Abstract
An anthrax sub-unit vaccine, comprising recombinant Protective Antigen (rPA83) and aluminium hydroxide adjuvant (Alhydrogel®) is currently being developed. Here a series of biophysical techniques have been applied to free and adjuvant bound antigen. Limited proteolysis and fluorescence identified no changes in rPA83 tertiary structure following binding to Alhydrogel and the bound rPA83 retained two structurally-important calcium ions. For adsorbed rPA83, differential scanning calorimetry revealed a small reduction in unfolding temperature but a large decrease in unfolding enthalpy whilst urea unfolding demonstrated unchanged stability but a loss of cooperativity. Overall, these results demonstrate that interactions between rPA83 and Alhydrogel have a minimal effect on the folded protein structure and suggest that antigen destabilisation is not a primary mechanism of Alhydrogel adjuvancy. This study also shows that informative structural characterisation is possible for adjuvant bound sub-unit vaccines.
Keywords: Anthrax, sub-unit vaccine, alhydrogel, fluorescence, calorimetry, PA83
Introduction
The classical approach to vaccine manufacture generally employs whole cell preparations or live attenuated micro-organisms. Although these vaccines have been effective in producing host immunity, they are not without their problems and can result in adverse reactions when administered to vaccinees whilst attenuated live organisms can also revert to virulent forms [1]. To overcome such issues, there has been a recent trend toward safer, more defined, sub-unit vaccine formulations using recombinant proteins as antigens [2]. However, a drawback of sub-unit vaccines is that recombinant proteins by themselves are often poor immunogens, unable to induce the desired level of protection [3]. To improve their potency, such antigens are combined with an adjuvant that augments the immune response by enhancing antigen delivery to antigen presenting cells and by activating cells of the immune system [4]. With these vaccines, it is essential to characterise the formulation and particularly the antigen for the following reasons: (1) to show that they can be consistently manufactured to the same standards; (2) to show that they are stable during prolonged storage; (3) to understand the interaction of their basic elements and (4) to comprehend the mechanism of their biological activity. Accordingly, with the development of new sub-unit vaccine against anthrax [5], we employed a set of modified biophysical methods to investigate the interaction between rPA83 and Alhydrogel with particular emphasis upon potential changes in protein structure.
The adjuvant concept was introduced in 1925 by Gaston Ramon who showed that combination of diphtheria or tetanus toxoids with bread crumbs, agar, starch, and oil produced higher levels of immunity [6]. Since then many more immune enhancing substances have been identified but as yet few compounds have been licensed for human use. By far the most widely used are the aluminium based hydrated gels, namely aluminium hydroxide and aluminium phosphate [7]. This technology dates back to the work of Glenny (1926) on alum precipitation of diphtheria toxoid [8] and although extensive research is being conducted to develop novel adjuvants, aluminium based hydrated gels are still predominant as a consequence of their regulatory position and extensive safety record.
Several mechanisms of action for aluminium based adjuvants have been proposed [9]. These include: a repository of antigen in tissues that slowly releases the antigen, stimulating the immune system for a prolonged period of time, i.e. the depot effect; an enhanced delivery of the antigen to antigen presenting cells, resulting in more efficient antigen presentation [10–12] an activation of inflammatory dendritic cells by uric acid [13]; and a structural destabilisation of the antigen protein, resulting in enhanced antigen presentation due to more efficient proteolytic processing of the antigen by the antigen presenting cells [14]. In reality it is probable that aluminium based adjuvants work by more than one mechanism, the relative importance of each being vaccine specific, depending on the antigen and the aluminium adjuvant formulation.
There have been a number of biophysical analyses specifically looking at the structure of model antigens bound to alhydrogel, which have revealed subtle effects upon the bound protein[14–16]. However, for the vast majority of adjuvant containing commercial vaccines, the antigen – adjuvant interactions have not been well characterised. In part this is due to the particulate nature of such vaccines [17], which is problematic for many protein analytical techniques. Indeed, because of such difficulties, proteins are often desorbed from the adjuvant for analysis thus losing important structural information on the protein bound state. To properly understand the effects of formulation it is essential to characterise the intact sub-unit vaccine. We have recently shown that electron microscopy can reveal the tertiary and quaternary structure of adjuvant bound proteins [18]. For a full picture of the effects of adjuvant adsorption on proteins it is necessary to investigate their thermodynamic stability [14]. It is in this context that we have investigated antigen-adjuvant interactions in a developmental vaccine against anthrax.
In recent years, particularly following the anthrax attack in the United States in 2001, a substantial effort has been made to develop an anthrax subunit vaccine. The existing U.S. licensed anthrax vaccine (AVA; Biothrax) has variable reactogenicity and a protracted immunization schedule [19] which could be avoided by a safer, more potent and well characterised vaccine. The anthrax sub-unit vaccine currently under development uses purified Protective Antigen (PA83) of Bacillus anthracis as the only antigen. PA83 is a non-toxic component of the anthrax tripartite toxin which binds to receptors on host cells and subsequently translocates the toxic lethal and edema factors into the cytosol. PA83 is an 83 kDa protein, rich in β-strands and consisting of 4 domains with different functions [20]. Domain 1 contains two structurally important calcium ions [21] and a site for cleavage by the activating pro-protein convertase, furin, that removes a 20 kDa N-terminal fragment (PA20). This allows the remaining 63 kDa molecule (PA63) to oligomerise as a heptamer [22]. Domain 2 forms the transmembrane channel which translocates toxic factors into the host cell [23], Domain 3 controls oligomerisation [24] and Domain 4 binds to the anthrax receptor [25, 26].
To provide a well characterised and safe antigen, recombinant Protective Antigen (rPA83), produced in non-pathogenic Escherichia coli, has been used as the drug substance. In the formulated vaccine, this drug substance is complexed with Alhydrogel adjuvant, consisting of fine particles with an average dimension of 4.5 × 2.2 × 10 nm [27]. The particles are made of corrugated layers of aluminium oxyhydroxide that are held together by hydrogen bonding. The adjuvant has a relatively large surface area of 514 ± 36 m2/g and can bind as much as 3 grams of protein per gram of adjuvant [27, 28]. In aqueous solution the particles aggregate into 1–10 μm clusters [29] and with a pI of 11.1, these are positively charged at physiological pH and adsorb negatively charged proteins, including rPA83 (pI = 5.6), electrostatically [30]. This combination of rPA83 and Alhydrogel has been shown to be protective in animal models [5] and safe and immunogenic in man [31].
Despite the efficacy of the vaccine, there is a paucity of data on the structure and physical properties of rPA83 adsorbed to Alhydrogel. Therefore, a series of biophysical methods have been developed which extend previous approaches [14–17] used to investigate the protein bound state. Here we characterise a commercially relevant subunit vaccine and show that the interactions between the rPA83 antigen and Alhydrogel adjuvant have little or no effect on the protein structure, whilst substantially altering the thermodynamic properties. These findings are of fundamental importance to the biopharmaceutical industry as well as relevant to those researching proteins adsorbed to solid surfaces.
Materials and Methods
Suppliers
All chemicals and reagents were purchased from either Sigma-Aldrich Company Ltd (UK) or Melford Laboratories (UK), unless otherwise stated. Recombinant protective antigen (rPA83) was provided by Avecia Biologics, Billingham, (UK). Aluminium hydroxide gel (Alhydrogel®) was purchased from Brenntag Biosector (Denmark).
Adsorption of rPA83 to Alhydrogel®
The concentration of rPA83 protein was determined by measuring absorbance at 280 nm using a 1 cm path-length quartz cuvette (Hellma, Germany) in a UV-1800 spectrophotometer (Shimadzu, Japan). A value of 1.176 was used as the absorbance at 280 nm of a 0.1 % w/v rPA83 solution. Adsorption of rPA83 to Alhydrogel was performed by adding the protein solution to the adjuvant suspension at ambient temperature. The working vaccine sample contained 50 μg/ml rPA83, 1 mg/ml Alhydrogel®, 30 mM MOPS, 100 mM NaCl, pH 7.0 unless otherwise stated. To ensure thorough adsorption, the mixture was inverted several times and allowed to stand on a bench for at least 10 minutes prior to any experiment. Throughout this article the term ‘absorbed rPA83’ will be used to describe the rPA- Alhydrogel suspension. To determine the quantity of adsorbed rPA83, the rPA83-Alhydrogel mixture was centrifuged at 3000 × g for 3 minutes and the protein concentration of the supernatant determined. The amount of bound rPA83 was calculated by subtracting the amount of protein in the supernatant from the total protein in the sample [14]. For kinetic studies 100 μg of rPA83 was added to 1.3 mg Alhydrogel and incubated from 10 sec to 1.5 hours before the amount of adsorbed protein was measured. A single measurement was made at each time point.
Langmuir linear regression
Solutions containing 0 – 2.35 mg rPA83 were mixed with 1 mg of Alhydrogel and the amount of adsorbed protein was measured. The binding capacity and adsorption coefficient were then determined by Langmuir linear regression, given by (1):
| (1) |
where c is the aqueous concentration of protein, Γ is the amount of adsorbed protein, Γmax is the maximum amount of adsorbed protein and K is the Langmuir equilibrium constant [32]. In the plot of c/Γ versus c, the binding capacity was calculated as 1/slope and adsorption coefficient as 1/Y-axis intercept.
Fluorescence measurements
Fluorescence measurements were carried out at an ambient temperature using a 1 cm path-length quartz cuvette (Hellma, Germany) in a Cary Eclipse spectrofluorometer (Varian, USA). The working vaccine sample contained 50 μg/ml rPA83, ± 1 mg/ml Alhydrogel®, 30 mM MOPS, 100 mM NaCl, pH 7.0. All samples were stirred while recording fluorescence. Heat denatured samples were prepared by incubating the sample at 95 °C for 3 minutes (and cooling down to ambient temperature) prior to measuring fluorescence. The tryptophan emission spectrum was recorded between 300 nm and 400 nm at an excitation wavelength of 280 nm. The Nile-red fluorescence emission spectrum was recorded between 570 nm and 800 nm at an excitation wavelength of 540 nm. Nile-red dye, final concentration 2.6 μM, was added to the sample immediately before measuring fluorescence.
For determination of calcium concentration the emission spectrum of Calcium Green 5N dye (Invitrogen, UK) was recorded between 515 nm and 600 nm at an excitation wavelength of 506 nm. Prior to measuring fluorescence, all rPA83-Alhydrogel and heat denatured samples were centrifuged at 3000 × g for 3 minutes. The supernatant was removed and mixed with Calcium Green 5N (final concentration 0.7 μM) and the calcium content of the supernatant was determined from a standard curve, ranging from 0 to 24 μM Ca2+.
The barycentric means of tryptophan and Nile red fluorescence emission spectra were calculated according to equation (2):
| (2) |
where, λbcm is the barycentric mean, λ is the wavelength, Fλ is the fluorescence intensity at wavelength λ [33]. For tryptophan fluorescence m = 300 nm and n= 400 nm and Nile Red m = 570 nm and n= 800 nm. For solvent accessibility measurements tryptophan fluorescence (described above) was quenched by 0.025 – 0.25 M solution of sodium iodide. The tryptophan emission spectra were then integrated (as above) and the area was plotted according to the Stern-Volmer relationship (3):
| (3) |
where F is the fluorescence intensity, F0 is the fluorescence intensity without quencher, Q is the molar concentration of quencher and KSV is the Stern-Volmer quenching constant [34].
Fluorescence measurements of thermal denaturation were performed in a Cary Eclipse spectrofluorometer (Agilent) equipped with a Peltier temperature controller. A 1 cm path-length quartz cuvette (Hellma, Germany) with a Teflon lid was used. While recording fluorescence all samples were stirred using a small magnetic stirring bar placed inside the cuvette.
Non-equilibrium protein thermal unfolding was monitored by measuring fluorescence at 340 nm at an excitation wavelength of 280 nm while scanning from 25 °C to 65 °C at the rate of 1 °C min−1. Steady state protein thermal unfolding measurements exploited the irreversibility of rPA83 thermal denaturation [21]. The protein sample was incubated at fixed temperature values (2.5 °C intervals) for 5 min each, cooled to room temperature (23 °C) and assessed by tryptophan fluorescence. Each measurement was made with a new sample and the barycentric mean was calculated for each. Analysis of samples was performed in replicates of three and the mean value was reported. Where appropriate, error bars show the standard deviation. Fluorescence curves/scans were reported as an average curve/scan of three independent replicates.
Unfolding of rPA83 in urea
Samples of standard and adsorbed rPA83 were mixed with 0 – 8 M urea and incubated at ambient temperature for 24 hours. The barycentric mean of the tryptophan fluorescence was calculated for each scan. The barycentric mean values were normalised, assuming that the protein was fully folded (normalised value of 0) at 0 M urea and fully unfolded (normalised value of 1) at 8 M urea. The standard free energy change of each unfolding transitions was calculated in Origin software by fitting a two state unfolding equilibrium model [35] with a non-linear least squares fitting routine kindly supplied by Dr. D. Hough, University of Bath, UK. To check whether rPA83 was still adsorbed to Alhydrogel in urea, samples were centrifuged (to sediment Alhydrogel) and the supernatant was measured by tryptophan fluorescence. There was no protein in the supernatant (see supplementary data). Unfolding of rPA83 protein was repeated at least twice over the full range of urea concentrations. Curves presented here are from one experiment containing approximately 80 individual readings.
Differential scanning calorimetry
Differential scanning calorimetry was performed in a VP-DSC micro-calorimeter (MicroCal, UK). Prior to analysis, all samples were de-gassed in a ThermoVac unit (MicroCal, UK). Samples were scanned between 25 °C and 60 °C at a rate of 1 °C min−1. DSC endotherms were deconvoluted using a non-2-state model with 1 transition in the MicroCal Origin software. Working samples contained: 0.5 mg/ml rPA83, 1 mg/ml Alhydrogel®, 30 mM MOPS, 100 mM NaCl, pH 7.0. The reference material was either sample buffer or 1 mg/ml Alhydrogel prepared in 30 mM MOPS, 100 mM NaCl, pH 7.0 buffer. The analysis was performed in duplicate.
SDS-polyacrylamide gel electrophoresis
SDS-polyacrylamide gel electrophoresis was performed on a 10 % polyacrylamide gel in a discontinuous Laemmli system using the Protean III electrophoresis system (BioRad, UK) at 50 mA. All samples were diluted 1 in 2 with a 200 mM phosphate buffer and then mixed with an equal volume of 2 × SDS loading buffer, and incubated at 95 °C for 5 min. Prior to electrophoresis, all samples were centrifuged using a bench top centrifuge for 5 minutes at maximum speed. Subsequently the alhydrogel free supernatant was loaded on the gel. Gels were stained with Coomassie brilliant blue G-250. The molecular weight of protein bands was estimated by using the Precision plus protein standard (BioRad, UK).
Limited proteolysis
A solution of either standard rPA83 or adsorbed rPA83 was mixed with a solution of protease and allowed to react for 1 hour at an ambient temperature. The mixture contained 0.2 mg rPA83, 0.5 mg Alhydrogel and 1 × 10−3 Units of protease. (trypsin, chymotrypsin, pronase E, proteinase K, V8 and Subtilisin Carlsberg). Proteolysis was terminated by heat denaturation; 25 μl of the reaction mixture was mixed with 25 μl of 200 mM phosphate buffer and 50 μl of 2 × SDS loading buffer, and incubated at 95 °C for 5 min. Products of limited proteolysis were separated by SDS-PAGE. Limited proteolysis was performed several times for each enzyme; images of gels show typical cleavage patterns.
Results
Adsorption of rPA83 to Alhydrogel: time course and Langmuir linear regression
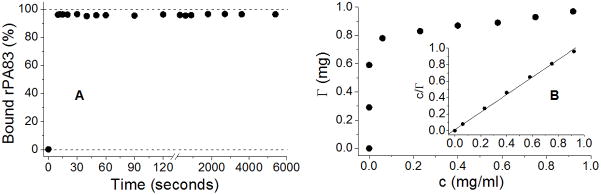
An adsorption time course was determined by measuring the amount of adsorbed rPA83 as a function of time following mixing with Alhydrogel®, (Figure 1A). More than 95 % of the protein was adsorbed to the adjuvant in as little as 10 seconds with no evidence of desorption in the subsequent 90 min tested. This result shows that adsorption occurs spontaneously and is time-independent for protein/adjuvant quantities used. Langmuir linear regression revealed the binding capacity and adsorption coefficient of rPA83 in 30 mM MOPS, 100 mM NaCl, pH 7.0 buffer to be 0.94 mg rPA83/mg Alhydrogel and 83.47 ml/mg, respectively (Figure 1B).
Figure 1.
Binding of rPA83 to Alhydrogel®. A) Adsorption time course, showing the percentage of rPA83 adsorbed to Alhydrogel over various incubation times. On average 96 ± 0.44 % of protein was adsorbed at any given time B) Binding isotherm and the Langmuir linear regression plot (insert). In 30 mM MOPS, 100 mM NaCl, pH 7.0 buffer, the rPA83 protein had a binding capacity of 0.94 mg rPA83/mg Alhydrogel and an adsorption coefficient of 83.47 ml/mg. The isotherm was measured twice, is highly reproducible and one representative example is shown.
Analysis of rPA83 by tryptophan and Nile-red fluorescence
The tryptophan emission spectrum of adsorbed rPA83 was similar to that of standard rPA83 (Figure 2A). Both spectra had identical barycentric mean values of 340.5 ± 0.1 nm. The tryptophan emission spectrum of heat-denatured rPA83 was red-shifted compared to either standard or adsorbed rPA83; this spectrum exhibited much weaker intensity and had a barycentric mean value of 347.0 ± 0.2 nm.
Figure 2.
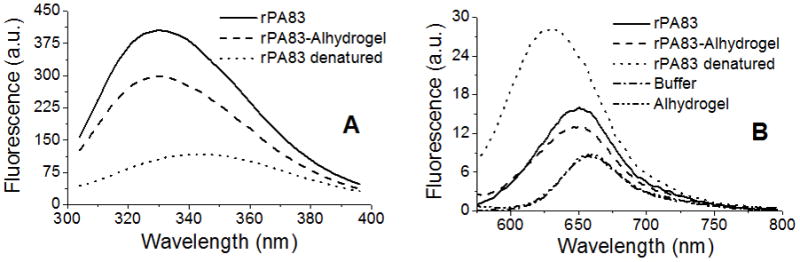
Tryptophan and Nile-red fluorescence of rPA83. A) Tryptophan emission spectrum 50 μg/ml rPA83 ± 1 mg/ml Alhydrogel®, 30 mM MOPS, 100 mM NaCl, pH 7.0 Excitation wavelength = 280 nm and B) Nile-red emission spectrum (samples as above ± 2.6μM Nile Red) Excitation wavelength = 540 nm
Nile-red fluorescence showed that adsorbed rPA83 and soluble rPA83 have similar emission spectra, with barycentric mean values of 654.5 ± 0.1 and 655.6 ± 0.1 nm respectively (Figure 2B) the differences being due to light scattering effects in the bound sample. For comparison the Nile-red emission spectrum of heat-denatured rPA83 was significantly blue-shifted in comparison to either soluble or adsorbed rPA83 and had a substantial enhancement in fluorescent intensity. Its barycentric mean value was 641.0 ± 0.4 nm.
In both tryptophan and Nile-red fluorescence, the emission spectrum of adsorbed rPA83 had a weaker intensity than that of standard rPA83 probably due to solution turbidity decreasing the light penetration and uneven distribution of fluorophores per given unit of volume [36]. Buffer and Alhydrogel controls indicated that Alhydrogel does not bind Nile-red dye. Overall these results demonstrate that the tertiary structure of rPA83 does not change upon adsorption to Alhydrogel®.
Measuring the degree of tryptophan exposure in rPA83
To measure the exposure of tryptophan residues in rPA83, tryptophan fluorescence was quenched with sodium iodide and analysed according to the Stern-Volmer relationship. The magnitude of the slope in the plot (i.e. Stern-Volmer constant) corresponds to the degree of exposure and demonstrated that solvent exposure of tryptophan residues in adsorbed rPA83 was the same as that in soluble rPA83 (Figure 3). The derived Stern-Volmer constants were 0.84 ± 0.04 M−1 and 0.85 ± 0.05 M−1 for absorbed rPA83 and soluble rPA83, respectively. Apo-rPA83, which is a calcium depleted, stable folding intermediate, with solvent-exposed tryptophan residues [21], had a Stern-Volmer constant of 1.64 ± 0.04 M−1. This experiment shows that th local environment of the tryptophan residues remains unchanged when rPA83 is adsorbed to Alhydrogel®.
Figure 3.
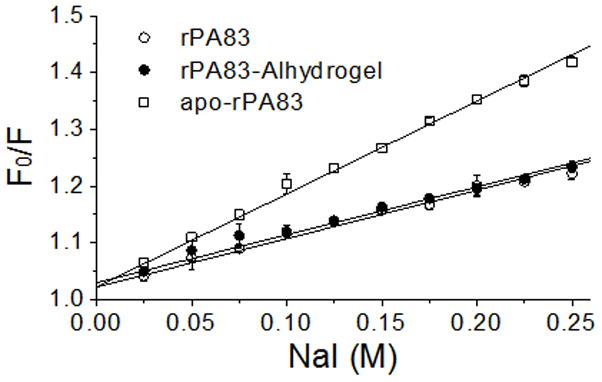
Stern-Volmer plot of soluble rPA83, adsorbed rPA83 and solvent exposed Apo-rPA83 at increasing sodium iodide concentrations. Points represent mean (±SEM) values of three independent measurements.
Measuring the calcium content of rPA83
A sample of 6 μM rPA83, which would theoretically contain 12 μM of calcium, was absorbed onto Alhydrogel and the calcium content of the sample was determined. As demonstrated in Table 1, both soluble and Alhydrogel adsorbed rPA83 retained 2 mol of calcium ions per mol of protein; the calcium only being released upon heat denaturation. The control sample showed that Alhydrogel does not sequester calcium. These data demonstrated that binding of rPA83 to Alhydrogel did not result in loss of bound calcium ions from domain 1.
Table 1.
Quantitative analysis of free calcium content in rPA83 samples, mean (±SEM) of three independent measurements.
| Sample | Calcium (μM) |
|---|---|
| rPA83 | −0.36 ± 0.08 |
| rPA83 heat-denatured | 11.77 ± 0.16 |
| rPA83-Alhydrogel | −0.46 ± 0.07 |
| rPA83-Alhydrogel heat-denatured | 11.70 ± 0.19 |
| Alhydrogel + 12 μM Calcium | 11.26 ± 0.40 |
Thermal unfolding of rPA83
Thermal unfolding of absorbed and soluble rPA83 was evaluated using tryptophan fluorescence. Protein samples were either thermally equilibrated (Figure 4A), or subjected to a temperature gradient increasing at the rate of 1 °C min−1 (Figure 4B). Soluble rPA83 unfolded rapidly over a 45 – 55 °C temperature range, with a transition temperature of 50.4 °C. In contrast, unfolding of adsorbed rPA83 occurred gradually, between 30 °C and 65 °C, with a transition temperature of 47.9 °C. Since calcium ions are structurally important to the rPA83 protein [20], the thermal denaturation curves were repeated but in the presence of 5 mM Ca2+. As shown in Figure 4B, the presence of 5 mM Ca2+ rendered soluble rPA83 more thermostable, increasing the transition temperature by almost 3 °C. In contrast the inclusion of 5 mM Ca2+ had little effect on the stability of adsorbed rPA83. Overall these studies demonstrate that the absorbed rPA83 is restrained from undergoing the normal co-operative unfolding. Moreover, the data indicate that the adsorbed protein is not in equilibrium with calcium ions in solution.
Figure 4.
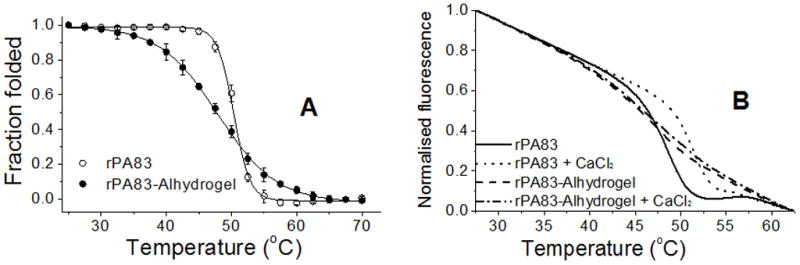
Thermal unfolding of rPA83. A) Tryptophan fluorescence and barycentric mean analysis was used to calculate the fraction of folded rPA83 at the indicated temperature after thermal equilibrium has been reached. Points represent mean (±SEM) values of three independent measurements.
B) Unfolding of rPA83 was monitored by tryptophan fluorescence at 340 nm while increasing temperature at the rate of 1 °C min−1. The protein unfolding was investigated with and without 5 mM calcium.
Unfolding of rPA83 with urea
Chaotrope-induced denaturation of soluble and absorbed rPA83 was also monitored using tryptophan fluorescence. Unfolding of soluble rPA83 occurred via a biphasic mechanism, with transitions taking place near 1 M and 4 M urea respectively (Figure 5A) [21, 37]. Adsorbed rPA83 also unfolded via a biphasic mechanism but the unfolding transitions lacked cooperativity. Broadening of unfolding transitions suggested that adsorbed rPA83 adopted various conformational states as shown also by electron microscopy [18]. Note, in soluble rPA the two transitions are distinct and there is a stable folding intermediate, characterised by the steady fluorescence between 1.5 and 3.5 M urea. In adsorbed rPA, this intermediate state is blurred so that the two transitions overlap, arguing for both increased and decreased conformational stability. However, overall the adsorbed rPA was less stable to urea denaturation than soluble rPA. At ambient temperature and pH 7.3, the standard free energy change (ΔG0H2O) of the two transitions of soluble rPA83 was 5.87 ± 0.27 kcal mol−1 and 7.36 ± 0.13 kcal mol−1 respectively. The free energy changes of the transitions of adsorbed rPA83 were 2.80 ± 1.17 kcal mol−1 and 5.11 ± 0.21 kcal mol−1 respectively. As with the thermal denaturation, the urea unfolding of absorbed rPA83 revealed that the binding to the colloid altered the stability of the rPA83 protein without altering the folded state itself.
Figure 5.
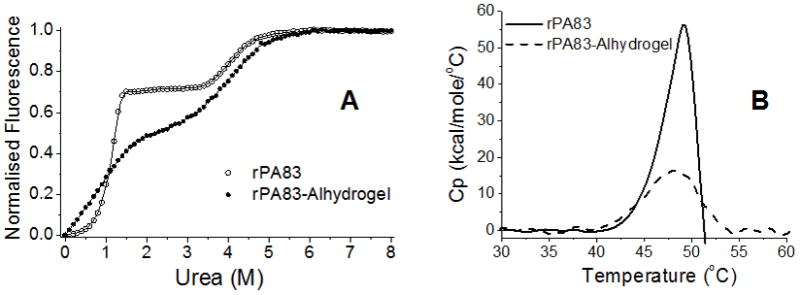
A) The unfolding of soluble and absorbed rPA83 in urea determined by tryptophan fluorescence; B) DSC endotherms of rPA83, showing the difference between adsorbed rPA83 and soluble rPA83. Note that adsorbed rPA83 does not undergo the precipitation/aggregation step shown by the rapid fall in Cp in the soluble rPA83 sample.
Differential Scanning Calorimetry of rPA83
Differential scanning calorimetry revealed that the mid-point of the thermal unfolding transition (Tm) and the calorimetric enthalpy (ΔHcal) of adsorbed rPA83 (47.8 °C; 116 ± 2 kcal mol−1) are less than those of soluble rPA83 (49.2 °C; 219 ± 11 kcal mol−1). (Figure 5B). The calculated Van’t Hof enthalpy (ΔHVH) of adsorbed rPA83 was 118 ± 2 kcal mol−1 and that of soluble rPA83 was 206 ± 13 kcal mol−1. Considering that the protein was still folded whilst bound to Alhydrogel®, these results suggest that the interactions between the protein and adjuvant alter the protein’s conformational freedom and the nature of the unfolded state. In order to obtain a useable signal the level of protein bound to the alhydrogel is 0.5 mg protein per mg alhydrogel which compares to 0.05 mg/mg in the vaccine. It is however below saturation levels and all the protein will be bound the alhydrogel. The agreement with the fluorescence data, which was obtained from rPA at the low density used in the vaccine, supports the use of this higher loading for DSC.
Limited proteolysis of rPA83
Limited proteolysis, to probe conformational features of adsorbed and soluble rPA83, was performed with the following proteases: trypsin, chymotrypsin, pronase E, and proteinase K, V8 and Subtilisin Carlsberg proteases. The loading density was 0.4 mg/mg alhydrogel i.e. slightly less than for DSC. Following limited digestion, the proteolytic products were separated by SDS polyacrylamide gel electrophoresis. Treatment of adsorbed and soluble rPA83 by Chymotrypsin, proteinase K, V8 and Subtilisin Carlsberg (supplementary data) each produced two polypeptides with an apparent molecular weight of 50 and 40 kDa (Figure 6A) [38]. Pronase E also gave similar cleavage patterns, producing the same 50 and 40 kDa bands with soluble and absorbed rPA83; however a 63 kDa band was also seen in both digests (Figure 6B). Compared to soluble rPA83, the trypsin cleavage pattern of adsorbed rPA83 contained an additional 50 kDa band (Figure 6C), indicating that binding to the Alhydrogel resulted a modification to the protein conformation.
Figure 6.
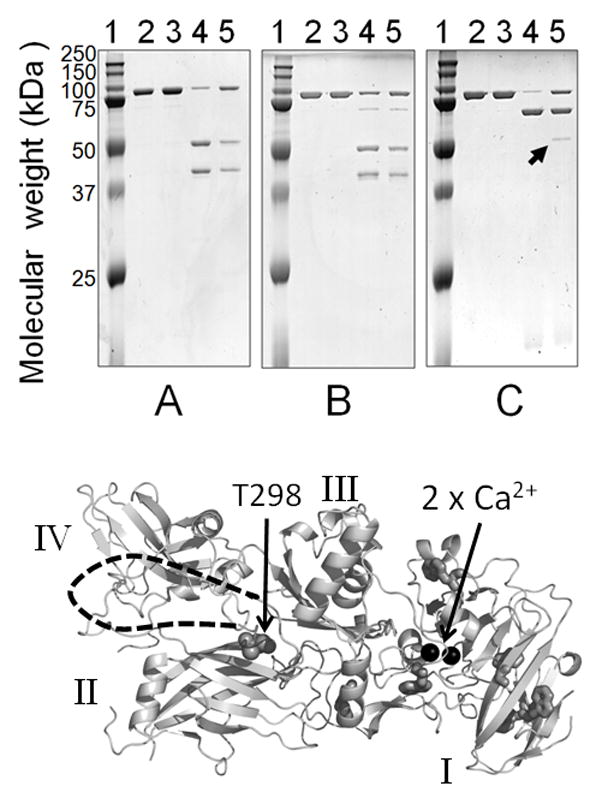
Limited proteolysis of rPA83, Upper –SDS PAGE showing cleavage patterns A) Chymotrypsin, B) Pronase E and C) Trypsin; indicated by the arrow is an additional band produced by trypsin in adsorbed rPA83. Lane 1 = protein standards, lane 2 = rPA83, lane 3 = adsorbed rPA83, lane 4 = soluble rPA83 + protease, and lane 5 = adsorbed rPA83 + protease. Lower- structure of PA82 (PDB;1ACC). Additional cleavage site marked at T296 with unresolved loop indicated by dashed line as in [20]. Also shown are the calcium ions as black spheres, tryptophan residues as sticks and domain structure in roman numerals.
Edman sequencing analysis was performed on the additional tryptic band and identified the N-terminal sequence: THTSEVHGNA showing that the new proteolytic site was located between R297 and T298 at the start of a large flexible loop in domain 2 [20].
Discussion
The rPA83-Alhydrogel complex was formulated by simply mixing the two components and the adsorptive capacity of Alhydrogel (0.94 mg of rPA83 per 1 mg of Alhydrogel®) was far in excess of that required to formulate the vaccine (50μg rPA83/mg Alhydrogel). Since the various methods used had different sensitivities, the study used different levels of loading from 50 μg rPA83/mg Alhydrogel for fluorescence to 500μg rPA83/mg for DSC. The stability data obtained from these two methods are in good agreement and support the use of DSC in such studies [14]. Our previous electron microscopy studies of proteins bound to alhydrogel showed no preferential protein orientation even on the surface of fully saturated protein/Alhydrogel formulations [18] and thus we believe that even the higher density formulations used in DSC are relevant to the drug product.
Alhydrogel formulations are suspensions of insoluble micro-particles that rapidly sediment under gravity [14, 18]. Such mixtures scatter light and have a heterogeneous distribution of chromophores per unit volume that disobeys a fundamental requirement of the Beer-Lambert law, causing flattening of the absorption spectrum [36]. Nevertheless, by gentle stirring to keep rPA83-Alhydrogel micro-particles in suspension, it was possible to measure intrinsic tryptophan fluorescence.
This allowed a range of structural studies, at 50 μg rPA83/mg Alhydrogel, which use the exposure of tryptophan residues normally buried in the folded state as a measure of unfolding. This was probed by emission wavelength changes due to increased water exposure and the use of water soluble quenchers which can reveal changes in surface exposure. However, in PA83 tryptophan residues are largely located in domain 1 and to obtain a more general measure of PA83 structure, an approach which measured the binding of a fluorescent dye (Nile-red) to exposed regions of the hydrophobic core was used. [39]. All the methods confirmed the preservation of the rPA tertiary structure in adsorbed rPA83.
PA contains two calcium ions in domain 1, essential for its structural stability and correct folding [20, 21]. In native rPA83 these ions are stably bound [40]. We showed that rPA83 retained both calcium ions upon binding to Alhydrogel®. In contrast, calcium was released by thermal denaturation of the adsorbed protein. Retention of structure was supported by limited proteolysis of free and bound rPA83. Free rPA83 produces three or four major bands on SDS-PAGE depending on the protease used. Thus there are few protease susceptible sites in the folded protein and, in such studies, unfolding normally exposes more sites with the appearance of extra bands on SDS-PAGE. Only with trypsin was there an extra band on the alhydrogel bound form and this maps to a large flexible loop in domain 2 that is unresolved in the crystal structure. Such flexible loops are known to be susceptible to proteolysis [41]; however it seems that upon binding this loop acquired a different conformation exposing a previously hidden tryptic site. Thus although the secondary/tertiary structure of the protein may be unaffected by binding to Alhydrogel®, already flexible regions may adopt novel conformations.
Thus is little evidence for a change in rPA83 structure upon binding to Alhydrogel®. However, the immobilisation affects protein stability. This was probed by thermal denaturation and urea induced unfolding [35]. Neither method showed large deceases in stability but the bound form was significantly less cooperative in its behaviour. This agrees with it binding in a variety of orientations [18] which will affect its local environment in subtly different ways. The most important change was in thermal unfolding where the broader unfolding transition observed by fluorescence showed that it is 10% unfolded at 37 °C and, unlike soluble rPA83, adsorbed rPA83 showed no gain in its thermal stability with increasing calcium concentration. The enhanced thermal stability of soluble rPA83 in 5 mM calcium was attributed to an increase in the proportion of the more stable calcium bound form [21]. The fact that adsorbed rPA83 could not interact with free calcium ions in solution suggested that upon heating the loss of calcium ions was irreversible.
A more evident difference between bound and adsorbed rPA83 was seen with the calorimetric analysis which showed that the heat capacity of adsorbed rPA83, compared to soluble rPA83, was markedly reduced. Because heat capacity depends on the number of ways that there are of distributing any added heat energy to the system [42], this suggests that the interactions between the protein and the adjuvant limit the protein conformational freedom, as would be expected from a protein anchored to a solid surface [43].
While the presented data do not preclude the possibility that Alhydrogel acts as a depot, enhances the uptake of the antigen due to its particulate nature, or directly stimulates the immune system, it did address the question of whether protein destabilisation enhances immunogenicity [14, 44]. The adsorbed protein retains all its structure at room temperature and drops to 90% of its folded structure at 37 °C. Thus immediate antigen destabilisation upon binding does not appear to explain the adjuvancy effect of Alhydrogel on rPA83. Nevertheless development of methods to observe the structural behaviour of the adsorbed antigen after injection at body temperature would provide the final picture.
Conclusions
The results of this study are in line with previous observations that binding of model proteins to aluminium hydroxide adjuvant does not adversely affect protein structure [15, 16]. However, we show that this conclusion also applies to an antigen that is intended for use in a licensed vaccine. The analytical techniques used here provide useful tools for the characterisation of sub-unit vaccines both during manufacture and within formulations.
Supplementary Material
Acknowledgments
This work was supported by NIH NIAID Grants N01 AI-25492 & N01 AI-30052, NIH NIAID Challenge Grant Award 1UC1AI67223-01 and a BBRSC Case Award with Avecia Biotechnology and Pharmathene. We thank Helen Ridley for excellent technical assistance and Chris Lakey for implementing the urea unfolding data fitting.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turnbull PCB. Anthrax Vaccines - Past. Present and Future Vaccine. 1991;9:533–539. doi: 10.1016/0264-410x(91)90237-z. [DOI] [PubMed] [Google Scholar]
- 2.Plotkin SA. Vaccines: past, present and future. Nat Med. 2005;11:S5–S11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liljeqvist S, Stahl S. Production of recombinant subunit vaccines:protein immunogens, live delivery systems and nucleic acid vaccines. Journal of Biotechnology. 1999;73:1–33. doi: 10.1016/s0168-1656(99)00107-8. [DOI] [PubMed] [Google Scholar]
- 4.O’Hagan DT, MacKichan ML, Singh M. Recent developments in adjuvants for vaccines against infectious diseases. Biomolecular Engineering. 2001;18:69–85. doi: 10.1016/s1389-0344(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 5.Williamson ED, Hodgson I, Walker NJ, Topping AW, Duchars MG, Mott JM, Estep J, LeButt C, Flick-Smith HC, Jones HE, Li H, Quinn CP. Immunogenicity of recombinant protective antigen and efficacy against aerosol challenge with anthrax. Infect Immun. 2005;73:5978–5987. doi: 10.1128/IAI.73.9.5978-5987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelman R. Vaccine Adjuvants. Reviews of Infectious Diseases. 1980;2:370–383. doi: 10.1093/clinids/2.3.370. [DOI] [PubMed] [Google Scholar]
- 7.Lindblad EB. Aluminium compounds for use in vaccines. Immunology and Cell Biology. 2004;82:497–505. doi: 10.1111/j.0818-9641.2004.01286.x. [DOI] [PubMed] [Google Scholar]
- 8.Glenny AT, Pope CG, Waddington H, Wallace U. The antigenic value of toxoid precipitated by potassium alum. Journal of Pathology and Bacteriology. 1926;29:38–45. [Google Scholar]
- 9.Hunter RL. Overview of vaccine adjuvants:present and future. Vaccine. 2002;20:S7–S12. doi: 10.1016/s0264-410x(02)00164-0. [DOI] [PubMed] [Google Scholar]
- 10.Mannhalter JW, Neychev HO, Zlabinger GJ, Ahmad R, Eibl MM. Modulation of the human immune-response by the non-toxic and non-pyrogenic adjuvant aluminum hydroxide - effect on antigen uptake and antigen presentation. Clin Exp Immunol. 1985;61:143–151. [PMC free article] [PubMed] [Google Scholar]
- 11.Morefield GL, Sokolovska A, Jiang DP, Hogenesch H, Robinson JP, Hem SL. Role of aluminum-containing adjuvants in antigen internalization by dendritic cells in vitro. Vaccine. 2005;23:1588–1595. doi: 10.1016/j.vaccine.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Flach TL, Ng G, Hari A, Desrosiers MD, Zhang P, Ward SM, Seamone ME, Vilaysane A, Mucsi AD, Fong Y, Prenner E, Ling CC, Tschopp J, Muruve DA, Amrein MW, Shi Y. Alum interaction with dendritic cell membrane lipids is essential for its adjuvanticity. Nat Med. 2011;17:479–487. doi: 10.1038/nm.2306. [DOI] [PubMed] [Google Scholar]
- 13.Kool M, Soullie T, van Nimwegen M, Willart MAM, Muskens F, Jung S, Hoogsteden HC, Hammad H, Lambrecht BN. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J Exp Med. 2008;205:869–882. doi: 10.1084/jem.20071087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones LS, Peek LJ, Power J, Markham A, Yazzie B, Middaugh CR. Effects of adsorption to aluminum salt adjuvants on the structure and stability of model protein antigens. Journal of Biological Chemistry. 2005;280:13406–13414. doi: 10.1074/jbc.M500687200. [DOI] [PubMed] [Google Scholar]
- 15.Bai SF, Dong AC. Effects of immobilization onto aluminum hydroxide particles on the thermally induced conformational behavior of three model proteins. Int J Biol Macromol. 2009;45:80–85. doi: 10.1016/j.ijbiomac.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Dong A, Jones LS, Kerwin BA, Krishnan S, Carpenter JF. Secondary structures of proteins adsorbed onto aluminum hydroxide: Infrared spectroscopic analysis of proteins from low solution concentrations. Anal Biochem. 2006;351:282–289. doi: 10.1016/j.ab.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Capelle MAH, Brügger P, Arvinte T. Spectroscopic characterization of antibodies adsorbed to aluminium adjuvants: correlation with antibody vaccine immunogenicity. Vaccine. 2005;23:1686–1694. doi: 10.1016/j.vaccine.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 18.Soliakov A, Harris JR, Watkinson A, Lakey JH. The structure of Yersinia pestis Caf1 polymer in free and adjuvant bound states. Vaccine. 2010;28:5746–5754. doi: 10.1016/j.vaccine.2010.05.074. [DOI] [PubMed] [Google Scholar]
- 19.Turnbull PCB, Broster MG, Carman JA, Manchee RJ, Melling J. Development of antibodies to protective antigen and lethal factor components of anthrax toxin in humans and guinea-pigs and their relevance to protective immunity. Infect Immun. 1986;52:356–363. doi: 10.1128/iai.52.2.356-363.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petosa C, Collier JR, Klimpel KR, Leppla SH, Liddington RC. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 21.Chalton DA, Kelly IF, McGregor A, Ridley H, Watkinson A, Miller J, Lakey JH. Unfolding transitions of Bacillus anthracis protective antigen. Arch Biochem Biophys. 2007;465:1–10. doi: 10.1016/j.abb.2007.04.030. [DOI] [PubMed] [Google Scholar]
- 22.Pimental RAL, Christensen KA, Krantz BA, Collier RJ. Anthrax toxin complexes: heptameric protective antigen can bind lethal factor and edema factor simultaneously. Biochem Biophys Res Commun. 2004;322:258–262. doi: 10.1016/j.bbrc.2004.07.105. [DOI] [PubMed] [Google Scholar]
- 23.Miller CJ, Elliott JL, Collier RJ. Anthrax protective antigen: Prepore-to-pore conversion. Biochemistry. 1999;38:10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 24.Mogridge J, Mourez M, Collier RJ. Involvement of domain 3 in oligomerization by the protective antigen moiety of anthrax toxin. J Bacteriol. 2001;183:2111–2116. doi: 10.1128/JB.183.6.2111-2116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varughese M, Teixeira AV, Liu SH, Leppla SH. Identification of a receptor-binding region within domain 4 of the protective antigen component of anthrax toxin. Infect Immun. 1999;67:1860–1865. doi: 10.1128/iai.67.4.1860-1865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosovitz MJ, Schuck P, Varughese M, Chopra AP, Mehra V, Singh Y, McGinnis LM, Leppla SH. Alanine-scanning mutations in domain 4 of anthrax toxin protective antigen reveal residues important for binding to the cellular receptor and to a neutralizing monoclonal antibody. Journal of Biological Chemistry. 2003;278:30936–30944. doi: 10.1074/jbc.M301154200. [DOI] [PubMed] [Google Scholar]
- 27.Wang SL, Johnson CT, Bish DL, White JL, Hem SL. Water-vapor adsorption and surface area measurement of poorly crystalline boehmite. Journal of Colloid and Interface Science. 2003;260:26–35. doi: 10.1016/s0021-9797(02)00150-9. [DOI] [PubMed] [Google Scholar]
- 28.Johnston CT, Wang SL, Hem SL. Measuring the surface area of aluminum hydroxide adjuvant. Journal of Pharmaceutical Sciences. 2002;91:1702–1706. doi: 10.1002/jps.10166. [DOI] [PubMed] [Google Scholar]
- 29.Yau PK, Schulze DG, Johnston CT, Hem SL. Aluminum hydroxide adjuvant produced under constant reactant concentration. Journal of Pharmaceutical Sciences. 2006;95:1822–1833. doi: 10.1002/jps.20692. [DOI] [PubMed] [Google Scholar]
- 30.Seeber S, White JJL, Hem SL. Predicting the adsorption of proteins by aluminium-containing adjuvants. Vaccine. 1991;9:201–203. doi: 10.1016/0264-410x(91)90154-x. [DOI] [PubMed] [Google Scholar]
- 31.Dyson EH, Irving MS, Goldwater R, Stolz R, Duchars M, Lockert AE, SAJH Ascending dose study to assess the safety and tolerability of an rPA anthrax vaccine, and compare its immunogenicity with that of anthrax vaccine adsorbed (AVA). ICAAC; 2006; San Francisco. 2006. [Google Scholar]
- 32.Langmuir I. The constitution and fundamental properties of solids and liquids: Part 1. Solids. Journal of The American Chemical Society. 1916;38:2221–2295. [Google Scholar]
- 33.Lakey JH, Ptak M. Fluorescence indicates a calcium-dependent interaction between the lipopeptide antibiotic LY146032 and phospholipid membranes. Biochemistry. 1988;27:4639–4645. doi: 10.1021/bi00413a009. [DOI] [PubMed] [Google Scholar]
- 34.Lehrer SS. Solute Perturbation of Protein Fluorescence - Quenching of Tryptophyl Fluorescence of Model Compounds and of Lysozyme by Iodide Ion. Biochemistry. 1971;10:3254. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- 35.Pace CN, Shirley BA, Thompson JA. Measuring the conformational stability of a protein. In: Creighton TE, editor. Protein structure; a practical approach. IRL Press; Oxford: 1989. pp. 311–330. [Google Scholar]
- 36.Bustamante C, Maestre MF. Statistical effects in the absorption and optical activity of particulate suspensions. PNAS. 1988;85:8482–8486. doi: 10.1073/pnas.85.22.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta PK, Chandra H, Gaur R, Kurupati RK, Chowdhury S, Tandon V, Singh Y, Maithal K. Conformational fluctuations in anthrax protective antigen: a possible role of calcium in the folding pathway of the protein. FEBS Lett. 2003;554:505–510. doi: 10.1016/s0014-5793(03)01226-2. [DOI] [PubMed] [Google Scholar]
- 38.Novak JM, Stein MP, Little SF, Leppla SH, Friedlander AM. Functional-characterization of protease treated bacillus-anthracis protective antigen. Journal of Biological Chemistry. 1992;267:17186–17193. [PubMed] [Google Scholar]
- 39.Sackett DL, Wolff J. Nile red as a polarity sensitive fluorescent probe of hydrophobic protein surfaces. Anal Biochem. 1987;167:228–234. doi: 10.1016/0003-2697(87)90157-6. [DOI] [PubMed] [Google Scholar]
- 40.Gao-Sheridan S, Zhang S, Collier RJ. Exchange characteristics of calcium ions bound to anthrax protective antigen. Biochem Biophys Res Commun. 2003;300:61–64. doi: 10.1016/s0006-291x(02)02771-7. [DOI] [PubMed] [Google Scholar]
- 41.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochimica Polonica. 2004;51:299–321. [PubMed] [Google Scholar]
- 42.Cooper A. Thermodynamics of protein folding and stability. In: Allan G, editor. Protein: Aa Comprehensive Treatise (Physical and Chemical Properties of Proteins) 2. JAI Press Inc; Stamford: 1999. [Google Scholar]
- 43.Renate Forch HS, Tobias A, Jenkins A, editors. Surface Design: Application in Bioscience and Nanotechnology. Wiley VCH Verlag GmbH & Co. KGaA; Weinheim: 2009. [Google Scholar]
- 44.Thai R, Moine G, Desmadril M, Servent D, Tarride JL, Menez A, Leonetti M. Antigen stability controls antigen presentation. Journal of Biological Chemistry. 2004;279:50257–50266. doi: 10.1074/jbc.M405738200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.