220 (2009). [Google Scholar]
- Walker T. N. & Hughes W. O. H. Adaptive social immunity in leaf-cutting ants. Biol. Lett 5, 446–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoso T., Moreira D. & Samuels R. Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biology Letters In press, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber N. A. Gardening ants the attines. (The American Philosophical Society: Philadelphia, 1972). [Google Scholar]
- Rohlfs M. & Kürschner L. Saprophagous insect larvae, Drosophila melanogaster, profit from increased species richness in beneficial microbes. J. Appl. Entomol. (2009). doi:10.1111/j.1439-0418.2009.01458.x [Google Scholar]
- Boomsma J. J. & Aanen D. K. Rethinking crop-disease management in fungus-growing ants. P. Natl. Acad. Sci. USA 106, 17611–17612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein L. M., Sul W. J. & Blackwood C. B. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75, 5428–5433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs J. L., Mueller U. G., Wilcox T. P. & Bull J. J. The evolution of cooperation. Q. Rev. Biol. 79, 135–160 (2004). [DOI] [PubMed] [Google Scholar]
- Sachs J. L., Essenberg C. J. & Turcotte M. M. New paradigms for the evolution of beneficial infections. Trends Ecol. Evol. 26, 202–209 (2011). [DOI] [PubMed] [Google Scholar]
- Kim M. K. et al. Solirubrobacter soli sp. nov., isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 57, 1453–1455 (2007). [DOI] [PubMed] [Google Scholar]
- Reddy G. S. N. & Garcia-Pichel F. Description of Patulibacter americanus sp. nov., isolated from biological soil crusts, emended description of the genus Patulibacter Takahashi et al. 2006 and proposal of Solirubrobacterales ord. nov. and Thermoleophilales ord. nov. Int. J. Syst. Evol. Microbiol. 59, 449 (2009). [DOI] [PubMed] [Google Scholar]
- Singleton D. R. et al. Solirubrobacter pauli gen. nov., sp. nov., a mesophilic bacterium within the Rubrobacteridae related to common soil clones. Int. J. Syst. Evol. Microbiol. 53, 485–490 (2003). [DOI] [PubMed] [Google Scholar]
- Ishak H. D. et al. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb. Ecol. 61, 821–831 (2011). [DOI] [PubMed] [Google Scholar]
- Gros O. et al. Serological and molecular characterization of Mesoplasma seiffertii strains isolated from hematophagous dipterans in France. Int. J. Syst. Bacteriol 46, 112–115 (1996). [DOI] [PubMed] [Google Scholar]
- Jaenike J., Unckless R., Cockburn S. N., Boelio L. M. & Perlman S. J. Adaptation via symbiosis: recent spread of a drosophila defensive symbiont. Science 329, 212–215 (2010). [DOI] [PubMed] [Google Scholar]
- Bastian F. O. et al. Spiroplasma spp. from transmissible spongiform encephalopathy brains or ticks induce spongiform encephalopathy in ruminants. J. Med. Microbiol. 56, 1235–1242 (2007). [DOI] [PubMed] [Google Scholar]
- Funaro C. F. et al. Army ants harbor a host-specific clade of Entomoplasmatales bacteria. Appl. Environ. Microbiol. online first, AEM.01896-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E. et al. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8, 43–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway T. R. et al. Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing. Poultry Sci. 88, 298–302 (2009). [DOI] [PubMed] [Google Scholar]
- Smith D. M. et al. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med. Genomics 3, 41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. T. et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 78, 1509–1519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontcharova V. et al. Black box chimera check (B2C2): a Windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol. J. 4, 47–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37, D141–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. & Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- Schloss P. D. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE 4, e8230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Lozupone C. & Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J. E. et al. The Bioperl Toolkit: Perl Modules for the Life Sciences. Genome Res. 12, 1611–1618 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset
Supplementary Tables S1, S2, S4, S8
Supplementary Table S7


