Abstract
Microglial cells are difficult to track during development due to the lack of specific reagents for myeloid sub-populations. To further understand how myeloid lineages differentiate during development to give rise to microglial cells, we investigated CX3CR1 and CCR2 transcription unit activation in Cx3cr1+/GFPCCR2+/RFP knock-in fluorescent protein reporter mice. The principal findings include: 1) CX3CR1+ cells localized to the AGM region, and visualized at E9.0 in the yolk sac and neuroectoderm, 2) At E10.5 CX3CR1 single positive microglial cells were visualized penetrating the neuroepithelium, 3) CX3CR1 and CCR2 distinguished infiltrating macrophages from resident surveillant or activated microglia within tissue sections and by flow cytometric analyses. Our results support the contribution of the yolk sac as source of microglial precursors. We provide a novel model to monitor chemokine receptor expression changes in microglia and myeloid cells early (E8.0-E10.5) in development and during inflammatory conditions, which have been challenging to visualize in mammalian tissues.
Introduction
Microglia, the resident tissue macrophages and intrinsic ‘immune effector’ cells of the central nervous system (CNS), constitute about 20% of the total glial population and are distributed throughout the brain, spinal cord and optical tissues(1). In CNS leukocyte preparations, CD45 is used to separate the CD45hi hematogenous population from CD45lo resident microglia by flow cytometry(2,3). Characterization of MHC-II expression and up regulation of activation markers such as CD40, CD44, and CD86 has provided insights into the effector functions of activated microglia during CNS inflammation(4). However, there are still debates regarding the alterations of CD45 expression by monocytes and CNS –born microglial cells. Up to date there is no simple combination of markers to facilitate the identification and differentiation of resident surveillant or activated microglia from hematogenous monocytes.
The discovery of the chemokine receptor CX3CR1 in 1994, and its unique ligand fractalkine in 1997 represented a major advancement in the understanding of microglial and monocyte function(5-8). Fractalkine is a distinct chemokine, expressed as a transmembrane glycoprotein on neurons and peripheral –but not CNS– endothelial cells(5,9). The fractalkine receptor (CX3CR1) is present on microglia and circulating monocytes, dendritic cells, NK cells and T cells(10-12). Membrane-bound fractalkine on endothelial cells mediates adhesion of CX3CR1+ leukocytes, and proteolytic cleavage releases the soluble domain that confers fractalkine its chemotactic properties(5). Importantly, CX3CR1 distinguished a monocyte subset in peripheral blood so-called ‘resident’, phenotypically identified as(13) LFA-1+, L-Sel−, Ly6C−, CCR2−, CX3CR1hi, whereas CCR2 -the monocyte chemoattractant protein 1 receptor (MCP-1 or CCL2)-, marked ‘inflammatory’ monocytes(14).; LFA-1−, L-Sel+, Ly6C+, CCR2+, CX3CR1lo(14-16). Circulating ‘resident’ monocytes patrol healthy tissues through long-range crawling on the healthy endothelium(13) playing critical role in immune-surveillance. In contrast, CCR2+CX3CR1−Ly6C+ monocytes are typically found in inflamed tissues(17). The generation of CX3CR1/GFP knock-in mice, in which the CX3CR1 was disrupted by insertion of green fluorescent protein (GFP), clarified the expression pattern and has facilitated the tracking of CX3CR1+ leukocytes in vivo. In the CNS of Cx3cr1+/gfp and Cx3cr1gfp/gfp mice, CX3CR1 transcription unit was exclusively active in microglial cells; as GFP expression was undetectable in neurons, NG2+ glia and GFAP+ astrocytes.
Due to the lack of appropriate anti-CCR2 antibodies, most CCR2 expression analyses have been limited to mRNA levels(18). It has been proposed that microglia activation during inflammatory settings up regulate CCR2(19,20). However, microglial CCR2 protein expression has been challenging to prove in situ. The recent generation of red fluorescent protein (RFP) - CCR2 knock-in mice has provided the tools to potentially fractionate CX3CR1 and CCR2 activation in microglial and peripheral monocytes and study their trafficking, molecular signatures and biological functions(21). CCR2-RFP mice showed an accurate correlation of CCR2 and RFP expression in blood monocytes both at the protein and mRNA levels. Characterization of Ccr2rfp/rfp mice revealed that in response to thioglycolate CCR2 was required for in vivo migration of monocytes to the inflamed peritoneum. In mice with experimental autoimmune encephalomyelitis (EAE), CCR2 was critical for efficient accumulation of LyC6hi/CCR2hi monocytes. Interestingly, during CNS inflammation, monocytes that infiltrate the CNS maintained their signature chemokine receptor profiles as found in circulating monocytes(21). Importantly, microglia in the adult naïve and inflamed CNS were found as CX3CR1hiCCR2− using the described Cx3cr1+/gfpCcr2+/rfp reporter mice. CNS infiltrating monocytes appeared as CCR2hiCX3CR1lo or CCR2loCX3CR1hi. Notably, CX3CR1 expression by microglia was significantly higher when compared to blood CX3CR1hi monocytes.
Given the fact that CX3CR1 is predominantly expressed by ‘resident tissue monocytes’ we sought to investigate CX3CR1 and CCR2 transcription unit activation pattern in microglial precursors as they colonize the developing CNS. For this, CCR2-RFP and CX3CR1-GFP mice were crossed and Cx3cr1+/gfp-Ccr2+/rfp embryos analyzed at various stages during embryonic development. The results indicate that CX3CR1 is active in early microglial precursors, and sustained throughout adulthood. In contrast, RFP as reporter of activation of the CCR2 transcription unit was not significantly detected in developing microglia. Therefore, we propose CX3CR1 and CCR2 as markers to easily differentiate, infiltrating macrophages from resident surveillant or activated microglia in tissue sections and by flow cytometric analyses. Importantly, the results support the contribution of the yolk sac as source of microglial precursors. These results bring potential applications to monitor neuronal-glial interactions throughout development and to further the understanding of microglial/monocytes/macrophage and dendritic cell plasticity in vivo.
Materials and Methods
Mice
Ccr2RFP/RFP mice and Cx3cr1GFP/GFP mice were crossed at the Biological Resources Unit, Cleveland Clinic and the Laboratory Animal resources Unit at the University of Texas at San Antonio. Timed pregnancies were set up to isolate embryos at stages E8.5-13.5. All experiments were performed in accordance with NIH guidelines and approved by the Cleveland Clinic and the University of Texas at San Antonio Institutional Animal Care and Use Committees.
Tissue processing
Females were sacrificed by CO2 asphyxiation and embryos dissected and kept in cold PBS. A small tissue (< 1mm) biopsy was taken from the caudal part of the embryo for genotyping purposes. Embryos (E8.5-9.5) were fixed in 4% PFA overnight (ON) and whole embryos were permeabilized ON in 3% triton. Alternatively, after fixation, embryos (>E10.5) were transferred sequentially to 15%, 20% and 25% sucrose solutions and 10 μm cryo sections obtained, or 60 μm free floating sections collected by sectioning embryos on a sliding microtome(12).
Generation of bone marrow chimeric mice
Recipient Cx3cr1+/GFP/Ccr2+/RFP CD45.2 mice (5-6 weeks old) irradiated with a dose of 900 rad were reconstituted with 20 × 106 cells isolated from C57BL/6 CD45.1 donor mice as previously described (22). Six weeks after reconstitution mice were bled via the submandibular vein and heparinized blood was lysed and efficiency of reconstitution determined by flow cytometry using anti-CD45.2 and CD45.1 antibodies. Chimeric mice were allowed to reconstitute for 10 weeks after bone marrow transfer prior to EAE induction.
Microscopic analysis
Whole embryos of selected sections were rinsed in 1X PBS and covered with 2 ml of a solution containing 450 nM DAPI in PBS and incubated for 10 min at RT. Excess DAPI was absorbed on paper towels and slides rinsed in PBS. To visualize the vasculature, separate tissue slides were permeabilized ON in 3% triton in PBS and incubated at 4°C for 2 days with biotin-conjugated isolectin-B4 (IB4). Streptavidin-Cy5, was used to detect the bound biotin-IB4. DAPI-counterstained tissues were visualized under a fluorescent microscope. Confocal images were obtained as projections of 20 μm thick sections in the z-axis scanned at 1.5 μm steps. In some experiments brain tissues were dissected from adult mice at peak of EAE disease and tissues sections incubated at 4°C with 7-4 antibody (Cedarlane, Ontario, Canada), and imaged at 40× magnification on a Zeiss 510 confocal microscope as previously described (21).
EAE induction
Active EAE was induced by subcutaneous immunization with MOG35-55 peptide in complete Freund's adjuvant as previously described (23). Mice were weighed and examined daily for EAE symptoms and scored as follows: (0) no signs of neurological disease, (1) lack of tail tone, (2) abnormal gait, hind limb weakness (2.5) partial hindlimb paralysis, (3) complete hindlimb paralysis, (3.5) ascending paralysis, (4) tetraplegia, (5) death. Mice were sacrificed when they reached a score of 2.5-3.0(22). Brain tissues were obtained from PFA perfused mice and free-floating 30 μm tissues obtained after cryoprotection and confocal images obtained.
Isolation of brain mononuclear cells and flow cytometry
For flow cytometry analysis perfused brain and spinal cord tissues were dissected, and mononuclear cells separated over discontinuous 70%/30% percoll gradients as previously described(24) and cellular pellets resuspended in cell staining buffer (Biolegend, San Diego, CA). Blood was collected from the submandibular vein, red blood cells depleted by hypotonic lysis, and washed in staining buffer. Isolated cells were incubated on ice for 5 min with anti-mouse CD16/CD32 (Clone 2.4G2, BD Pharmingen) to block Fc receptors, and then incubated on ice for 30 min with a mix of fluorochrome-conjugated anti-mouse antibodies: CD45.1-Pacific blue (Clone A20, eBioscience), CD45.2-APC (Clone 104, eBioscience) and CD11b-PerCP (Clone M1/70, Biolegend), or CD45.1-PeCy7 (Clone A20, eBioscience), CD45.2-APC (Clone 104, eBioscience), CD11b-PerCP (Clone M1/70, Biolegend) and I-A/I-E- Pacific blue (Clone M5/114.15.2). After washes, cells were resuspended in 2% paraformaldehyde and acquired in a LSR-II (BD biosciences, Franklin Lakes, NJ). Samples were analyzed with FlowJo software (Tree Start, Ashland, OR).
Results
CX3CR1-GFP reporter expression co-localizes to early microglial cells that colonize the developing CNS
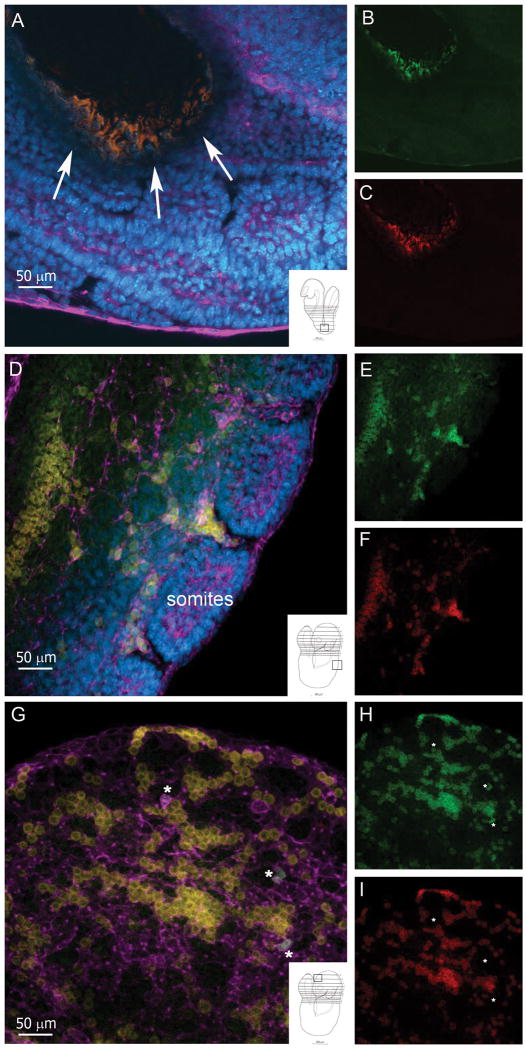
Whole mounted embryos during E8.5-E10.5 embryonic stages, were dissected as they easily displayed the overall mental map of the embryo and allowed grossly localization of CX3CR1-green/CCR2-red cells based on landmark histological structures. Unturned embryos at E8.5 stage showed clusters of double positive CX3CR1GFP/CCR2RFP cells co-localizing to the gut region, and sparsely visualized throughout the embryo (Figure 1A-C). At stage E8.5-9.0 (turned embryo) cells revealing fluorescence intensity appeared as double positive within tracks in the developing somites (Figure 1D-F) and also present in neural tissues, within the telencephalic vesicle (Figure 1 G-I). Isolectin–B4 (IB4; Figure 1; purple) was utilized to highlight the vasculature but also allowed the visualization of cells with features of developing microglia that colonize the neural tissue appearing as CX3CR1GFP+ (Figure 1G; asterisks). The exact origin of these cells is not clear. However, visualization of yolk sac tissues at E9.5 revealed the presence of sparsely located GFP+ cells with large cell bodies suggesting that a population of CX3CR1+ cells develops in the embryonic circulation (Supplementary Figure 1). Double positive cells were also found in foci at mid and caudal regions of the embryo in close proximity to the peritoneal cavity co-localizing to the midline dorsal aorta with tracks of cells distributing throughout the developing embryo (Supplementary Figure 1).
FIGURE 1. Ccr2+/RFP/Cx3cr1+/GFP positive cells are visualized at E8.5 stage of development.
Whole embryos were incubated with with IB4 –Cy5 to visualize the vasculature (magenta) and tissues (A-F) were counterstained with DAPI (blue). Merged image from an unturned embryo (A) at the level of gut region (A, arrows and inset) shows clusters of CX3CR1+ (B, green) and CCR2+ (C, red) double positive cells. Images from a turned embryo at the level of the somites (D-F) and telencephalic vesicle (G-I) show abundant numbers of CX3CR1+ (E and H, green) and CCR2+ cells (F and I, red) present throughout the tissues. Notably, a population consisting of CX3CR1hi single positive cells were detected in the developing brain which appear IB4+ (G-I, asterisks). Insets in panels A, D and G, show a schematic of the embryo with a small square highlighting the source of the image.
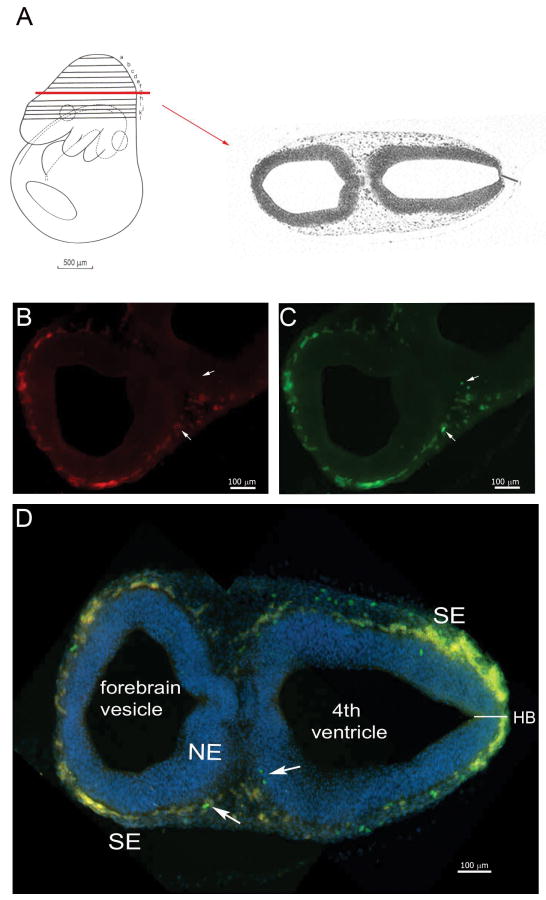
To closely monitor the morphology and neural colonization of CX3CR1+ cells, cross-sections (Figure 2) and sagittal slices (Supplementary Figure 2) from E9.5-10.5 tissues were analyzed. Double positive cells were found in both posterior and anterior ends of the neural tube (Figure 2), mostly confined to the surface ectoderm (SE) and tightly opposed to the outer layer of the neuroepithelium (NE). A distinct population of CX3CR1-GFPbright and CCR2-RFPdim cells was visualized in the junction of the surface ectoderm and neuroepithelium (Figure 2).
FIGURE 2. Ccr2+/RFP/Cx3cr1+/GFP cells are localized in the neuroectoderm and visualized in developing E9.5 tissues.
(A) Shows the plane of section in schematic form. An E9.5 tissue shows that both receptors are expressed by myeloid precursors localized between the developing neuroepithelium and surface ectoderm. Tissues were stained with DAPI to visualize the nuclei (blue). (B) Distribution of CCR2-RFP+ cells, and (C) CX3CR1-GFP+ cells. (D) Merged image shows a dense band of myeloid precursors that appear mostly double positive. CX3CR1 single positive microglial precursor cells were clearly detected in the surface ectoderm (arrows).
CX3CR1 but not CCR2 is expressed by developing microglial cells
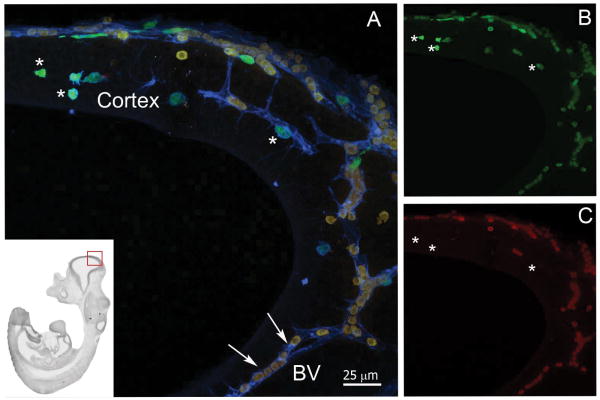
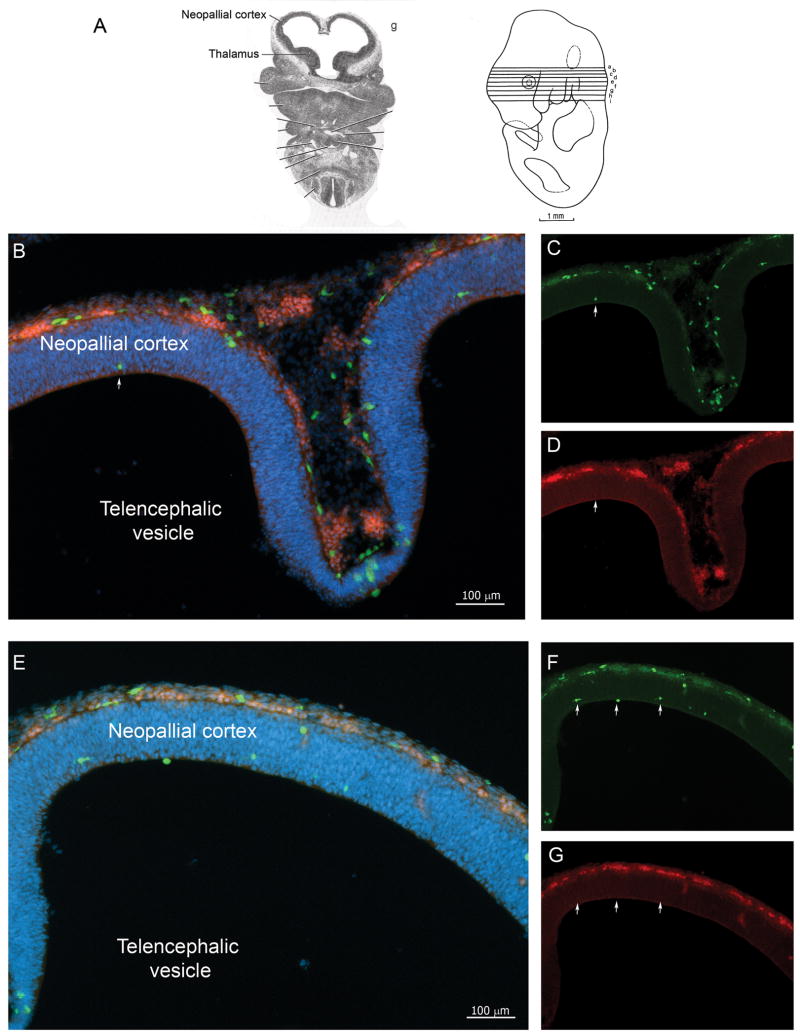
Figure 3 shows a sagittal section at stage E11.5 highlighting IB4+ vasculature, with presence of double positive cells inside blood vessels. Scarce CX3CR1 single positive cells localized to the developing cortical tissue without direct association with vasculature. These CX3CR1+ cells displayed amoeboid morphology typical of early microglial precursors (Figure 3). At stage E11.5 the neural tissue revealed a thicker neopallial cortex regions and mixed populations of cells showing various degrees of CX3CR1-GFP and CCR2-RFP intensities were visualized. Clusters of small/round cells appearing as CX3CR1-GFPDim/CCR2-RFPbright were detected in the surface ectoderm or meningeal membranes (Figure 3 and 4). Interestingly, single positive CX3CR1-GFPbright cells appeared localized within the neuroepithelium, migrating from the surface ectoderm reaching the inner part of the developing cortex (Figure 4). The number of CX3CR1GFP+ cells expands dramatically at E11.5 stage and they colonize not only the developing cortex but the optic vesicles and developing spinal cord (Supplementary Figure 2).
FIGURE 3. Developing microglial cells appear as CX3CR1bright and CCR2−.
(A) Low power of the whole embryo is shown in the inset to locate the origin of the image (red square). E11.5 tissues show that CX3CR1 but not CCR2 is expressed by developing microglial cells (A, merged image). Free-floating sagital -30 um sections in thickness- were obtained and incubated with IB4 (blue) to visualize the vasculature. Confocal image of the neopallial cortex shows CX3CR1+ (A and B, green) and CCR2+ (A and C, red) expressing cells. Developing amoeboid microglia appear CX3CR1bright (asterisks) and CCR2- (asterisks), while circulating mononuclear phagocytes exhibit a CX3CR1loCCR2bright phenotype and appear within blood vessels (BV, arrows).
FIGURE 4. Distribution of cells in Ccr2+/RFP/Cx3cr1+/GFP E11.5 tissues.
(A) Shows the plane of section in schematic form. (B-G) Confocal images of free-floating 30um cross-sections of E11.5 tissues counterstained with DAPI (B and E, merged imaged, DAPI;blue) show CX3CR1+/CCR2+ double positive cells in the surface ectoderm (C and F, CX3CR1-GFP;green) and (D and G, CCR-RFP; red). Developing amoeboid microglia appear CX3CR1bright and localized in deep layers of neuroepithelium (arrows).
Detailed visualization of E13.5 embryos revealed a significant expansion of CX3CR1+ cells detected throughout the whole embryo. E13.5 neural tissue showed a similar pattern to E10.5 stage, with an increased number of single GFP+ cells populating the developing CNS. The developing spinal cord is easily visualized at this stage and neural CX3CR1+ cells showed the characteristic microglial phenotype with bigger cell bodies and extended processes, IBA-1 immunoreactivity and lack of CCR2-RFP reporter expression (Supplementary Figure 2).
The previous observations suggest a scenario in which the cells that first colonize neural tissues are double CX3CR1/CCR2 positive and accumulate at the surface ectoderm. Cells that remain in the surface ectoderm -meningeal areas- down regulate CX3CR1 and appear predominantly as CCR2+. Due to the fact that most cells in the surface ectoderm are double positive, is it likely that cells entering the deep layers of the neuroepithelium down regulate CCR2 appearing as single positive CX3CR1+. However, we do not rule out the possibility of discrete single positive cells that reach the surface ectoderm, proliferate and give rise to the population that colonizes and expand within the developing CNS tissue. Therefore, early stages of development (E8.5) showed 1) cells with the capacity of becoming peripheral monocytes and 2) cells that colonize the perivascular and cortical regions as CNS microglia. Both populations appeared distinct based on the activity of the CCR2 and CX3CR1 transcription units respectively.
At P0, CCR2 expression remained confined to a population of cells that localize to meningeal membranes and also visualized within blood vessels. All CCR2+ cells appeared as CX3CR1Dim. As the cortical tissue developed, migrating microglial cells that seeded parenchymal tissues exhibited a rapidly expansion and appeared as single CX3CR1+ (Figure 5A-F). In the developed CNS, RFP+ cells appeared confined to meningeal areas and absent in brain parenchyma, indicating that neither astrocytic, microglial cells, oligodendroglial nor neuronal cell types express significant amounts of CCR2 protein to be visualized by expression of the RFP-reporter (Figure 5).
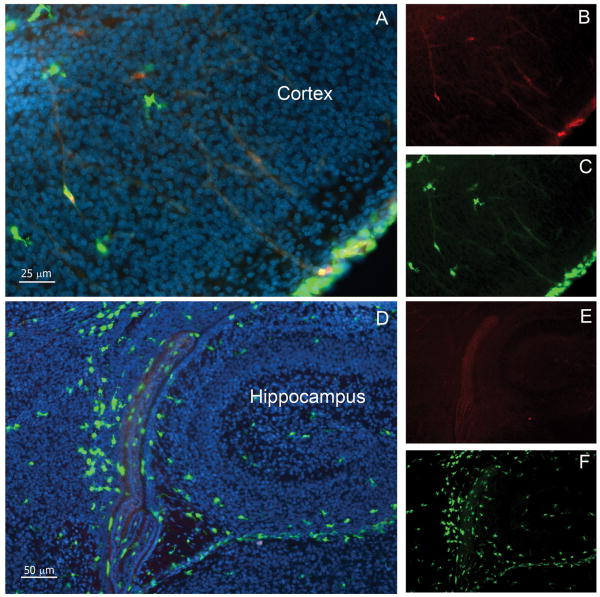
FIGURE 5. CX3CR1 is confined to microglial cells during development and expression is sustained throughout adulthood.
Confocal images were obtained from brain tissues at P0 and counterstained with DAPI (blue). Cortical (A-C) and hippocampal (D-F) images shows that parenchymal cells are mostly CX3CR1bright (A and C, D and F; green) and fewer CCR2+ cells (A and B, and D and E; red) remain localized in the meningeal region.
These observations suggest that CX3CR1 and CCR2 expression is timely orchestrated during development. It has been established that neither CX3CR1 nor CCR2 are required for the development of microglia as knock out model for both receptors show normal microglia in the healthy adult CNS. However, the tight regulation of both receptors provides a key phenotypic approach to characterize and distinguish monocytes/macrophages from resident microglia.
Adult microglia exhibit a CX3CR1-GFP+ CCR2-RFP– expressing pattern and microglia do not show significant CCR2RFP expression upon inflammation
To get insights into the CCR2 expression pattern in microglial cells during inflammation, Cx3cr1+/GFP/Ccr2+/RFP mice were immunized with MOG(35-55). CX3CR1 and CCR2 expression was analyzed by flow cytometry in brain leukocyte preparations. Results reveal that brain inflammatory lesions contain cells with distinct levels of CX3CR1 and CCR2 expression. Notably, microglia did not up regulate RFP expression and flow cytometry analyses distinctly showed CCR2-RFP expression in hematogenous CD45hi leukocytes and CCR2-RFP signal was absent in the CD45lo microglial population (Figure 6A,B). Two populations of CCR2-RFP+ cells are visualized, and we have previously characterized them by flow cytometry (21) and shown that CCR2-deficiency depletes inflammatory Ly6Chi monocytes. Importantly, absence of CCR2hiCX3CR1−Ly6Chi monocytes delays EAE progression. To have a better understanding of CCR2 and CX3CR1 expression in myeloid cells, brain tissues were imaged and CCR2-RFP and CX3CR1-GFP cells visualized (Supplementary Figure 3). In addition we implemented a three color imaging using the 7-4 marker that identifies Ly6Chi monocytes and neutrophils, but not NK or T cells. In the lesions we observed activated microglial cells with shorter and thicker processes and these cells clearly appear as CX3CR1 single positive (Supplementary Figure 3), whereas classical infiltrating monocytes appear CX3CR1lo or negative and CCR2+. Analyses of skin and cardiac tissues (Supplementary Figure 4) show that in naïve tissues long lived macrophage populations appear CX3CR1hi/CCR2-RFPneg similar to the phenotype of resident microglial cells.
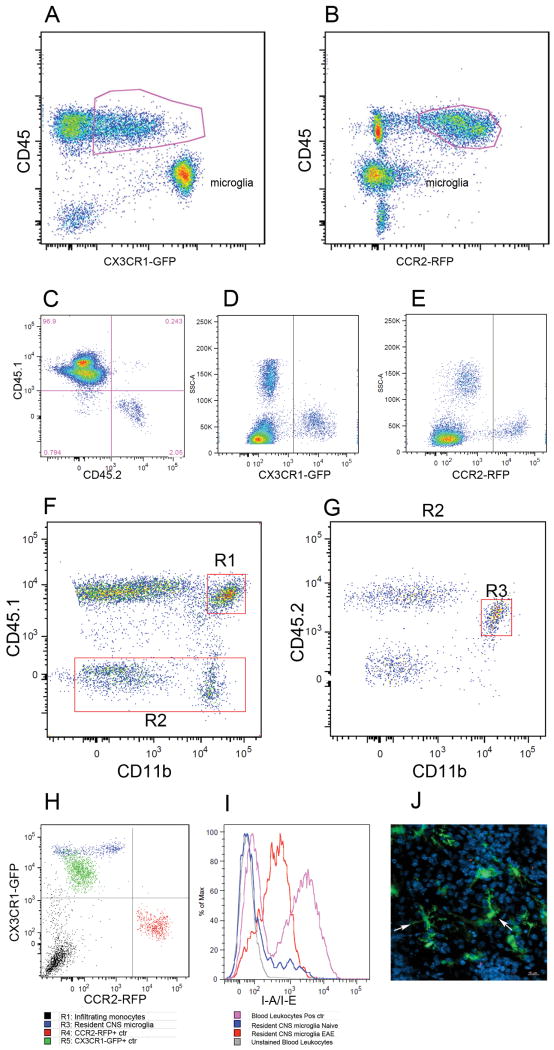
FIGURE 6. Analyses of monocyte subsets in brain lesions of EAE CX3CR1+/GFP CCR2+/RFP mice at peak disease.
Flow cytometry analysis of mononuclear cells isolated from adult brain at peak EAE disease shows that the CD45lo population contains CX3CR1bright microglial cells (A), contrasting their lack of CCR2-RFP expression (B). Degree of bone marrow recosntitution was evaluated by staining PBMCs with CD45.1 and CD45.2 antibodies (C). CX3CR1-GFP (D) and CCR2-RFP (E) positive controls were obtained from Cx3cr1GFP/+/CCR2+/+ or Cx3cr1+/+/CCR2RFP/+ mice respectively. Brain leukocytes were analyzed for CD45.1 and CD11b expression (F) and peripherally derived myeloid cell (R1) were distinguished from resident CD45.2+ CD11b+ cells via expression of the congenic markers (G, R3). Analyses of CX3CR1 and CCR2 based on activation of the transcription unit and expression of the GFP and RFP reporters (H) shows that resident microglial cells appear as CX3CR1-single positive. (I) activated resident cells up regulated MHC-II expression upon EAE induction as compared with naïve brains, and confocal imaging from diseased mouse brains shows morphologically activated microglia as CX3CR1-GFP bright cells (J). Results show one representative set of data from one mouse out of 8 analyzed by flow cytometry and histology.
During EAE microglial cells become activated and using conventional monocytes markers become indistinguishable from infiltrating myeloid cells. Therefore, we sought to clarify the relationship between CCR2, CX3CR1 and microglial cells using congenic bone marrow transfer in CX3CR1-GFP/CCR2-RFP recipient mice. For this, Cx3cr1+/GFP/Ccr2+/RFP double heterozygous mice (in a CD45.2 congenic background) were reconstituted with congenic wild type CD45.1 bone marrow cells. Upon EAE induction, these chimeric mice would allow us to distinguish the CD45.1 infiltrating population from radioresistant CNS resident CD45.2+ cells, among them microglial cells carrying CX3CR1-GFP/CCR2-RFP reporter proteins. Upregulation of the CCR2 transcription unit measured by virtue of red fluorescence will be then analyzed. First, we determined the efficiency of bone marrow reconstitution; PBMCs from chimeric mice were analyzed 6 weeks after bone marrow transfer for CD45.1 and CD45.2 expression. The protocol produced 96.8 ± 0.75 % reconstitution (Figure 6C). At peak of EAE disease, brain leukocytes were isolated and stained with antibodies against CD45.1, CD45.2 and CD11b to distinguish peripherally derived myeloid cells (CD45.1) from resident microglial cells (CD45.2). Single positive controls (Figure 6D and E) for CX3CR1-GFP and CCR2-RFP expression where used to compensate data acquired from chimeric mice and to compare levels of expression in the infiltrating versus recipient myeloid populations. Brain leukocytes were initially analyzed based on expression of CD45.1 and CD11b to identify myeloid cells of peripheral origin (Figure 6F, R1 gate). Disease severity is evident by the massive infiltration of peripherally derived leukocytes (CD45.1+ cells, Figure 6F) which accounted for 90 ± 3.5% of the total CD45hi population. Next, the CD45.1 negative population (Figure 6F, R2 gate) was analyzed for CD45.2 expression to identify the population of resident microglial cells (Figure 6G, R3 gate). Even though bone marrow reconstitution was >95% efficient, peripheral CD45.2 recipient cells that remained gave rise to a CD45.2hi population that infiltrated the CNS tissue and accounted for about 9.3 ± 3.5% of the inflammatory infiltrated. Comparison of the CX3CR1-GFP and CCR2-RFP fluorescence intensities (Figure 6H) shows that the CD45.2+ myeloid population from recipient mice is negative for CCR2-RFP. Mean fluorescent intensities from the resident microglial population were 9860± 632.6 for GFP, and 120.7± 18.38 for RFP, n = 5 mice. Furthermore, the CD11b population although activated based on MHC-II up regulation expression (Figure 6I) appears with a CD45.2lo phenotype in flow cytometric analysis. Confocal images of brain tissues of chimeric mice confirmed that the activated microglial population as noted by the shorter processes and bigger cell bodies appear as CX3CR1-GFP single positive (Figure 6J, arrows).
Discussion
Surveillant parenchymal microglial cells are extremely plastic and provide the first line of defense within the CNS. Resident microglial cells are morphologically, and functionally distinct from other mononuclear CNS populations, such as perivascular macrophages, supraependymal macrophages, epiplexus cells of the choroids plexus and meningeal macrophages(25,26). In the naïve brain, microglia display small cell bodies with thin, long and branched processes(1). Although microglial functions are intended to be protective, is documented that dysregulated microglial responses lead to neurotoxicity in vitro and in vivo(12). Recent data have clearly shown that activation of microglia might be beneficial in some pathological settings. More specifically, absence of CX3CR1 in two different models of Alzheimer's disease correlated reduced β-amyloid deposition due to enhanced phagocytosis by activated microglia(27,28). Upon activation, due to inflammation of neuronal damage, microglial cells undergo programmed molecular and morphological changes that include cytoskeleton rearrangements, increased in cell body area exhibiting shorter and thicker processes that ultimately make amoeboid microglia phenotypically indistinguishable from peripheral macrophages.
Another challenging area regarding microglial biology relates to their origin which has been a subject of controversy for many years. It is now well accepted that microglia arise from mesodermal progenitors that colonize the nervous tissues during embryonic development and fetal periods, and become fully established during postnatal life(29-32). However, the exact mechanisms that control the accumulation of microglial progenitors to the developing CNS are still enigmatic. Del Rio-Hortega in the 1910's classified microglia into two morphological distinct types, which are still current; ramified and amoeboid microglia. Ramified microglial cells are fully established postnatally, persist in the adult, and represent the surveying population that constantly scans the surrounding environment via extension and retraction of their processes to perceive activating or inhibitory signals. It is hypothesized that during development amoeboid microglia are initially highly proliferative cells. Subsequently with decreased in plan plasticity microglial proliferation slows and the remaining cells differentiate into ramified microglia(33,34). Supporting these initial findings, Juba in 1933 carried out the first comprehensive study of microglia in a series of human embryos 23-280 crown-rump length (measurement of the length of human embryos and fetuses from the top of the head (crown) to the bottom of the buttocks (rump) (35,36). He identified the various cell forms with developmental progression, ranging from amoeboid to mature branched varieties. Juba considered microglia to be derived from invading mesodermal elements as microglia-like cells were also detected within connective tissue of the head, which were closely related to embryonic vascular elements. Since early 1930's, it has been maintained that microglia were ‘transformed blood elements’ or ‘wandering mesenchymal elements’ that migrated into the nervous tissue from the lumen of blood vessels. Although recent studies have elegantly identified microglia as an ontogenically distinct population in the mononuclear phagocyte system(37), we still lack a precise lineage-restricted marker for microglial progenitors that is a key to clarify the debates as to the transformation of these cells in vivo.
The presence of CX3CR1+ cells in the yolk sac at E9.5-E11.3 stages of developments is of great interest. The yolk sac is an important structure defined as the birth place of hematopoietic cells in mammals where primitive macrophages and erythrocytes originate, and therefore likely to harbor microglial progenitors. It was proposed that microglia have a dual origin, first coming from the yolk sac macrophages during the nonvascularized prenatal stage followed by a second engraftment from a population that represents a developmental and transitory form of fetal macrophage (derived from blood-borne precursors, possibly monocytes), which may be related to the amoeboid microglial population seen in the postnatal period in rodents(35). This hypothesis has been supported by other groups, showing that in the embryo, microglia originate from monocytes that enter the CNS before development of the blood brain barrier(38,39). However, our data supports the notion that resident CNS tissue macrophages arise from yolk sac and as suggested previously, tightly regulated molecular cues dictate the final localization.
The current study also brings a novel contribution in the characterization of monocyte subsets. The long standing limitation of differentiating microglia and peripheral macrophages in inflamed tissue sections has been partially addressed in chimeric mice, and through flow cytometric analyses utilizing antibodies against various activation-associated cell surface markers(40-43). CX3CR1GFP mice have been widely used to monitor various aspects of monocytes biology and inflammatory reactions associated with heart disease, pain, and cancer(44,45). Although CX3CR1 and CCR2 deficient mice show normal pattern of adult microglia, it is uncertain whether the timely and organized targeting of early microglia precursors in the developing CNS is altered in the absence of CX3CR1 or CCR2; aspects that are under currently under investigation. The chimeric mice generated in this study provided close to 97% reconstitution. However, the 3% that remained from recipient accounted for a small proportion (<3%) of non-myeloid inflammatory cells (CD45.2hiCD11b-) in the diseased brain. This model has valuable applications to dissect function and mechanisms of turnover of myeloid cell populations under normal and inflammatory settings. Our data reports for the first time the temporal expression of CX3CR1 during development with important implications in understanding neuronal-glial communication during early stages of development. Our results show that CX3CR1 is expressed in early microglial progenitors, and its expression is sustained throughout adulthood. The GFP reporter in CX3CR1GFP mice represents a valuable marker to clarify the seeding cues of microglia within the developing CNS. In contrast, CCR2 is absent in all CNS resident cell types and microglia do not significantly up regulate CCR2 during inflammation using the CCR2-RFP mice. These data brings a new set of phenotypical features not only to monitor surveying microglia but also to analyze the composition of cellular infiltrates in various CNS pathologies.
Supplementary Material
Acknowledgments
We thank, Dr. Bruce Trapp (Cleveland Clinic) for providing the IBA-1 antibody, Jenny A. Garcia (UTSA) for secretarial assistance, Difernando Vanegas (UTHSCSA) for assistance on the processing of skin tissues, the RCMI confocal core facility and advanced imaging center at UTSA, and to Elsevier Academic Press for permission to reproduce the schematic of embryos from the Atlas of mouse Development by M.H. Kaufmann.
Abbreviations used in this article
- CNS
central nervous system
- EAE
experimental autoimmune encephalomyelitis
- GFP
green fluorescent protein
- MOG
myelin oligodendrocyte glycoprotein
- NE
neuroectoderm
- RFP
red fluorescent protein
- SE
surface ectoderm
Footnotes
This work was supported in part by the National Institutes of Health (NIH R01NS32151 to RMR) and the National Multiple Sclerosis Society (TA3021-A1/T to AEC).
References
- 1.del Rio-Hortega P. Microglia. In: Penfield W, editor. Cytology and cellular pathology of the nervous system. Hoeber; New York: 1932. pp. 481–584. [Google Scholar]
- 2.Carson MJ, Reilly CR, Sutcliffe JG, Lo D. Mature microglia resemble immature antigen-presenting cells. Glia. 1998;22:72–85. doi: 10.1002/(sici)1098-1136(199801)22:1<72::aid-glia7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 3.Ford AL, Goodsall AL, Hickey WF, Sedgwick JD. Normal adult ramified microglia separated from other central nervous system macrophages by flow cytometric sorting. Phenotypic differences defined and direct ex vivo antigen presentation to myelin basic protein-reactive CD4+ T cells compared. J Immunol. 1995;154:4309–4321. [PubMed] [Google Scholar]
- 4.Ponomarev ED, Shriver LP, Maresz K, Dittel BN. Microglial cell activation and proliferation precedes the onset of CNS autoimmunity. J Neurosci Res. 2005;81:374–389. doi: 10.1002/jnr.20488. [DOI] [PubMed] [Google Scholar]
- 5.Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 6.Rossi DL, Hardiman G, Copeland NG, Gilbert DJ, Jenkins N, Zlotnik A, Bazan JF. Cloning and characterization of a new type of mouse chemokine. Genomics. 1998;47:163–170. doi: 10.1006/geno.1997.5058. [DOI] [PubMed] [Google Scholar]
- 7.Combadiere C, Gao J, Tiffany HL, Murphy PM. Gene cloning, RNA distribution, and functional expression of mCX3CR1, a mouse chemotactic receptor for the CX3C chemokine fractalkine. Biochemical & Biophysical Research Communications. 1998;253:728–732. doi: 10.1006/bbrc.1998.9849. [DOI] [PubMed] [Google Scholar]
- 8.Cook DN, Chen SC, Sullivan LM, Manfra DJ, Wiekowski MT, Prosser DM, Vassileva G, Lira SA. Generation and analysis of mice lacking the chemokine fractalkine. Mol Cell Biol. 2001;21:3159–3165. doi: 10.1128/MCB.21.9.3159-3165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatori K, Nagai A, Heisel R, Ryu JK, Kim SU. Fractalkine and fractalkine receptors in human neurons and glial cells. J Neurosci Res. 2002;69:418–426. doi: 10.1002/jnr.10304. [DOI] [PubMed] [Google Scholar]
- 10.Harrison JK, Jiang Y, Chen S, Xia Y, Maciejewski D, McNamara RK, Streit WJ, Salafranca MN, Adhikari S, Thompson DA, Botti P, Bacon KB, Feng L. Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia. Proc Natl Acad Sci U S A. 1998;95:10896–10901. doi: 10.1073/pnas.95.18.10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardona A, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, Lee J, Cook DN, Jung S, Lira S, Littman DR, Ransohoff RM. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- 13.Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 14.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 15.Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 16.Geissmann F, Auffray C, Palframan R, Wirrig C, Ciocca A, Campisi L, Narni-Mancinelli E, Lauvau G. Blood monocytes: distinct subsets, how they relate to dendritic cells, and their possible roles in the regulation of T-cell responses. Immunol Cell Biol. 2008;86:398–408. doi: 10.1038/icb.2008.19. [DOI] [PubMed] [Google Scholar]
- 17.King IL, Dickendesher TL, Segal BM. Circulating Ly-6C+ myeloid precursors migrate to the CNS and play a pathogenic role during autoimmune demyelinating disease. Blood. 2009;113:3190–3197. doi: 10.1182/blood-2008-07-168575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafler DA, Slavik JM, Anderson DE, O'Connor KC, De Jager P, Baecher-Allan C. Multiple sclerosis. Immunol Rev. 2005;204:208–231. doi: 10.1111/j.0105-2896.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- 19.Eltayeb S, Berg AL, Lassmann H, Wallstrom E, Nilsson M, Olsson T, Ericsson-Dahlstrand A, Sunnemark D. Temporal expression and cellular origin of CC chemokine receptors CCR1, CCR2 and CCR5 in the central nervous system: insight into mechanisms of MOG-induced EAE. J Neuroinflammation. 2007;4:14. doi: 10.1186/1742-2094-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Shi XQ, Echeverry S, Mogil JS, De KY, Rivest S. Expression of CCR2 in both resident and bone marrow-derived microglia plays a critical role in neuropathic pain. J Neurosci. 2007;27:12396–12406. doi: 10.1523/JNEUROSCI.3016-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CL, Ransohoff RM, Charo IF. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One. 2010;5:e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardona AE, Sasse ME, Mizutani M, Cardona SM, Liu L, Savarin C, Hu T, Ransohoff RM. Scavenging roles of chemokine receptors: chemokine receptor deficiency is associated with increased levels of ligand in circulation and tissues. Blood. 2008 doi: 10.1182/blood-2007-10-118497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DR, Wang J, Kivisakk P, Rollins BJ, Ransohoff RM. Absence of monocyte chemoattractant protein 1 in mice leads to decreased local macrophage recruitment and antigen-specific T helper cell type 1 immune response in experimental autoimmune encephalomyelitis. J Exp Med. 2001;193:713–726. doi: 10.1084/jem.193.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardona AE, Huang D, Sasse ME, Ransohoff RM. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc. 2006;1:1947–1951. doi: 10.1038/nprot.2006.327. [DOI] [PubMed] [Google Scholar]
- 25.Ransohoff RM, Liu L, Cardona AE. Chemokines and chemokine receptors: multipurpose players in neuroinflammation. Int Rev Neurobiol. 2007;82:187–204. doi: 10.1016/S0074-7742(07)82010-1. [DOI] [PubMed] [Google Scholar]
- 26.Ransohoff RM, V, Perry H. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- 27.Fuhrmann M, Bittner T, Jung CK, Burgold S, Page RM, Mitteregger G, Haass C, LaFerla FM, Kretzschmar H, Herms J. Microglial Cx3cr1 knockout prevents neuron loss in a mouse model of Alzheimer's disease. Nat Neurosci. 2010;13:411–413. doi: 10.1038/nn.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Varvel NH, Konerth ME, Xu G, Cardona AE, Ransohoff RM, Lamb BT. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer's disease mouse models. Am J Pathol. 2010;177:2549–2562. doi: 10.2353/ajpath.2010.100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashwell K. The distribution of microglia and cell death in the fetal rat forebrain. Brain Res Dev Brain Res. 1991;58:1–12. doi: 10.1016/0165-3806(91)90231-7. [DOI] [PubMed] [Google Scholar]
- 30.Boya J, Calvo J, Prado A. The origin of microglial cells. J Anat. 1979;129:177–186. [PMC free article] [PubMed] [Google Scholar]
- 31.Cuadros MA, Navascues J. The origin and differentiation of microglial cells during development. Prog Neurobiol. 1998;56:173–189. doi: 10.1016/s0301-0082(98)00035-5. [DOI] [PubMed] [Google Scholar]
- 32.Cuadros MA, Navascues J. Early origin and colonization of the developing central nervous system by microglial precursors. Prog Brain Res. 2001;132:51–59. doi: 10.1016/S0079-6123(01)32065-4. [DOI] [PubMed] [Google Scholar]
- 33.Kaur C, Ling EA, Wong WC. Origin and fate of neural macrophages in a stab wound of the brain of the young rat. J Anat. 1987;154:215–227. [PMC free article] [PubMed] [Google Scholar]
- 34.Leong SK, Ling EA. Amoeboid and ramified microglia: their interrelationship and response to brain injury. Glia. 1992;6:39–47. doi: 10.1002/glia.440060106. [DOI] [PubMed] [Google Scholar]
- 35.Rezaie P, Patel K, Male DK. Microglia in the human fetal spinal cord--patterns of distribution, morphology and phenotype. Brain Res Dev Brain Res. 1999;115:71–81. doi: 10.1016/s0165-3806(99)00043-7. [DOI] [PubMed] [Google Scholar]
- 36.Juba A. Untersuchungen über die Entwicklung der Hortegaschen Mikroglia des Menschen. Arch Psychiatr. 1933;101:577–592. [Google Scholar]
- 37.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan WY, Kohsaka S, Rezaie P. The origin and cell lineage of microglia: new concepts. Brain Res Rev. 2007;53:344–354. doi: 10.1016/j.brainresrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Andjelkovic AV, Nikolic B, Pachter JS, Zecevic N. Macrophages/microglial cells in human central nervous system during development: an immunohistochemical study. Brain Res. 1998;814:13–25. doi: 10.1016/s0006-8993(98)00830-0. [DOI] [PubMed] [Google Scholar]
- 40.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Bruck W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 41.Prinz M, Priller J. Tickets to the brain: Role of CCR2 and CX(3)CR1 in myeloid cell entry in the CNS. J Neuroimmunol. 2010 doi: 10.1016/j.jneuroim.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Soulet D, Rivest S. Bone-marrow-derived microglia: myth or reality? Curr Opin Pharmacol. 2008;8:508–518. doi: 10.1016/j.coph.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Rivest S. Regulation of innate immune responses in the brain. Nat Rev Immunol. 2009;9:429–439. doi: 10.1038/nri2565. [DOI] [PubMed] [Google Scholar]
- 44.Cardona AE, Ransohoff RM. Chemokine Receptor CX3CR1. UCSD-Nature Molecule Pages. 2009 doi: 10.1038/mp.a000633.01. [DOI] [Google Scholar]
- 45.Cardona AE, Ransohoff RM. Chemokine Ligand CX3CL1. UCSD-Nature Molecule Pages. 2010 doi: 10.1038/mp.a000955.01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.