Abstract
Hindbrain rhombomere 1 (r1) is located caudal to the isthmus, a critical organizer region, and rostral to rhombomere 2 in the developing mouse brain. Dorsal r1 gives rise to the cerebellum, locus coeruleus, and several brainstem nuclei, whereas cells from ventral r1 contribute to the trochlear and trigeminal nuclei as well as serotonergic and GABAergic neurons of the dorsal raphe. Recent studies have identified several molecular events controlling dorsal r1 development. In contrast, very little is known about ventral r1 gene expression and the genetic mechanisms regulating its formation. Neurons with distinct neurotransmitter phenotypes have been identified in ventral r1 including GABAergic, serotonergic, and cholinergic neurons. Here we show that PITX2 marks a distinct population of GABAergic neurons in mouse embryonic ventral r1. This population appears to retain its GABAergic identity even in the absence of PITX2. We provide a comprehensive map of markers that places these PITX2-positive GABAergic neurons in a region of r1 that intersects and is potentially in communication with the dorsal raphe.
Keywords: hindbrain, development, transcription factor
1. Introduction
The hindbrain region of the central nervous system is responsible for coordination of motor control and regulation of autonomic processes such as respiration, heart rate, blood pressure, and arousal (Saper, 2000). The developing hindbrain is subdivided along the rostral-caudal axis into eight rhombomeres (r1–r8) which are distinguished by boundaries of gene expression, patterns of cell differentiation, and morphology (Lumsden and Keynes, 1989). Rhombomere 1 (r1), located caudal to the midbrain and isthmus and rostral to r2, is easily distinguishable morphologically in the mouse brain by E10.5. Dorsal r1 gives rise to the cerebellum and neurons of the locus coeruleus whereas ventral r1 contributes to the sensory vestibular nuclei, trigeminal and trochlear nuclei, pedunculopontine tegmental nucleus, parabrachial nucleus, Kölliker-Fuse nucleus, dorsal nucleus of the lateral lemniscus and neurons of dorsal raphe (Alder et al., 1996; Aroca et al., 2006; Chatonnet et al., 2007; Ding et al., 2003; Eddison et al., 2004; Jensen et al., 2008; Lin et al., 2001; Machold and Fishell, 2005; Marin and Puelles, 1995; Wingate and Hatten, 1999).
Much attention has been directed toward understanding the molecular markers and mechanisms of development of the cerebellum, the most prominent r1 derivative (Herrup and Kuemerle, 1997; Wang and Zoghbi, 2001). The cerebellum is one of the first brain structures to differentiate and one of the last to mature; it is estimated to contain as many as 80–85% of all human neurons and is an important center for many processes critical for life (Wang and Zoghbi, 2001). In contrast, the definition and origins of the neuronal populations in ventral r1 have received relatively little attention. This lack of molecular information on developing r1 has impeded progress in the characterization of mouse mutants and therapies for individuals with hindbrain defects.
Here, we test the hypothesis that Pitx2, a paired-like homeodomain transcription factor, specifies distinct GABAergic neurons that derive from ventral r1, using loss-of-function, conditional, and Cre knock-in alleles. We also present data from analyses of gene expression patterns in embryonic mouse ventral r1. Our results suggest that PITX2-positive neurons in r1 are GABAergic and can be divided into two unique subpopulations based on expression of unique combinations of transcription factors, including LHX1/5, NKX6.1/6.2, PAX2, and SOX2. Both PITX2-positive GABAergic populations occupy specific bilaterally symmetric regions of ventro-medial r1. The GABAergic identity of these cells is not disrupted by loss of PITX2. These observations provide a framework for ongoing studies aimed at exploring the functional molecular genetic pathways that regulate r1 neuronal development.
2. Materials and Methods
2.1 Mice
C57BL/6J mice were obtained from the Jackson Laboratory (JAX 000664). GAD67-GFP embryos were generated by crossing GAD67-GFP males with C57BL/6J females (Tamamaki et al., 2003). Pitx2Cre/+ mice (Liu et al., 2002) were crossed with FlpeR mice (JAX 003946) to excise the neomycin cassette. Dbx1Cre;R26YFP tissues were obtained by Frédéric Causeret by crossing Dbx1Cre/+ mice (Bielle et al., 2005) with a ROSA26loxP-stop-loxP-YFP strain (Srinivas et al., 2001). Pitx2Cre/+;ZsGrn embryos were generated by crossing Pitx2Cre/+ mice to ZsGrn reporter mice obtained from Jackson Laboratories (JAX 007006) (Madisen et al., 2010). Pitx2+/− mice were as previously described (Gage et al., 1999). Nestin-Cre transgenic (Tronche et al., 1999) mice were bred to Pitx2tlz/+ mice which are heterozygous for a null allele that expresses β-galactosidase under the control of Tau (manuscript in preparation). Nestin-Cre;Pitx2tlz/+ mice were then bred to Pitx2flox/flox (Gage et al., 1999) mice to generate embryos for analysis.
2.2 Tissue Preparation
Timed pregnancies were established with the morning of plug identification designated as E0.5. Embryos were dissected into PBS from pregnant females following cervical dislocation and hysterectomy. Embryos were fixed and processed for antibody staining or paraffin in situ hybridization histochemistry as previously described (Novitch et al., 2001; Skidmore et al., 2008). For frozen in situ hybridization, sections were fixed in 4% PFA for 40 minutes, washed in PBS, then incubated in TEA/acetic anhydride for 10 minutes (50 ml DPC-H20, 580 µl TEA 0.1M pH 8, 150 µl acetic anhydride). The remaining steps were performed as previously described (Martin et al., 2002).
Embryos were fixed in 4% paraformaldehyde for 1–2 hours depending on age and genotype. For frozen sections, embryos were cryoprotected overnight in 30% sucrose, flash frozen in O.C.T. embedding compound (Tissue Tek, Torrance, CA), and stored at −80°C until sectioning at 12–30 µm. For paraffin sections, tissues were embedded in paraffin and sectioned at 7 µm thickness. From each embryo and pup, an amniotic sac or tail was retained for genotyping. All procedures were approved by the University Committee on Use and Care for Animals at the University of Michigan.
2.3 Immunofluorescence and in situ hybridization
Immunofluorescence on paraffin embedded tissues was done as previously described (Martin et al., 2002; Martin et al., 2004). Immunofluorescence on frozen sections was done as previously described (Novitch et al., 2001). Antibodies used were guinea pig anti-phosphohistone H3 at 1:200 (Upstate Biotechnology, Inc., Lake Placid, NY), rabbit anti-PITX2 at 1:8000 (provided by Dr. Thomas Jessell, Columbia University), rabbit anti-PITX2 at 1:4000 (Capra Science, Ängelholm, Sweden), rabbit anti-VGLUT2 at 1:1000 (Millipore), rabbit anti-GABA at 1:1000 (Sigma), rabbit anti-5-hydroxytryptamine (5-HT) at 1:5000 (Sigma), goat anti-ChAT at 1:100 (Millipore), rabbit anti-LBX1 at 1:10000 (provided by Thomas Müller, Max-Delbrück Center of Molecular Medicine, Berlin), guinea pig anti-LMX1B at 1:5000 (provided by Dr. Thomas Müller), rabbit anti-SOX2 (Millipore) at 1:250, guinea pig anti-NKX6.2 at 1:8000 (provided by Dr. Thomas Jessell), guinea pig anti-BHLHB5 at 1:32,000 (provided by Dr. Ben Novitch), and the following mouse antibodies from Developmental Studies Hybridoma Bank at 1:100–1:500: anti-PAX7, anti-LHX1/5 (4F2), anti EVX1 (3A2), anti-EN1 (4G11), anti-NKX6.1 (F64A6B4), and anti-ISL1 (39.4D5). In situ hybridization on frozen and paraffin sections was done as previously described (Martin et al., 2002; Martin et al., 2004) using cRNA probes for Pitx2, Gbx2, Hoxa2, Otx2, Fgf8, Phox2a, Phox2b, and Lmx1a.
2.4 Microscopy and cell counts
Confocal fluorescent images were taken using a Leica TCS SP5 X Supercontinuum Confocal System with Upright Fluorescent Microscope. For single in situ and X-gal-stained slides, sections were photographed in brightfield. For pseudocolored neighboring merged images, sections were photographed in brightfield and converted into pseudo-fluorescent color, then overlaid in Photoshop. Digital images were processed with Adobe Photoshop CS2 v9.0 software. For quantification of double labeled cells, cells in r1 were counted in a minimum of 3 sections from E12.5 (NKX6.1, PAX2, SOX2, LHX1/5) or E14.5 (GABA) embryos.
3. Results
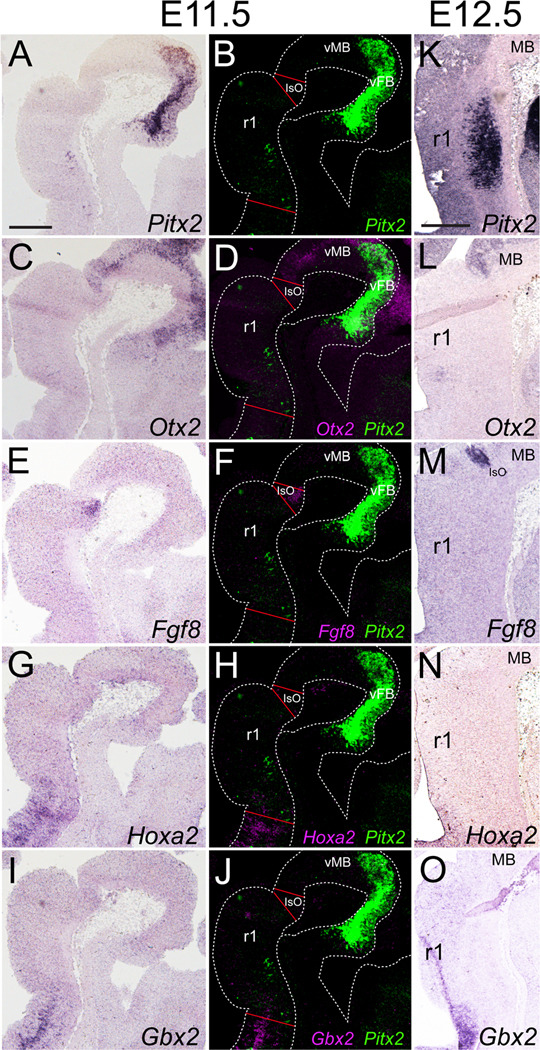
Pitx2-positive neuronal populations are localized in the forebrain, midbrain, hindbrain, and spinal cord (Mucchielli et al., 1996; Zagoraiou et al., 2009). At E11.5, Pitx2-positive cells were located in ventral r1 which was bordered rostrally by the Otx2-positive midbrain (Fig. 1C, D) and Fgf8-positive isthmic organizer (Fig. 1E, F) and caudally by Hoxa2-expressing r2 (Fig. 1G–H). Interestingly, at E11.5 most Pitx2-expressing cells were located in r1, however a few Pitx2-positive cells appeared to localize at the r1/r2 boundary (Fig. 1A). By E12.5, all Pitx2-positive cells were in a single population in mid-r1 (Fig. 1K). At this timepoint, r1 continued to be bordered rostrally by the Otx2-positive midbrain and Fgf8-positive isthmic organizer (Fig. 1L, M) and caudally by the Hoxa2/Gbx2-positive r2 (Fig. 1N, O).
Figure 1. Pitx2-expressing cells localize to rhombomere 1.
E11.5 (A–J) or E12.5 (K–O) sagittal sections processed for (A, K) Pitx2, (C, L) Otx2, (E, M) Fgf8, (G, N) Hoxa2, or (I, O) Gbx2 single in situ hybridization. Pseudocolored and merged images of neighboring slides processed for in situ hybridization to detect mRNA for Pitx2 and either Otx2 (D), Fgf8 (F), Hoxa2 (H), or Gbx2 (J). Scale bars in A and K are 250 µm and apply to panels A–J and K–O, respectively.
Adult ventral r1 contains several different cell populations, including trochlear motor neurons, branchiomotor neurons of the trigeminal nucleus, locus coeruleus neurons derived from the alar plate, GABAergic neurons of the pedunculopontine nucleus and laterodorsal tegmental nucleus, and the GABAergic, serotonergic, and glutamatergic neurons of the dorsal raphe (Aroca et al., 2006; Fu et al., 2010; Jensen et al., 2008; Marin and Puelles, 1995; Martin et al., 2002; Wang and Morales, 2009). As an early step toward characterizing the transcriptional profiles of ventral r1 neurons, we focused on cells expressing the paired-like transcription factor PITX2, which prior studies suggested were GABAergic interneurons (Martin et al., 2002). Here, we asked whether some PITX2-positive cells in r1 adopt neurotransmitter fates other than GABAergic, and whether their neurotransmitter identity requires functional PITX2.
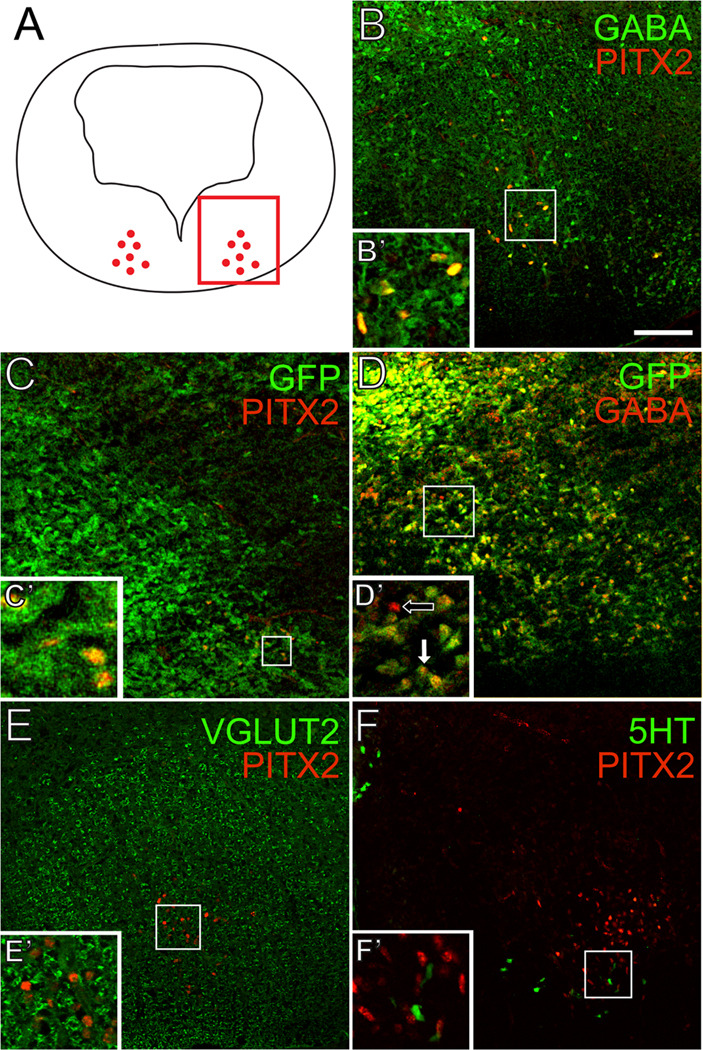
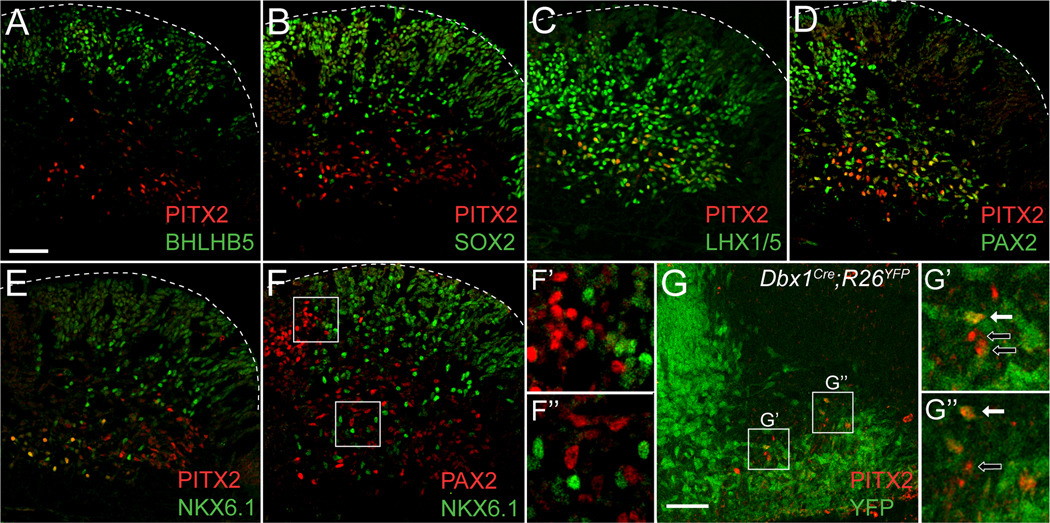
Through double immunofluorescence, we found that 100% (+/−0%) of E14.5 PITX2-positive cells were positive for GABA and GAD67-GFP (Fig. 2B–C’). Importantly, most GAD67-GFP-positive neurons in r1 were also positive for GABA immunoreactivity (Fig. 2D, D’), confirming that GABA is an accurate marker of GABAergic neurons in this tissue. PITX2-positive cells were negative for VGLUT2 immunofluorescence (Fig. 2E, E’), which likely marks glutamatergic tracts passing through the hindbrain, such as ponto-tegmental afferents (Geisler et al., 2007). There was no co-localization between PITX2 and 5-HT (5-hydroxytryptamine) (Fig. 2F), suggesting that r1 PITX2-positive neurons do not contribute to serotonergic cells of the raphe nuclei or locus coeruleus. Interestingly, some serotonin- and PITX2-positive cells intermingle at the ventromedial limits of Pitx2 expression, and these may represent GABAergic neurons that contribute to the raphe nuclei, pedunculopontine nucleus, or laterodorsal tegmental nucleus (Fig. 2F) (Torterolo et al., 2000; Wang and Morales, 2009). The close proximity of PITX2-positive cells to the 5-HT-positive population also raises the possibility that these neurons may communicate with each other (Jensen et al., 2008).
Figure 2. PITX2-positive GABAergic neurons in r1 are distinct from serotonergic, glutamatergic, and cholinergic neurons.
Double immunofluorescence of E14.5 mouse brain tissues sectioned transversely at the level of r1. Schematic in A indicates transverse orientation for panels B–F. Square inset in A shows the area represented in B–F. PITX2 co-localizes with GABA (B–B’) and GAD67-GFP (C–C’). Most GFP-positive cells are also GABA-positive (D–D’). PITX2-positive cells are negative for VGLUT2 (E–E’) and 5-HT (F–F’). Scale bar in B is 100 µm and applies to panels B–F. All images were taken using confocal microscopy.
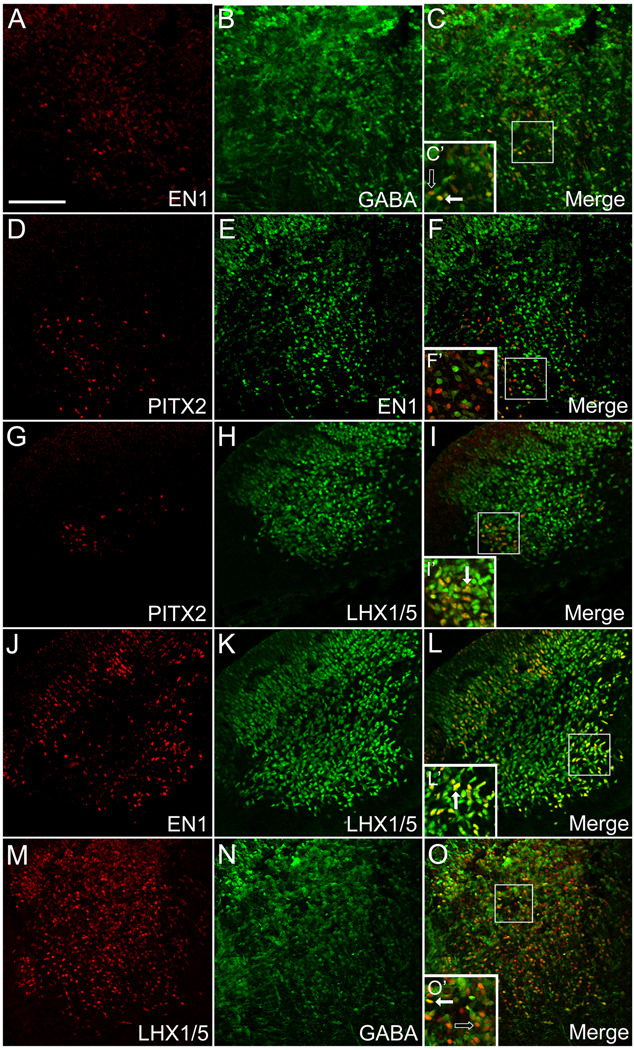
To better understand the distribution of developing neurons in the ventral hindbrain, we performed an extensive analysis of gene expression in the E12.5 mouse brain, a period of active neurogenesis (Figs. 3 and 4). The Engrailed homeobox genes En1/2 are expressed in a broad region encompassing the midbrain and anterior hindbrain and are critical for midbrain and dorsal r1 development (Sgaier et al., 2007; Zervas et al., 2005). We found that, similar to PITX2-positive cells, many EN1-positive cells in E12.5 ventral r1 were also GABAergic (Fig. 3A–C’). At E12.5 there was no significant co-expression of PITX2 and EN1 in ventral r1 cells (Fig. 3D–F’). These data suggest a mixed population of PITX2-positive and EN1-positive GABAergic neurons.
Figure 3. GABAergic r1 neurons express PITX2, EN1, and LHX1/5.
Double immunolabeling of E12.5 mouse brain tissues sectioned transversely at the level of r1 (orientation as in Figure 2A) reveals distinct patterns of overlap among GABA and several transcription factors. Insets in C–O are enlarged in C’–O’ and show double (solid arrow) or single (open arrow) labeled cells. Scale bar in A is 100 µm and applies to panels A–O. All images were taken using confocal microscopy.
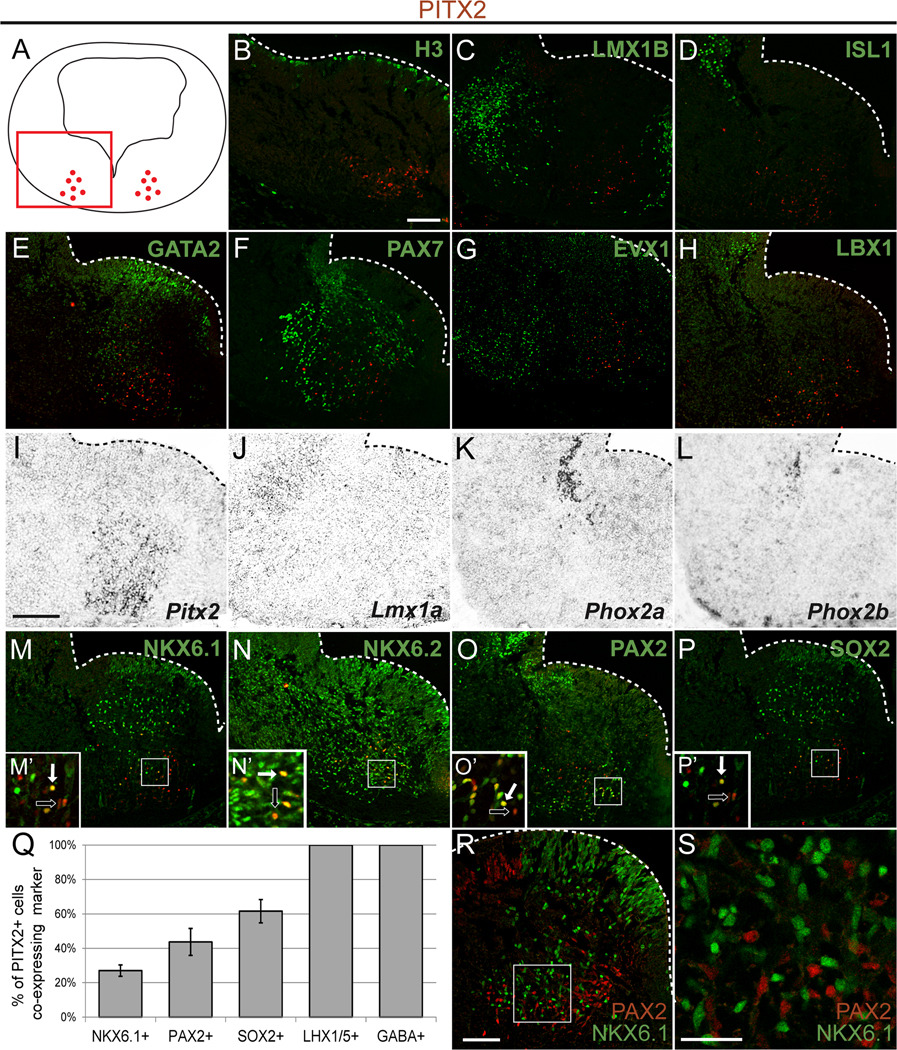
Figure 4. PITX2-positive GABAergic neurons occupy distinct regions of the ventral hindbrain.
Double immunolabeling (A–H, M–P, R, S) or in situ hybridization (I–L) of E12.5 mouse brain tissues sectioned transversely at the level of r1 (orientation as in panel A) reveals distinct patterns of overlap between PITX2 and several transcription factors. Insets in M–P are enlarged in M’–P’, and show double (solid arrow) or single (open arrow) labeled PITX2-positive cells. Graph in Q indicates the percentage of PITX2-positive cells which co-express the marker indicated (NKX6.1, PAX2, SOX2, and LHX1/5 are from E12.5 embryos; GABA is from E14.5 embryos). Error bars are +/− standard error of the mean of cell counts from N≥3 sections. (R, S) PAX2 and NKX6.1 mark separate cell populations in r1. Cells in the intermediate layer of panel R are enlarged in S. Scale bar in B is 100 µm and applies to panels B–H and M–P. Scale bar in I is 100 µm and applies to panels I–L. Scale bars in R and S are 75 µm and 30 µm, respectively. All immunofluorescent images were taken using confocal microscopy.
The LIM-homeodomain transcription factors are expressed by many neuronal subtypes throughout the developing central nervous system (Hunter and Rhodes, 2005). Lhx1 and Lhx5 are widely expressed in GABAergic cells throughout the midbrain, r1, and the developing cerebellum, and are required for normal cerebellar development (Morales and Hatten, 2006; Zhao et al., 2007). We sought to determine whether PITX2-positive cells in ventral r1 also express Lhx1/5 using an antibody that marks both LHX1 and LHX5, but does not distinguish between the two. We found that at E12.5, 100% (+/−0%) of the PITX2-positive cells were also LHX1/5-positive (Fig. 3G–I’ and Fig. 4Q), whereas LHX1/5-positive cells comprised a much larger population of cells, most of which were negative for PITX2. In addition, all EN1-positive cells were also positive for LHX1/5 (Fig. 3J–L’) and some were also positive for GABA (Fig. 3A–C’), providing evidence that r1 GABAergic neurons express unique and specific combinations of Pitx2, En1, and Lhx1/5. Additionally, many LHX1/5-positive cells were negative for GABA (Fig. 3M–O’). Thus, the LHX1/5-positive population of cells in ventral r1 appears to represent a heterogeneous group of neurons, and combinations of GABA, Pitx2 or En1 expression may be used to define distinct subpopulations within this group.
PITX2-positive cells in r1 at E12.5 occupy a region of the neural tube that contains non-mitotic cells that were negative for phospho-histone H3 (Fig. 4B). This is consistent with expression of PITX2 in post-mitotic developing neurons of the hypothalamus and midbrain (Martin et al., 2002). Early developing neurons in r1 can be identified by expression of several different transcription factors. LMX1B is a LIM homeodomain transcription factor involved in initiation and maintenance of the isthmic organizer and is important for midbrain and hindbrain patterning (Jacob et al., 2009; Matsunaga et al., 2002; Mishima et al., 2009). Lmx1b is expressed in the principal sensory nucleus of the trigeminal nerve, the Kölliker-Fuse nucleus, the dorsal raphe, and the parabrachial nuclei (Dai et al., 2008; Jacob et al., 2009; Matsunaga et al., 2002; Prakash et al., 2009; Zervas et al., 2005). LMX1B is also required for the differentiation of midbrain dopaminergic neurons and hindbrain serotonergic neurons (Ding et al., 2003; Jacob et al., 2009). We observed two groups of LMX1B-positive cells in E12.5 r1, one medial and one more laterally positioned (Fig. 4C). The PITX2-positive neurons were located between these two groups of LMX1B-positive cells (Fig. 4C).
Islet1 (Isl1) is expressed by motor neurons, including the oculomotor and trochlear nuclei that contain cell bodies of the third and fourth cranial nerves, respectively (Agarwala and Ragsdale, 2002; Prakash et al., 2009). In our study, ISL1-positive cells marking the trochlear nucleus were located at the dorsal-ventral axis border of r1 and lateral to PITX2-positive neurons (Fig. 4D). The lack of ISL1 co-localization with PITX2 suggests that PITX2-positive cells do not contribute to the r1-derived trochlear nucleus (see Fig. 9).
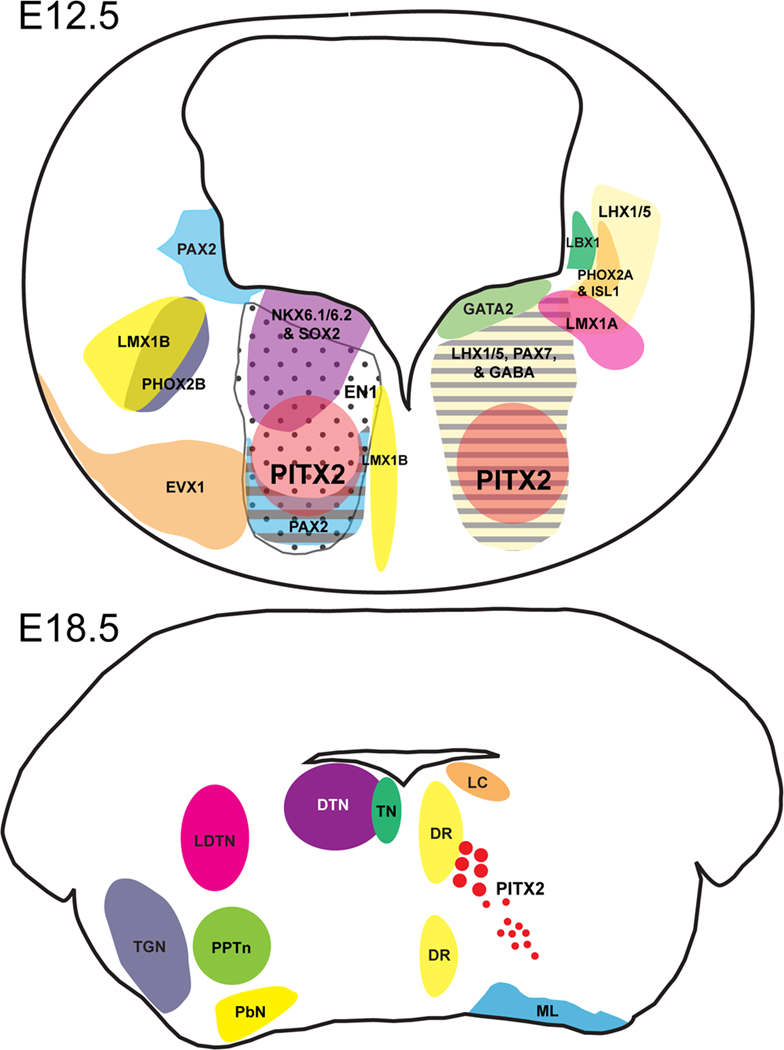
Figure 9. Schematic of transverse sections through the developing mouse brain at the level of rhombomere 1.
(A) E12.5 transverse section through r1 showing transcription factor and GABA patterning in relation to PITX2. (B) E18.5 transverse section through r1 showing nuclei patterning based on “Atlas of the Prenatal Mouse Brain” (Schambra et al., 1992) in relation to PITX2 (red circles). Abbreviations: DR, dorsal raphe nucleus; DTN, dorsal tegmental nucleus; LC, locus coeruleus; LDTN, laterodorsal tegmental nucleus; ML, medial lemniscus; PbN, parabrachial nucleus; PPTn, pedunculopontine nucleus; TN, trochlear nucleus; TGN, trigeminal motor nucleus.
GATA2, a transcription factor implicated in GABAergic development in both dorsal and ventral midbrain and required for serotonergic neuron development in the hindbrain, is highly expressed in a superficial ventral domain of r1 (Craven et al., 2004). In our studies, this GATA2-positive population was distinct from the PITX2-positive cell population (Fig. 4E). PAX7, a paired-like transcription factor, is widely expressed in ventral r1, but did not mark PITX2-positive cells (Fig. 4F). EVX1, a transcription factor that is highly expressed in spinal cord interneurons, was also expressed in ventral r1 cells in a region adjacent to but distinct from PITX2-positive cells (Fig. 4G). LBX1 marked a distinct population of neurons dorsal to the PITX2-expressing population (Fig. 4H). Thus, PITX2-positive neurons are negative for the transcription factors GATA2, PAX7, EVX1, and LBX1.
In addition to Isl1, we tested whether Pitx2 neurons express other markers of trochlear and parabrachial nuclei. Trochlear nucleus motor neurons in rostral r1 can also be identified by expression of Phox2a and Phox2b (Pattyn et al., 1997). We determined the expression patterns of Phox2a and Phox2b in E12.5 ventral r1 in relation to Pitx2-expressing cells (Fig. 4I–L). Analysis of neighboring sections showed no overlap between Phox2a or Phox2b with Pitx2 mRNA, providing further evidence that PITX2-positive cells do not contribute to the trochlear nucleus. Lmx1a encodes a LIM homeodomain transcription factor that is expressed in the parabrachial nucleus and rhombic lip and is necessary for proper cerebellar development (Mishima et al., 2009; Zou et al., 2009); however, its role in ventral r1 has not been explored. Lmx1a was weakly expressed lateral and dorsal to the LMX1B population of cells in the LHX1/5-positive region (Fig. 4I–J and Fig. 3H, K). Together, these data suggest that PITX2-positive r1 neurons do not express Phox2a, Phox2b, or Lmx1a and thus do not contribute to trochlear or parabrachial nuclei.
In the caudal hindbrain and spinal cord, NKX6.1 and NKX6.2 are expressed in visceral motor neurons and interneurons where they are required for neural identity (Briscoe et al., 2000; Pattyn et al., 2003; Sander et al., 2000). NKX6.1 is also implicated in serotonergic neuronal specification in r1 (Craven et al., 2004). NKX6.1 and NKX6.2 were both broadly expressed in ventral r1 and co-localized with some PITX2-positive cells (Fig. 4M, N). Quantitative analysis showed that 27.0% (+/−3.3%) of PITX2-positive cells were NKX6.1-positive (Fig. 4Q). PAX2, a paired-like homeodomain transcription factor and marker of early midbrain/hindbrain (Rowitch and McMahon, 1995), was expressed in ventral r1 in a domain that partially overlapped with PITX2 positive cells wherein 43.7% (+/−7.8%) of PITX2-positive cells were PAX2-positive (Fig. 4O). We also observed 61.6% (+/−6.8%) overlap between PITX2-positive cells and SOX2 (Fig. 4P). Co-expression of some PITX2-positive cells with NKX6.1, NKX6.2, and PAX2 is consistent with the idea that PITX2-positive cells may adopt interneuron fates. In the developing spinal cord, PAX2 and NKX6.1 mark separate post-mitotic populations with distinct progenitor populations (Lebel et al., 2001). Similarly, PAX2 and NKX6.1 in ventral r1 mark separate populations (Fig. 4R, S), suggesting the presence of at least two subpopulations of PITX2-positive cells that arise from separate progenitor populations.
In order to identify whether Pitx2 is expressed in progenitor populations, we looked for early Pitx2 expression in r1 at E10.5. Although Pitx2 is expressed in the E10.5 midbrain, there was no detectable Pitx2 mRNA in r1 at E10.5 (data not shown). At E11.5, PITX2-positive cells were located lateral to the progenitor zone and negative for the transcription factors BHLHB5, which marks V1 and V2 populations in the spinal cord (Liu et al., 2007), and SOX2 (Fig. 5A, B). Like the E12.5 PITX2-positive population, PITX2- positive E11.5 cells were also positive for LHX1/5, PAX2, and NKX6.1 (Fig. 5C–E). Again, PAX2 and NKX6.1 marked separate r1 populations with PAX2-positive cells medially bordering the NKX6.1-positive population (Fig. 5F–F“), indicating PITX2-positive cells form at least two subpopulations. To determine whether PITX2-positive cells derive from a single progenitor population, we analyzed E11.5 Dbx1Cre;R26YFP embryos for PITX2 patterning. In the spinal cord, the Dbx1-lineage marks V0 populations and was chosen for analysis because Pax2 positive, En1-negative cells in the spinal cord are known to derive from V0 Dbx1-positive progenitors (Lanuza et al., 2004), whereas Nkx6.1-expressing cells are more ventral and constitute V2–V3 populations (Sander et al., 2000). Interestingly, most ventral r1 PITX2-positive cells were also positive for YFP, although there were several cells which were YFP-negative (Fig. 5G–G”), which likely indicates separate progenitor populations, but may also be due to inefficient Cre recombination (Teissier et al., 2010). Thus, r1 PITX2-positive cells appear to derive from at least two different ventral r1 progenitor populations that can be divided into subpopulations based on transcription factor expression patterns.
Figure 5. Early expression of PITX2 is similar to E12.5 patterning and includes Dbx1-lineage cells.
Immunolabeling of E11.5 (A–G”) Pitx2+/+ or Dbx1Cre;R26YFP embryos sectioned transversely at the level of r1 (orientated as in Figure 4A). (A–E) E11.5 PITX2-positive cells in r1 are negative for BHLHB5, some are SOX2-positive, and many are positive for LHX1/5, PAX2, and NKX6.1. (F–F”) In the E11.5 r1, PAX2 and NKX6.1 mark separate cell populations. (G–G”) Many PITX2-positive cells are YFP-positive, although some are negative. Insets in F and G are enlarged in F’/F” and G’/G”, respectively. G’ and G” show double (solid arrow) or single (open arrow) labeled cells. Scale bar in A is 50 µm and applies to panels A–F. Scale bar in G is 50 µm. All immunofluorescent images were taken using confocal microscopy.
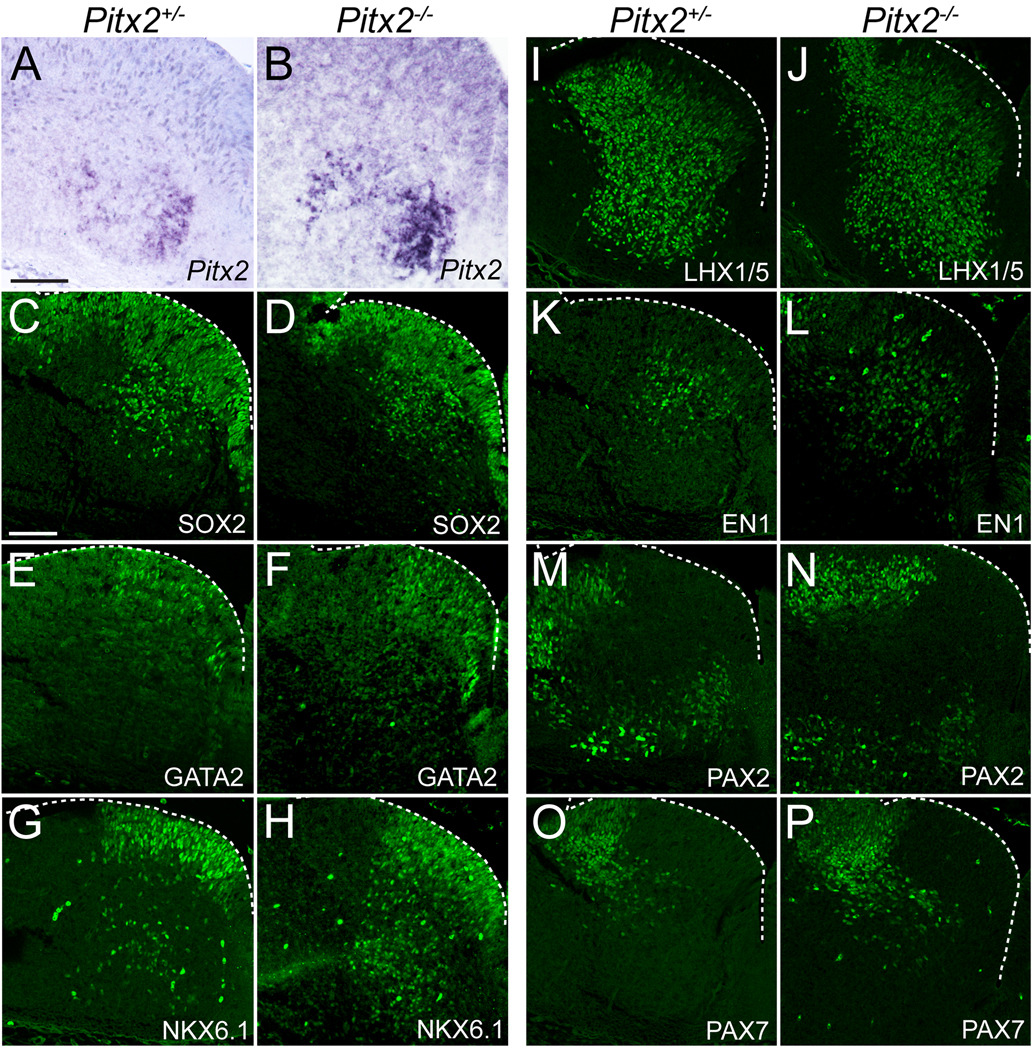
Next we asked whether loss of PITX2 affects neuronal distribution or early patterning by analyzing transcription factor expression in E12.5 Pitx2+/− and Pitx2−/− littermate embryos. Loss of Pitx2 did not affect overall distribution of Pitx2-expressing cells (Fig. 6A, B) as determined by in situ hybridization using a cRNA probe that is expressed from both the wild type and null alleles (Martin et al., 2004). The increased density of Pitx2 mRNA in null embryos compared to controls could signify increased transcription or mRNA stability. Additionally, expression of several transcription factors (SOX2, GATA2, NKX6.1, LHX1/5, EN1, PAX2, PAX7) in Pitx2−/− ventral r1 appeared normal as compared to Pitx2+/− littermate (Fig. 6) or wild type (Fig. 4) embryos. These data suggest that PITX2 is not required for the proper early migration of Pitx2-expressing cells or early patterning of ventral r1.
Figure 6. PITX2 is dispensable for early neuronal migration and ventral r1 patterning.
In situ hybridization (A–B) or immunolabeling (C–P) of E12.5 Pitx2+/− and Pitx2−/− mouse brain tissues sectioned transversely at the level of r1 (orientation as in Figure 4A) reveals normal transcription factor patterning. Scale bars in A and C are 100 µm and apply to panels A–B and C–P, respectively. All immunofluorescent images were taken using confocal microscopy.
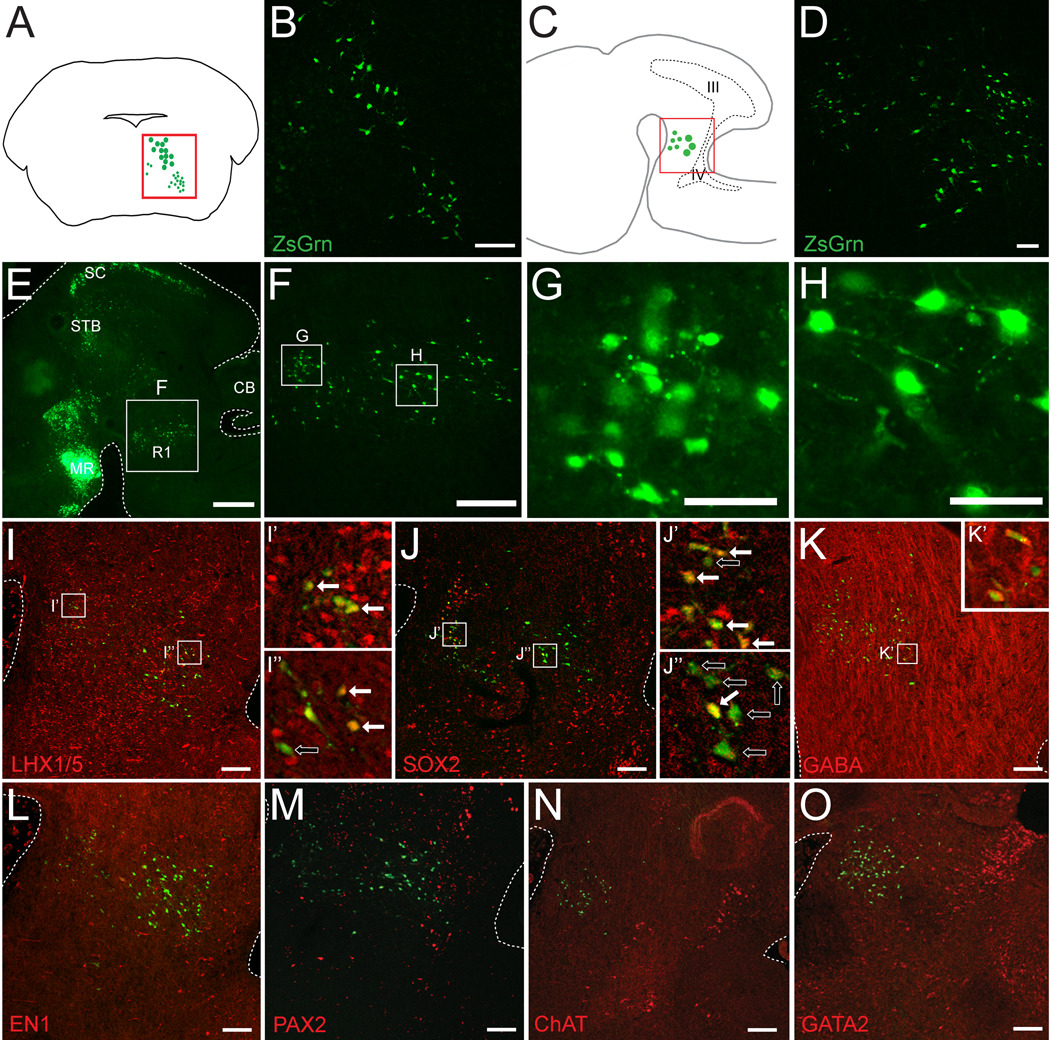
By E18.5, most neurons in the hindbrain have undergone initial stages of differentiation and many of the early transcription factors are no longer expressed. To assay Pitx2-lineage neuronal fates, we crossed Pitx2Cre/+ mice (Liu et al., 2002; Skidmore et al., 2008) with a ZsGrn Cre reporter strain. Interestingly, at E18.5 PITX2-positive neurons comprised two contiguous regions in ventral r1. Transverse sections revealed a deep PITX2-positive population with larger cell bodies and a more superficial population of smaller cells (Fig. 7B). ZsGrn staining showed short neurites in Pitx2Cre/+;ZsGrn embryos, further suggesting these cells may be interneurons. Analysis of sagittal sections of Pitx2Cre/+;ZsGrn also highlighted the cell localization and size differences between the superficial and deep populations of PITX2-positive cells in ventral r1 (Fig. 7D–H).
Figure 7. Two PITX2-positive populations span ventral r1.
E18.5 Pitx2Cre/+;ZsGrn transverse (B) and sagittal (D–O) sections processed for immunofluorescence with antibodies against transcription factors, ChAT, and GABA in the hindbrain. (A) Schematic of a transverse section identifying the region pictured in panel B. (C) Schematic of a sagittal section identifying the region pictured in panels D–O. Boxes in I–K are enlarged in I’–K” and show double (solid arrow) or single (open arrow) labeled PITX2-lineage cells. Panels are arranged medial (I, L) to lateral (K, O). Scale bar in B is 100 µm, in E is 500 µm, in F is 200 µm, and in G–H is 50 µm. Scale bars in D and I–O are 100 µm. All immunofluorescence images were taken using confocal microscopy. Abbreviations: III, third ventricle; IV, fourth ventricle; CB, cerebellum; MR, mammillary region; SC, superior colliculus; STB, subtectal band.
Previous studies in the spinal cord identified a requirement for LHX1 and LHX5 in the maintenance of inhibitory interneuron identity (Pillai et al., 2007). We observed LHX1/5-positive cells distributed throughout the rostral hindbrain and both PITX2-positive populations in r1 contained many LHX1/5-positive cells (Fig. 7I–I”). Interestingly, the more superficial population of PITX2-positive cells was mostly SOX2-positive, whereas only a few cells in the deep population expressed SOX2 (Fig. 7J–J”). Although PITX2-positive cells appear to constitute two separate r1 populations based on localization and morphology, they are both GABAergic at E18.5 (Fig. 7K–K’). PITX2-lineage E18.5 neurons did not express the transcription factor EN1 (Fig. 7L) similar to results at E12.5 (Fig. 3D–F). Interestingly, neither PITX2-positive r1 population was PAX2-positive at E18.5, in contrast to E12.5, when some PITX2-positive cells co-labeled with PAX2 (Fig. 4O and Fig. 7M). Additionally, PITX2-positive cells were negative for GATA2, which marks serotonergic neurons in r1 (Gavalas et al., 2003), and for choline acetyltransferase (ChAT), an enzyme produced in cholinergic populations (Fig. 7N, O). A few ChAT-positive cells were observed caudal to Pitx2-positive neurons but appeared to constitute distinct neuronal groups, such as the pedunculopontine, parabrachial, and microcellular tegmental nuclei (Machold and Fishell, 2005; Mizukawa et al., 1986).
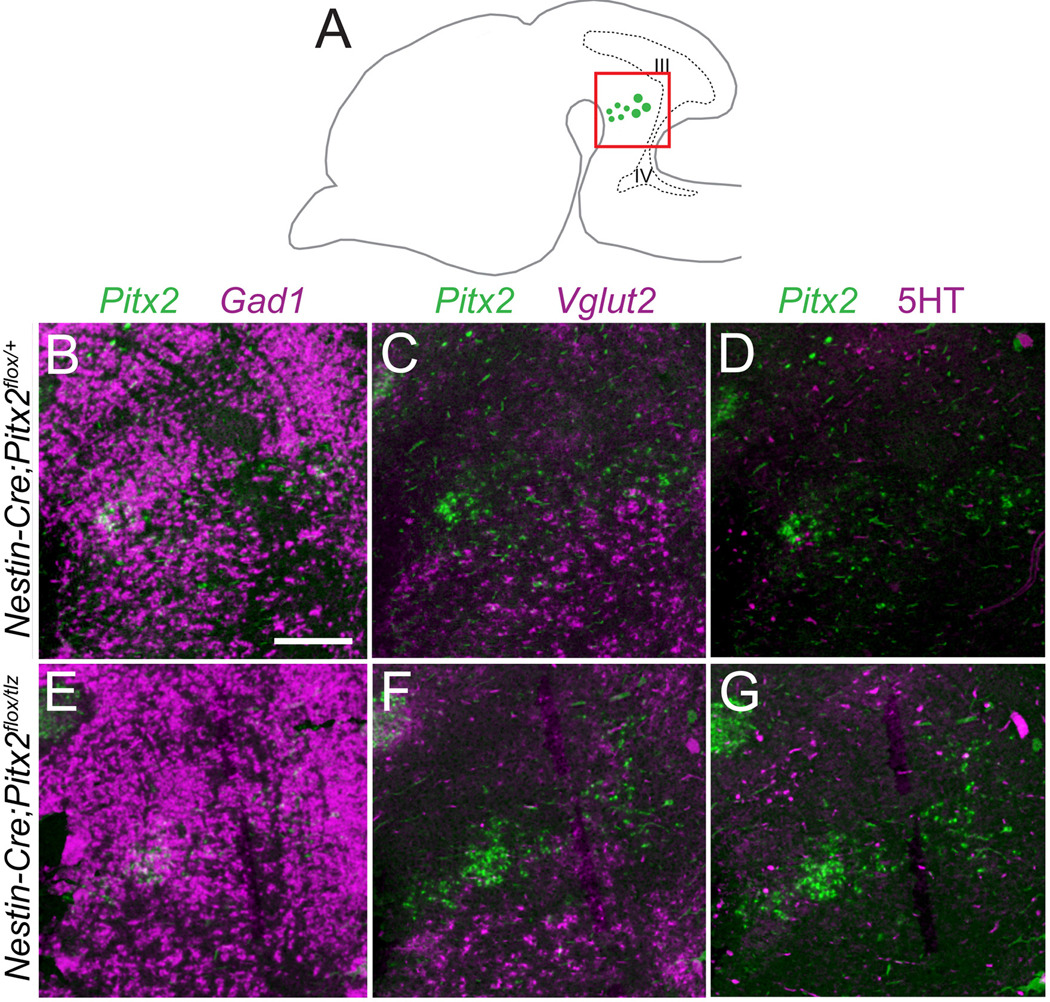
To determine whether loss of Pitx2 affects hindbrain neuronal fate specification, we analyzed E18.5 Nestin-Cre conditional Pitx2-knockout embryos for neurotransmitter identity in r1. Pitx2-positive cells in Nestin-Cre;Pitx2flox/+ embryos occupied a region that highly expresses Gad1 but not Vglut2 or 5-HT (Fig. 8B–D), consistent with GABAergic but not glutamatergic or serotonergic fates. Loss of Pitx2 in Nestin-Cre;Pitx2flox/tlz embryos did not disrupt Gad1, Vglut2, or 5-HT, suggesting that reduced Pitx2 dosage does not alter the GABAergic fate of PITX2-positive hindbrain neurons.
Figure 8. Ventral r1 GABAergic identity is PITX2-independent.
Sagittal E18.5 Nestin-Cre;Pitx2flox/+ and Nestin-Cre;Pitx2flox/tlz littermate brains processed for single in situ hybridization for Pitx2, Gad1, or Vglut2 or immunohistochemistry with antibodies against 5-HT. (A) Cartoon of a sagittal section showing the orientation of panels B–G. (B–G) Merged images of neighboring sections processed for in situ hybridization for Pitx2 and either Gad1 (B, E), Vglut2 (C, F), or 5-HT (D, G) immunohistochemistry and pseudocolored. Scale bar in B is 200 µm and applies to panels B–G.
4. Discussion
Here we show that PITX2, a paired-like homeodomain transcription factor, is expressed in GABAergic neurons in the ventral aspect of mouse embryonic r1. These PITX2-positive GABAergic neurons may comprise a population of inhibitory interneurons, based on their (a) co-expression with LHX1/5 and PAX2 (Pillai et al., 2007), (b) location in ventral r1, and (c) lack of co-expression with markers of glutamatergic, serotonergic, noradrenergic, and cholinergic neurons. We also show that subsets of GABAergic PITX2-positive neurons express the transcription factors SOX2, NKX6.1, and NKX6.2 at E12.5. Interestingly, Pitx2 loss of function does not disrupt the production or specification of these GABAergic neurons, suggesting compensatory mechanisms likely exist.
A summary of our marker analysis in wild type embryos is depicted in Fig. 9. PITX2-positive cells mark a distinct, bilaterally symmetric region of ventromedial r1 containing some cells that also express LHX1/5, NKX6.1, NKX6.2, PAX2, and SOX2 (Fig. 9). The EN1-positive domain spans this PITX2-positive region but extends further toward the ventricle than PITX2-positive cells. LHX1/5-positive cells comprise two separate populations in ventral r1, a medial population that contains PITX2-positive cells and a separate, more lateral population that is PITX2-negative.
4.1 Axial level and context determine PITX2-positive neuronal identity
PITX2-positive neuronal fate (as defined by neurotransmitter phenotype) appears to depend on rostro-caudal and dorso-ventral location along the neural tube. At rostral axial levels in the hypothalamus, PITX2-lineage neurons contribute to the glutamatergic subthalamic nucleus, where it is required for neuronal migration (Martin et al., 2004). In the midbrain, PITX2-positive superior colliculus GABAergic neurons also require Pitx2 for proper migration and differentiation (Martin et al., 2004; Waite et al., 2011). At more caudal levels in the spinal cord, PITX2-positive interneurons of the V0C subclass are cholinergic, whereas those of the V0G class are glutamatergic (Zagoraiou et al., 2009). These subclasses of PITX2-positive V0 interneurons are unevenly distributed along the rostro-caudal axis of the spinal cord, wherein cholinergic neurons occupy rostral lumbar levels while glutamatergic neurons occupy caudal lumbar areas (Zagoraiou et al., 2009). The axial dependence of PITX2-positive neuronal identity suggests that neurotransmitter fate specification is not likely to be determined by PITX2-mediated transcriptional regulation.
Previous studies have attempted to relate the dorsal-ventral developmental patterning in spinal cord to hindbrain patterning (Lebel et al., 2007). Our data suggest that both similarities and differences exist between ventral r1 and spinal cord development. The observation that a subpopulation of PITX2-positive cells in ventral r1 is PAX2/LHX1/5-positive, EN1-negative, and derives from Dbx1-positive progenitors suggests that these cells are homologous to V0 interneurons like their PITX2-positive spinal cord counterparts (Gray, 2008; Lanuza et al., 2004). V0 PITX2-positive cells in r1 and the V0 domain in spinal cord differentiate into several different types of neurons (cholinergic and glutamatergic in spinal cord, GABAergic in r1), suggesting that neural progenitors destined to express PITX2 are not necessarily pre-specified with respect to neurotransmitter fate but influenced by local factors dependent on axial level. We also show that a subpopulation of PITX2-positive cells was NKX6.1-positive and PAX2/LMX1B-negative, suggesting that some of the PITX2-positive r1 cells might be homologous to a spinal V2 interneuron population (Gray, 2008; Lebel et al., 2007; Sander et al., 2000). Thus, PITX2 expression in r1 may be progenitor derived lineage-independent and instead be regulated by planar positioning within the developing hindbrain.
Through comparison of the identities of r1 PITX2-positive cells to previously published spinal cord patterning maps, we found differences between cell populations in r1 versus spinal cord. In spinal cord, V2 neurons are GATA2-positive, whereas r1 PITX2-positive V2 neurons (based on Nkx6.1-positive and Lmx1b-negative expression similar to spinal V2 interneurons), did not express Gata2. Because our studies did not include GATA2-lineage tracing, we cannot distinguish between transient and absent expression of GATA2 by r1 PITX2-positive cells. It is also possible that V2 neurons have different molecular signatures in r1 and spinal cord, as is true for several of the dorsal populations, the pMNvs, and the V3 populations (Gray, 2008). Alternatively, previous studies have shown that several D–V populations in r1 may not exist in the developing spinal cord (such as the DA4 and DB2 populations), or may exist in developing spinal cord and caudal rhombomeres but not r1 (such as the DI2, DI3, DI5, and DI6) and that some populations have unique expression patterns between the hindbrain and spinal cord (such as DI1–DI3, pMN, and V3 populations) (Gray, 2008).
4.2 Transcriptional mechanisms of GABAergic neuronal differentiation
Throughout the central nervous system, distinct neuronal populations are distinguished by the complement of transcription factors they express as well as by characteristics such as neurotransmitter fate and unique projection patterns. Here we provide a description of several early r1 transcription factors and their mapping with respect to PITX2-positive populations. There is substantial overlap between PITX2 and LHX1/5 expression in GABAergic neurons, and some PITX2-positive cells also express PAX2 in ventral r1 at E12.5. PAX2 and LHX1/5, along with PAX5 and PAX8, function to regulate formation of spinal cord GABAergic inhibitory interneurons (Pillai et al., 2007). Further genetic studies are necessary to identify the factors that regulate formation of PITX2-positive GABAergic inhibitory neurons in r1.
In addition to PAX and LHX genes, several other transcription factor genes have been shown to regulate GABAergic neuronal differentiation. Interestingly, mutations of many of these transcription factor genes also disrupt (or augment) glutamatergic differentiation, although findings vary by neuronal population and rostro-caudal axial level (Cheng et al., 2005; Pillai et al., 2007). The basic helix-loop-helix (bHLH) transcription factor Ptf1a is required for proper formation of dorsal spinal cord, cerebellar, and retinal GABAergic inhibitory interneurons and loss of Ptf1a leads to an expansion of spinal cord glutamatergic neurons (Glasgow et al., 2005). The homeobox gene Lbx1 is also necessary (and sufficient) for spinal cord GABAergic neuronal differentiation (Cheng et al., 2005), but Lbx1 activity can be modified by the homeobox gene Tlx3, itself an important regulatory of GABAergic vs. glutamatergic spinal cord neuronal differentiation (Cheng et al., 2005). While Pitx2 does not appear critical for GABAergic fate specification, it may regulate expression of genes that modulate other, as yet unidentified aspects of GABAergic neuronal function or maintenance.
4.3 Potential roles for PITX2-positive GABAergic r1 neurons
There are several important caveats to consider in our interpretation of the functional and molecular identities of r1 PITX2-positive GABAergic interneurons. First, the molecular characteristics of r1 neurons are incompletely described in the literature, forcing us to rely on comparisons with spinal cord or other axial levels. For example, studies showing that Dbx1-lineage cells in more caudal rhombomeres are responsible for regulating breathing have excluded r1 from their analysis (Borday et al., 2006). Second, r1 is unique among rhombomeres in that it develops through signaling from the isthmic organizer, does not express any Hox genes, and has unique requirements for Shh signaling (Blaess et al., 2006; Irving and Mason, 2000; Lebel et al., 2007). Nonetheless, the potential V0 and V2 GABAergic neurotransmitter identities of PITX2-positive cells and their transcription factor profiles are important for future studies exploring the functions of PITX2-positive cells in r1.
Ventral r1 contains many neurons that participate in critical life processes such as control of respiration (Gray, 2008). Locomotion is also controlled in part by the activities of ventral hindbrain interneuron populations (Grossmann et al., 2010). Our studies provide evidence that r1 PITX2-positive GABAergic neurons are distinct from neurons of the trochlear motor nucleus, serotonergic neurons of the dorsal raphe, Lmx1a-positive parabrachial neurons, Phox2a/b-positive visceral motor neurons, locus coeruleus neurons, and trigeminal neurons. Several nuclei in ventral r1 are known to contain GABAergic neurons including the dorsal raphe, laterodorsal tegmental nucleus, and pedunculopontine nucleus (Mena-Segovia et al., 2009). In the ventral r1 field, the dorsal raphe is medial, the dorsal tegmental nucleus is in deep ventral r1 near the ventricle, while the pedunculopontine tegmental nucleus is more lateral and superficially localized (Martin, 2003; Schambra et al., 1992). Based on GABAergic identity and localization, we predict that PITX2-positive cells contribute to the pedunculopontine tegmental nucleus which is thought to be involved in local inhibition controlling locomotion, REM, alertness, and respiratory patterns (Datta et al., 2001; Kozak et al., 2005; Saponjic et al., 2005; Tsang et al., 2010). A modulatory role for PITX2-positive GABAergic neurons in respiration, alertness, or other important autonomic functions could help explain why Nestin-Cre conditional Pitx2 mutants fail to survive beyond the immediate postnatal period (Sclafani et al., 2006). Further studies should help clarify the physiological roles of PITX2-positive hindbrain neurons and their potential contributions to control of locomotion, respiration, or other autonomic functions.
Acknowledgements
We thank Martin Meyers for the GAD67-GFP and Ben Novitch for helpful discussions. MRW was supported by the NIH Cellular and Molecular Biology Training Grant (T32-GM007315), a Rackham Regents Fellowship, and a Rackham Predoctoral Fellowship. KS was supported by the University of Michigan Neuroscience Graduate Training Program and the Center for Organogenesis Training Grant (5-T32-HD007505). This work was supported by NIH RO1 grant NS054784 to DMM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mindy R. Waite, Email: mrwaite@umich.edu.
Kaia Skaggs, Email: kskaggs@umich.edu.
Parisa Kaviany, Email: pkaviany@umich.edu.
Jennifer M. Skidmore, Email: camelot@umich.edu.
Frédéric Causeret, Email: causeret.frederic@ijm.univparis-diderot.fr.
James F. Martin, Email: jmartin@ibt.tamhsc.edu.
Donna M. Martin, Email: donnamm@umich.edu.
References
- Agarwala S, Ragsdale CW. A role for midbrain arcs in nucleogenesis. Development. 2002;129:5779–5788. doi: 10.1242/dev.00179. [DOI] [PubMed] [Google Scholar]
- Alder J, Cho NK, Hatten ME. Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron. 1996;17:389–399. doi: 10.1016/s0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Aroca P, Lorente-Canovas B, Mateos FR, Puelles L. Locus coeruleus neurons originate in alar rhombomere 1 and migrate into the basal plate: Studies in chick and mouse embryos. J Comp Neurol. 2006;496:802–818. doi: 10.1002/cne.20957. [DOI] [PubMed] [Google Scholar]
- Bielle F, Griveau A, Narboux-Neme N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Borday C, Vias C, Autran S, Thoby-Brisson M, Champagnat J, Fortin G. The pre-Botzinger oscillator in the mouse embryo. J Physiol Paris. 2006;100:284–289. doi: 10.1016/j.jphysparis.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Chatonnet F, Wrobel LJ, Mezieres V, Pasqualetti M, Ducret S, Taillebourg E, Charnay P, Rijli FM, Champagnat J. Distinct roles of Hoxa2 and Krox20 in the development of rhythmic neural networks controlling inspiratory depth, respiratory frequency, and jaw opening. Neural Dev. 2007;2:19. doi: 10.1186/1749-8104-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Craven SE, Lim KC, Ye W, Engel JD, de Sauvage F, Rosenthal A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- Dai JX, Hu ZL, Shi M, Guo C, Ding YQ. Postnatal ontogeny of the transcription factor Lmx1b in the mouse central nervous system. J Comp Neurol. 2008;509:341–355. doi: 10.1002/cne.21759. [DOI] [PubMed] [Google Scholar]
- Datta S, Spoley EE, Patterson EH. Microinjection of glutamate into the pedunculopontine tegmentum induces REM sleep and wakefulness in the rat. Am J Physiol Regul Integr Comp Physiol. 2001;280:R752–R759. doi: 10.1152/ajpregu.2001.280.3.R752. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- Eddison M, Toole L, Bell E, Wingate RJ. Segmental identity and cerebellar granule cell induction in rhombomere 1. BMC Biol. 2004;2:14. doi: 10.1186/1741-7007-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Le Maitre E, Fabre V, Bernard JF, David Xu ZQ, Hokfelt T. Chemical neuroanatomy of the dorsal raphe nucleus and adjacent structures of the mouse brain. J Comp Neurol. 2010;518:3464–3494. doi: 10.1002/cne.22407. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Ruhrberg C, Livet J, Henderson CE, Krumlauf R. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development. 2003;130:5663–5679. doi: 10.1242/dev.00802. [DOI] [PubMed] [Google Scholar]
- Geisler S, Derst C, Veh RW, Zahm DS. Glutamatergic afferents of the ventral tegmental area in the rat. J Neurosci. 2007;27:5730–5743. doi: 10.1523/JNEUROSCI.0012-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gray PA. Transcription factors and the genetic organization of brain stem respiratory neurons. J Appl Physiol. 2008;104:1513–1521. doi: 10.1152/japplphysiol.01383.2007. [DOI] [PubMed] [Google Scholar]
- Grossmann KS, Giraudin A, Britz O, Zhang J, Goulding M. Genetic dissection of rhythmic motor networks in mice. Prog Brain Res. 2010;187:19–37. doi: 10.1016/B978-0-444-53613-6.00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Kuemerle B. The compartmentalization of the cerebellum. Annu Rev Neurosci. 1997;20:61–90. doi: 10.1146/annurev.neuro.20.1.61. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Rhodes SJ. LIM-homeodomain genes in mammalian development and human disease. Mol Biol Rep. 2005;32:67–77. doi: 10.1007/s11033-004-7657-z. [DOI] [PubMed] [Google Scholar]
- Irving C, Mason I. Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development. 2000;127:177–186. doi: 10.1242/dev.127.1.177. [DOI] [PubMed] [Google Scholar]
- Jacob J, Storm R, Castro DS, Milton C, Pla P, Guillemot F, Birchmeier C, Briscoe J. Insm1 (IA-1) is an essential component of the regulatory network that specifies monoaminergic neuronal phenotypes in the vertebrate hindbrain. Development. 2009;136:2477–2485. doi: 10.1242/dev.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, Dymecki SM. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak R, Bowman EM, Latimer MP, Rostron CL, Winn P. Excitotoxic lesions of the pedunculopontine tegmental nucleus in rats impair performance on a test of sustained attention. Exp Brain Res. 2005;162:257–264. doi: 10.1007/s00221-004-2143-3. [DOI] [PubMed] [Google Scholar]
- Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron. 2004;42:375–386. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Lebel M, Gauthier Y, Moreau A, Drouin J. Pitx3 activates mouse tyrosine hydroxylase promoter via a high-affinity binding site. J Neurochem. 2001;77:558–567. doi: 10.1046/j.1471-4159.2001.00257.x. [DOI] [PubMed] [Google Scholar]
- Lebel M, Mo R, Shimamura K, Hui CC. Gli2 and Gli3 play distinct roles in the dorsoventral patterning of the mouse hindbrain. Dev Biol. 2007;302:345–355. doi: 10.1016/j.ydbio.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Lin JC, Cai L, Cepko CL. The external granule layer of the developing chick cerebellum generates granule cells and cells of the isthmus and rostral hindbrain. J Neurosci. 2001;21:159–168. doi: 10.1523/JNEUROSCI.21-01-00159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liu Z, Chen T, Li H, Qiang B, Yuan J, Peng X, Qiu M. Selective expression of Bhlhb5 in subsets of early-born interneurons and late-born association neurons in the spinal cord. Dev Dyn. 2007;236:829–835. doi: 10.1002/dvdy.21061. [DOI] [PubMed] [Google Scholar]
- Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Puelles L. Morphological fate of rhombomeres in quail/chick chimeras: a segmental analysis of hindbrain nuclei. Eur J Neurosci. 1995;7:1714–1738. doi: 10.1111/j.1460-9568.1995.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Martin DM, Skidmore JM, Fox SE, Gage PJ, Camper SA. Pitx2 distinguishes subtypes of terminally differentiated neurons in the developing mouse neuroepithelium. Dev Biol. 2002;252:84–99. doi: 10.1006/dbio.2002.0835. [DOI] [PubMed] [Google Scholar]
- Martin DM, Skidmore JM, Philips ST, Vieira C, Gage PJ, Condie BG, Raphael Y, Martinez S, Camper SA. PITX2 is required for normal development of neurons in the mouse subthalamic nucleus and midbrain. Dev Biol. 2004;267:93–108. doi: 10.1016/j.ydbio.2003.10.035. [DOI] [PubMed] [Google Scholar]
- Martin JH. Neuroanatomy: text and atlas. McGraw-Hill Medical; 2003. [Google Scholar]
- Matsunaga E, Katahira T, Nakamura H. Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development. 2002;129:5269–5277. doi: 10.1242/dev.129.22.5269. [DOI] [PubMed] [Google Scholar]
- Mena-Segovia J, Micklem BR, Nair-Roberts RG, Ungless MA, Bolam JP. GABAergic neuron distribution in the pedunculopontine nucleus defines functional subterritories. J Comp Neurol. 2009;515:397–408. doi: 10.1002/cne.22065. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Lindgren AG, Chizhikov VV, Johnson RL, Millen KJ. Overlapping function of Lmx1a and Lmx1b in anterior hindbrain roof plate formation and cerebellar growth. J Neurosci. 2009;29:11377–11384. doi: 10.1523/JNEUROSCI.0969-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukawa K, McGeer PL, Tago H, Peng JH, McGeer EG, Kimura H. The cholinergic system of the human hindbrain studied by choline acetyltransferase immunohistochemistry and acetylcholinesterase histochemistry. Brain Res. 1986;379:39–55. doi: 10.1016/0006-8993(86)90253-2. [DOI] [PubMed] [Google Scholar]
- Morales D, Hatten ME. Molecular markers of neuronal progenitors in the embryonic cerebellar anlage. J Neurosci. 2006;26:12226–12236. doi: 10.1523/JNEUROSCI.3493-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucchielli ML, Martinez S, Pattyn A, Goridis C, Brunet JF. Otlx2, an Otx-related homeobox gene expressed in the pituitary gland and in a restricted pattern in the forebrain. Mol Cell Neurosci. 1996;8:258–271. doi: 10.1006/mcne.1996.0062. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Sander M, Ericson J. Complementary roles for Nkx6 and Nkx2 class proteins in the establishment of motoneuron identity in the hindbrain. Development. 2003;130:4149–4159. doi: 10.1242/dev.00641. [DOI] [PubMed] [Google Scholar]
- Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development. 2007;134:357–366. doi: 10.1242/dev.02717. [DOI] [PubMed] [Google Scholar]
- Prakash N, Puelles E, Freude K, Trumbach D, Omodei D, Di Salvio M, Sussel L, Ericson J, Sander M, Simeone A, Wurst W. Nkx6-1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development. 2009;136:2545–2555. doi: 10.1242/dev.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, McMahon AP. Pax-2 expression in the murine neural plate precedes and encompasses the expression domains of Wnt-1 and En-1. Mech Dev. 1995;52:3–8. doi: 10.1016/0925-4773(95)00380-j. [DOI] [PubMed] [Google Scholar]
- Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell TM, Rubenstein JL. Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes and Development. 2000;14:2134–2139. doi: 10.1101/gad.820400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB. Arousal, emotion, and behavioral homeostasis. In: Kandel ER, Schwartz JH, Jessell TM, editors. Principles of Neural Science. 4th edition. New York: McGraw-Hill; 2000. pp. 873–909. [Google Scholar]
- Saponjic J, Cvorovic J, Radulovacki M, Carley DW. Serotonin and noradrenaline modulate respiratory pattern disturbances evoked by glutamate injection into the pedunculopontine tegmentum of anesthetized rats. Sleep. 2005;28:560–570. doi: 10.1093/sleep/28.5.560. [DOI] [PubMed] [Google Scholar]
- Schambra U, Lauder J, Silver J. Atlas of the Prenatal Mouse Brain. London: Academic Press; 1992. [DOI] [PubMed] [Google Scholar]
- Sclafani A, Skidmore J, Ramaprakash H, Trumpp A, Gage P, Martin D. Nestin-Cre mediated deletion of Pitx2 in the mouse. Genesis. 2006;44:336–344. doi: 10.1002/dvg.20220. [DOI] [PubMed] [Google Scholar]
- Sgaier SK, Lao Z, Villanueva MP, Berenshteyn F, Stephen D, Turnbull RK, Joyner AL. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134:2325–2335. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore JM, Cramer JD, Martin JF, Martin DM. Cre fate mapping reveals lineage specific defects in neuronal migration with loss of Pitx2 function in the developing mouse hypothalamus and subthalamic nucleus. Mol Cell Neurosci. 2008;37:696–707. doi: 10.1016/j.mcn.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Teissier A, Griveau A, Vigier L, Piolot T, Borello U, Pierani A. A novel transient glutamatergic population migrating from the pallial-subpallial boundary contributes to neocortical development. J Neurosci. 2010;30:10563–10574. doi: 10.1523/JNEUROSCI.0776-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torterolo P, Yamuy J, Sampogna S, Morales FR, Chase MH. GABAergic neurons of the cat dorsal raphe nucleus express c-fos during carbachol-induced active sleep. Brain Res. 2000;884:68–76. doi: 10.1016/s0006-8993(00)02891-2. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tsang EW, Hamani C, Moro E, Mazzella F, Poon YY, Lozano AM, Chen R. Involvement of the human pedunculopontine nucleus region in voluntary movements. Neurology. 2010;75:950–959. doi: 10.1212/WNL.0b013e3181f25b35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite MR, Skidmore JM, Billi AC, Martin JF, Martin DM. GABAergic and glutamatergic identities of developing midbrain Pitx2 neurons. Dev Dyn. 2011;240:333–346. doi: 10.1002/dvdy.22532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Morales M. Pedunculopontine and laterodorsal tegmental nuclei contain distinct populations of cholinergic, glutamatergic and GABAergic neurons in the rat. Eur J Neurosci. 2009;29:340–358. doi: 10.1111/j.1460-9568.2008.06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Reviews Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- Wingate RJ, Hatten ME. The role of the rhombic lip in avian cerebellum development. Development. 1999;126:4395–4404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- Zagoraiou L, Akay T, Martin JF, Brownstone RM, Jessell TM, Miles GB. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64:645–662. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas M, Blaess S, Joyner AL. Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr Top Dev Biol. 2005;69:101–138. doi: 10.1016/S0070-2153(05)69005-9. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci U S A. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou HL, Su CJ, Shi M, Zhao GY, Li ZY, Guo C, Ding YQ. Expression of the LIM-homeodomain gene Lmx1a in the postnatal mouse central nervous system. Brain Res Bull. 2009;78:306–312. doi: 10.1016/j.brainresbull.2008.12.001. [DOI] [PubMed] [Google Scholar]