Abstract
Previously we reported that lipopolysaccharide (LPS) treatment of murine mammary carcinomas resulted in decreased growth of the tumors. Here we show the decreased growth following LPS treatment was mediated through effects downstream of TLR4 and Myd88. Perhaps more notably, simply reducing TLR4 or Myd88 levels was sufficient to slow tumor growth rates. Moreover, reduced levels of Myd88 correlated with a significant reduction in lung metastasis as well as decreased CCL2 and CCL5 expression. To determine whether inhibiting Myd88 function could also alter tumor growth and chemokine expression we used a Myd88 homodimerization inhibitory peptide. Indeed, inhibiting Myd88 function in four different murine mammary carcinomas as well as the human breast cancer cell line MDA-MB-231 led to decreased growth as well as CCL2 and CCL5 expression. These data imply that Myd88 is important for growth and metastasis of breast cancer, and expression of at least two proinflammatory chemokines.
Keywords: TLR, Myd88, breast cancer, metastasis, 4T1, MDA-MB-231
1. Introduction
With the resurgence in exploration of innate immunity coinciding with the cloning of Toll-like receptor 4 (TLR4) and many other pattern recognition receptors (PRRs), and the identification of pathogen-associated molecular patterns (PAMPs), there has been renewed interest in the relationship between pathogenic products and cancer. Although many of these studies have focused on the effects PAMPs have on white blood cells [1–3], it is now widely understood that tumors also express PRR and are capable of directly responding to PAMPs [4].
TLR are a family of evolutionarily conserved pattern recognition receptors that are expressed by a variety of cells and participate in interrelated signal transduction systems [5]. Deciphering how TLR agonists could be used to benefit patients with cancer has not been easy. In fact, studies have reported both beneficial and detrimental effects following treatment with TLR agonists. For instance, Hirabayashi et al., [6] reported that Poly(I:C), a TLR3 agonist, could slow the growth of a panel of tumors including the commonly used breast cancer cell lines MCF7, T47D, MDA-MB-468, MDA-MB-453 and SKBR3, whereas a panel of normal cell lines were unaffected, and Tormo et al., [7] reported that Poly(I:C) and CpG ODN in combination with an adenoviral vector encoding tyrosinase-related protein 2 (TRP2) could delay growth of the B16 melanoma. On the other hand, Huang et al., [8] reported that the MC26 murine colon carcinoma responded to LPS in a manner that inhibited anti-tumor immunity. Moreover, in several cases the findings have been harder to decipher. For instance, in a TLR2 dependent mechanism Listeria monocytogenes promoted H22 tumor progression when injected intratumorally, whereas intravenous injection had an inhibitory effect on tumor growth [9]. Also, flagellin, a TLR5 agonist, could delay growth of the mammary tumor D2F2 when given multiple times following tumor delivery, yet when given at the time of tumor delivery there was enhanced tumor growth [10]. Similarly, we found that 4T1 tumors would grow faster or slower depending upon the conditions of LPS treatment [11].
Here, rather than exploring new ways to utilize TLR agonists to modulate tumor growth, we wanted to investigate whether decreased growth of the murine mammary carcinoma 4T1 following treatment with LPS was dependent upon TLR4 and the adaptor molecule Myd88 [12]. For this reason we generated clones with reduced TLR4 or Myd88 expression using RNA interference and confirmed that the decreased growth of 4T1 following LPS treatment was dependent upon TLR4 and Myd88. Surprisingly, we also found that decreasing Myd88 expression was sufficient to slow tumor growth and metastasis, and resulted in decreased expression of the proinflammatory chemokines CCL2 and CCL5. We obtained similar data using a Myd88 homodimerization inhibitory peptide which inhibits Myd88 function while not altering its expression [13]. Treatment of the murine mammary carcinomas 4T1, 168, EMT6, and SM1 as well as the human breast cancer cell line MDA-MB-231 with the Myd88 inhibitor led to decreased growth of the tumor cells as well as a decrease in tumor-derived CCL2 and CCL5 expression. These data suggest that signaling through Myd88 is important for tumor growth and proinflammatory chemokine expression, and may provide a novel way to target breast cancer. Results from this project also have implications for patients with chronic inflammatory diseases and may lead to a better understanding of how DAMPs and PAMPs impact cell cycle progression and inflammation.
2. Materials and methods
2.1 Cells and mice
4T1, EMT6, 168, and SM1 murine mammary carcinomas and the human breast cancer cell line MDA-MB-231 were maintained in complete RPMI (cRPMI) (RPMI 1640, Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (Lonza), glutamine (2mM, Lonza), penicillin (100U/mL, Lonza), streptomycin (100ug/mL, Lonza), nonessential amino acids (Sigma, St. Lois, MO), 2-mercaptoethanol (5×10−5 M, Sigma), and sodium pyruvate (1mM, Lonza). Cells transfected with shRNA expression vectors were maintained in cRPMI containing 800ug/ml G418 (Gibco). Balb/c mice were bred on site and were housed in a thoren caging system (Thoren Caging Systems Inc., Hazelton, PA). Food and water were provided ad libitum. All mice were used in accordance with an Institutional Animal Care and Use Committee approved protocol that followed the guidelines for ethical conduct in care and use of animals.
2.2 siRNA screening and shRNA vector construction
For siRNA screening 4T1 were cultured in cRPMI (without antibiotics) in 24-well culture dishes at 1×104 cells/well. After 24 hours, the media was replaced. For siRNA delivery 0, 3 or 6ul of siRNA (20uM stock) was mixed with Optimem (Invitrogen, Carlsbad, CA) for a final volume of 50ul in a sterile microcentrifuge tube and incubated for 5 min. at room temperature. In a separate microcentrifuge tube 12ul oligofectamine (Invitrogen) was mixed with 3ul Optimem, and incubated at room temperature for 5 min. The contents of the tubes were mixed, incubated at room temperature for 20 min., and then added to the cells which were harvested 24 hours later to assess TLR4 and Myd88 expression using quantitative reverse transcriptase polymerase chain reaction (QRT-PCR). The siRNA were synthesized by Integrated DNA Technologies (Coralville, IA).
| lacZ | |
| siRNA | 5′-CCAACGUAACCUAUCCCAUUACGGT |
| GUGGUUGCAUUGGAUAGGGUAAUGCCA-5′ | |
| TLR4 | |
| siRNA1 | 5′-GGACUAACAAGUUUAGAGAAUCUGG |
| GACCUGAUUGUUCAAAUCUCUUAGACC-5′ | |
| siRNA2 | 5′-GAACAAACAGCCUGAGACACUUAGA |
| CCCUUGUUUGUCGGACUCUGUGAAUCU-5′ | |
| siRNA3 | 5′-GAACAAAUGACAUGUGCAACACCTG |
| AACUUGUUUACUGUACACGUUGUGGAC-5′ | |
| Myd88 | |
| siRNA1 | 5′-CCAACAGAAGCGACUGAUUCCUATT |
| CAGGUUGUCUUCGCUGACUAAGGAUAA-5′ | |
| siRNA2 | 5′-CCAGGAGUCCGAGAAGCCUUUACAG |
| UUGGUCCUCAGGCUCUUCGGAAAUGUC-5′ | |
| siRNA3 | 5′-GCCAGAAAUACUUAGGUAAGCAGCA |
| GACGGUCUUUAUGAAUCCAUUCGUCGU-5′ | |
The pBLOCK-it DEST vector system (Invitrogen) was used to create the shRNA expression vectors. DNA encoding TLR4, Myd88, and lacZ shRNA were synthesized by Integrated DNA Technologies and ligated into the pENTR/U6 vector which places a U6 promoter upstream and Pol III terminator downstream from the sequence. A ligation reaction was used to transfer the U6 RNAi cassette encoding the shRNA into the pBLOCK-it DEST vector according to manufacturer’s instructions. The pBLOCK-it DEST vector encodes neomycin resistance for selection of stable cell lines. Following construction of the vectors they were sequenced in both directions (Molecular Genetics Core Facility, Hershey Medical Center, Hershey, PA).
DNA encoding the top strand of lacZ specific shRNA
5′-CACCGCTACACAAATCAGCGATTTCGAAAAATCGCTGATTTGTGTAG-3′
DNA encoding the top strand of TLR4 specific shRNA
5′CACCGAACAAACAGCCTGAGACACTTAGACGAATCTAAGTGTCTCAGGCTGTTTGTTC-3′
DNA encoding the top strand of Myd88 specific shRNA
5′-ACCGCCAACAGAAGCGACTGATTCCTATTCGAAAATAGGAATCAGTCGCTTCTGTTGG-3′
2.3 Transfection and cell cloning
Prior to transfection, plasmid DNA encoding shRNA specific for TLR4, Myd88, or lacZ were purified using the Qiaprep DNA extraction kit (Qiagen, Valencia, CA). Next, 1×104 4T1 were plated/well in a 24-well plate in cRPMI without antibiotics. Twenty-four hours later 800ng vector DNA was mixed with Optimem for a final volume of 50ul in a sterile microcentrifuge tube and incubated for 5 min. at room temperature. In a separate microcentrifuge tube 2ul lipofectamine (Invitrogen) was mixed with 48ul Optimem, and incubated at room temperature for 5 min. The contents of the tubes were mixed, incubated at room temperature for 20 min., and then added to the cells. The following day cRPMI (without antibiotics) was replaced, and after another 24 hours the cells were trypsinized and moved to a T75cm2 tissue culture flask with cRPMI containing 800ug/ml G418. Following 4 days of selection in 800ug/ml G418, the cells were cloned at 0.5 cells/well in 96-well round bottom tissue culture plates.
2.4 Growth kinetics
To determine in vitro growth rates, tumor cells were plated at 1×104 cells/T25cm2 flask for 24 and 48 hours, or T75cm2 flask for 72 and 96 hours. At the specified time points the cells were trypsinized, washed, and counted. Each cell count was performed in duplicate. For studies with the Myd88 inhibitor 1×104 tumor cells were cultured in 24-well tissue culture plates with the Myd88 specific inhibitor (Imgenex, San Diego, CA) or a control peptide lacking the Myd88 binding domain (Imgenex). Prior to examining in vivo kinetics the cells were plated at 1×106 cells/T75cm2 flask in 10 ml cRPMI containing 100ng/ml LPS or an equivalent volume of HBSS as a control. Twenty-four hours later the cells were harvested, counted, washed three times in HBSS and adjusted to 5×105 cells/ml. Next, 5×104 cells in 100ul HBSS were delivered subcutaneously to 6–8 week old female BALB/c mice in the left hind flank. Starting at day 7, and every 2–3 days thereafter, the tumors were measured using vernier calipers and the tumor volume calculated (LxW2/2).
2.5 Analysis of metastasis
Following sacrifice, lungs and livers were harvested from mice. Visible metastatic nodules were recorded from the lungs which were then mechanically minced and digested in 5 mL of tissue digest solution containing 1 g/L collagenase type IV (Worthington Biomed, Lakewood, NJ) and 100 mg/L elastase (Worthington Biomed). The livers were minced and digested in an enyzme cocktail containing 1 mg/ml collagenase type I (Worthington Biomed), 1 mg/ml hyaluronidase (Sigma), and 0.01% bovine serum albumin. After one hour of mixing on a rotator (Appropriate Technical Resources, Laurel, MD) at room temperature, the cells were centrifuged at 450×g and resuspended in 10 mL of cRPMI. The cells were incubated in 10 cm tissue culture dishes at 37°C and 5% CO2 for fourteen days with cRPMI containing 60 μM thioguanine (Sigma). Finally, the cells were fixed with methanol (VWR International, West Chester, PA), stained with a 0.03% solution of methylene blue (Sigma), and the colonies representing metastasis were counted.
2.6 ELISA
To quantitate secretion of proinflammatory mediators chemokine specific ELISAs were used. For this purpose, 1 ml of cells at 1×106 cells/ml were added to each well in 24-well tissue culture plates, and after 24 hours, supernatants were harvested, centrifuged at 450×g to remove particulate materials, and stored at −20°C. For analysis, samples were assayed for CCL2 and CCL5 using Quantikine Sandwich ELISAs (R&D Systems).
2.7 QRT-PCR
Gene expression was analyzed by QRT-PCR. First, mRNA was isolated from 1×106 cells using a mRNA isolation kit (Invitrogen). All of the mRNA was precipitated, resuspended in 25ul DEPC treated H2O, and used for cDNA synthesis. Complementary DNA was generated using random hexamer primers (0.5ug, Promega, Madison, WI), dithiothreitol (2mM, Promega), dNTP (0.2mM each, Promega) and 200 units of M-MLV reverse transcriptase (Promega) in a 1 hour reaction at 42°C. An aliquot (0.5ul) of cDNA was amplified in a reaction with 1 x iQ SYBR Green Supermix (Bio-Rad Laboratories, Richmond, CA), and 200nM gene specific primers. Each reaction was run in duplicate or triplicate. The reaction conditions consisted of 40 cycles of a two-step PCR reaction with 94°C for 10 sec., and 68°C for 30 sec., on an iQ5 Real Time PCR Detection System (Bio-RAD). Gene specific primers for the mouse lines included gapdh left 5′-cttccgtgttcctacccccaatgt-3′, gapdh right 5′-gcctgcttcaccaccttcttgatgt-3′, TLR4 left 5′-agtgccccgctttcacctctg-3′, TLR4 right 5′-caataaccttccggctcttgtgga-3′, Myd88 left 5′-cctgacccc actcgcagtttgt-3′, Myd88 right 5′-tgcgcgacttcagctccttca-3′, CCL2 left 5′-tcatgcttctgggcctgctgt-3′, CCL2 right 5′-ctcattgggatcatcttgctggtg-3′, CCL5 left 5′-ccactccctgctgctttgccta-3′, and CCL5 right 5′-tggcacacacttggcggttc-3′. Gene specific primers for the human line included gapdh left 5′-actggcatggccttccgtgt-3′, gapdh right 5′-cgcctgcttcaccaccttcttg-3′, CCL2 left 5′-attccccaagggctcgctca-3′, CCL2 right 5′-tgggacacttgctgctggtga-3′, CCL5 left 5′-aaccgccaagtgtgtgccaac-3′, and CCL5 right 5′-tcaggttcaaggactctccatcctagc-3′. The primers were synthesized by Integrated DNA Technologies and analyzed for specificity with the NCBI Blast Program. Standard curves were used to examine efficiency and reproducibility of each reaction, and melt curves were used to validate amplification of single products. The housekeeping gene gapdh was used to establish normalized expression (ΔΔCT).
2.8 Western blot
To analyze protein expression 5×106 cells were washed 3 times with ice-cold PBS, resuspended in 150uL of buffer A (10mM Hepes (Sigma), 10mM KCl (Sigma), 0.1mM EDTA (Sigma), 0.1mM EGTA (Sigma)), supplemented with the protease inhibitors aprotinin, leupeptin, chymostatin, and pefabloc (Roche Molecular Biochemicals, Indianapolis, IN) and placed on ice. Following a 15 min. incubation, 10uL of 10% Nonidet P-40 (Sigma) was added. The samples were vortexed for 10 sec., and centrifuged 15,000×g at 4°C for 1 min. NuPAGE LDS sample buffer (Invitrogen) and dithiothreitol (1x, New England Biolabs, Ipswich, MA) were added and the samples were stored at −20°C. SDS PAGE gels (12.5%, Invitrogen) were loaded with 15uL of proteins, electrophoresed, and transferred to PVDF membranes (Invitrogen). The membranes were blocked at 4°C in PBS with 5% powdered milk and 0.05% Tween 20 (Sigma) overnight. Primary antibodies (10ug) specific for actin, TLR4, or Myd88 (Santa Cruz Biotechnology, Santa Cruz, CA) were added, and the blots were incubated at room temperature for 3 hours. After washing 2 times with blocking buffer, a horse-radish-peroxidase-conjugated secondary antibody (Santa Cruz) was added and the blots were incubated for 1 hour at room temperature. Following 4 washes, proteins were visualized by enhanced chemiluminesence on an Alpha Innotech Gel Documentation System (Alpha Innotech Corp., San Leandro, CA).
3. Results
3.1 Inhibition of TLR4 and Myd88
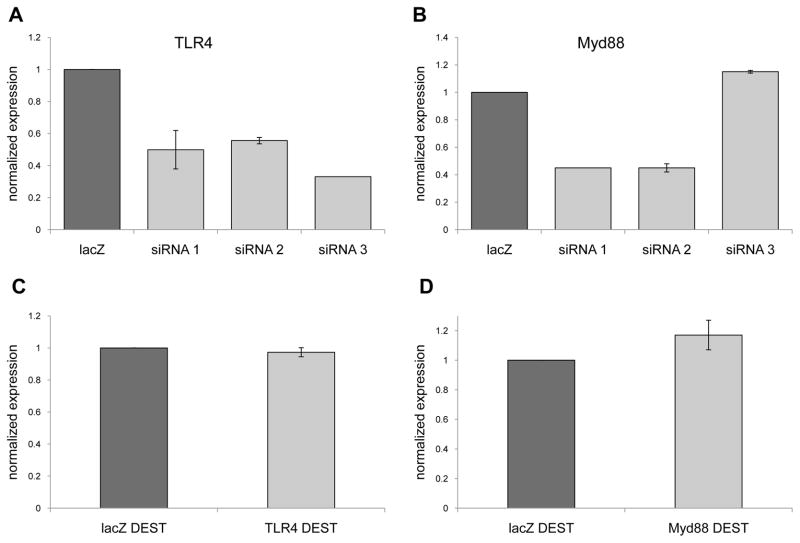
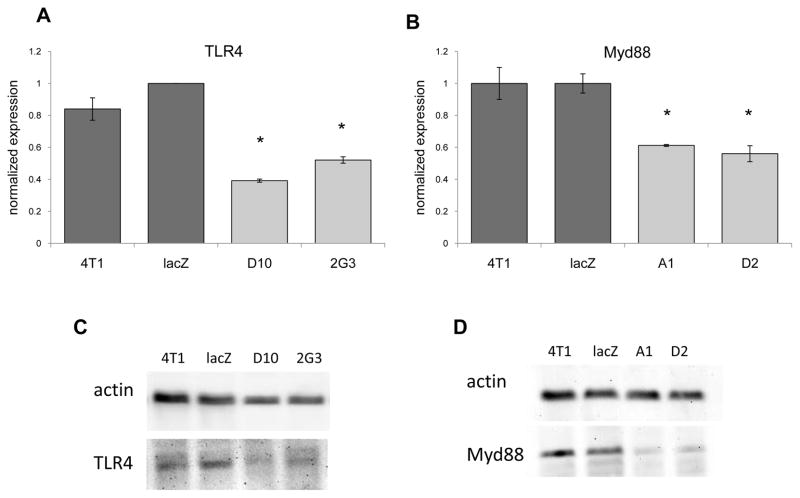
Initially three different siRNA specific for TLR4 or Myd88 were screened to identify siRNA sequences that could inhibit gene expression (Figs. 1A, 1B). Because we wanted to inhibit expression in a stable manner we used the siRNA sequences that reduced TLR4 or Myd88 expression to create shRNA expression vectors. However, following transfection and G418 selection of bulk transfected 4T1 cells we found no inhibition of TLR4 or Myd88 gene expression relative to cells transfected with a vector expressing lacZ specific shRNA (Figs. 1C, 1D). As a consequence of the lack of inhibition of TLR4 or Myd88 expression with bulk transfected 4T1 we turned to cloning the transfected cells, and identified four clones that exhibited a reduction in TLR4 or Myd88 expression that was stable for over four weeks in culture. With respect to TLR4, clone D10 exhibited a 61% reduction in mRNA expression, while clone 2G3 exhibited a 48% reduction in mRNA expression relative to the lacZ control (Fig. 2A). With respect to Myd88, clone A1 exhibited a 39% reduction in mRNA expression, while clone D2 exhibited a 44% reduction in mRNA expression relative to the lacZ control (Fig. 2B). Western blots were used to determine whether the decrease in mRNA expression correlated with reduced protein levels. Indeed, clones D10 and 2G3 exhibited reduced TLR4 protein levels, and clones A1 and D2 exhibited reduced Myd88 protein levels relative to the lacZ control (Figs. 2C, 2D). Collectively, these data revealed the generation of clones with reduced levels of TLR4 or Myd88 expression.
Fig. 1.
Targeting TLR4 and Myd88 using RNA interference. Messenger RNA was isolated from 4T1 cells 24 hours after treatment with different siRNA specific for TLR4 (A) or Myd88 (B), and screened by QRT-PCR for decreased gene expression using gapdh as the reference gene, and lacZ siRNA treated cells as the control. 4T1 were transfected with a shRNA expression vector specific for TLR4 (C) or Myd88 (D) and screened two weeks after G418 selection by QRT-PCR for decreased gene expression using gapdh as the reference gene, and cells transfected with a lacZ specific shRNA expression vector as the control. All data represent the average and standard error of 3 separate experiments.
Fig. 2.
4T1 clones with reduced levels of TLR4 and Myd88. 4T1 was cloned following transfection with shRNA expression vectors specific for TLR4 (A) or Myd88 (B). The clones were screened for TLR4 or Myd88 expression by QRT-PCR using gapdh as the reference gene, and cells transfected with a lacZ specific shRNA expression vector as the control. The data represent the average and standard error of 3 separate experiments. Where indicated (*) p < 0.05 using Student’s t-Test relative to the lacZ control. (C, D) Western blots were used to determine whether protein expression correlated with RNA expression. One of 3 separate experiments is shown.
3.2 Reduced responsiveness to a TLR4 agonist
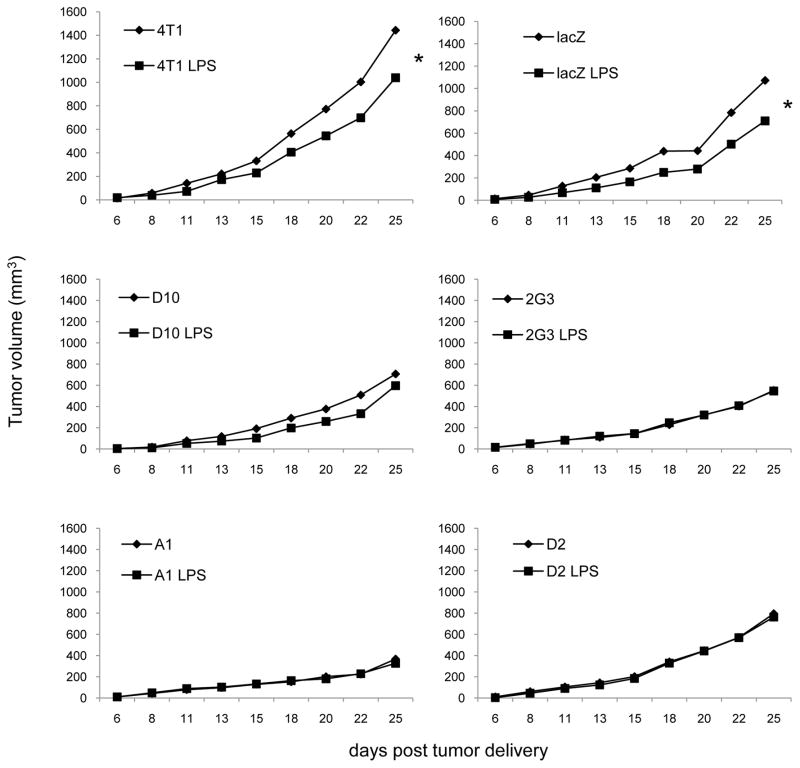
Previously we reported that 4T1 treated with LPS grow slower in vivo, but not in vitro [12]. We used the clones to determine whether the decreased growth rate of LPS treated 4T1 was dependent upon TLR4 or Myd88. For this reason we treated 4T1, lacZ controls, and the clones D10, 2G3, A1, and D2 with LPS, delivered the tumors to mice, and monitored tumor growth (Fig. 3). Notably, the decreased growth rate evident in 4T1, and the lacZ control following LPS treatment was ablated by reducing TLR4 or Myd88 expression (Fig. 3). After 25 days of tumor growth only 4T1 and lacZ controls treated with LPS showed a small, yet significant reduction in growth, while none of the clones were similarly affected by LPS treatment. These data indicate that LPS mediates effects on 4T1 in a TLR4 and Myd88 dependent manner, and that the decreased growth rate of LPS treated 4T1 requires Myd88 signaling.
Fig. 3.
Decreased growth of LPS treated tumor cells is dependent upon TLR4 and Myd88. Cells were treated with 100ng/ml LPS or an equivalent volume of HBSS as a control for 24 hours then delivered to mice subcutaneously in the left hind flank. The data represent the average of 3–4 separate experiments with 5–6 mice/experiment for each tumor. Where indicated (*) p < 0.05 using Student’s t-Test relative to non-treated cells.
3.3 Inhibiting Myd88 expression decreases tumor growth and chemokine expression
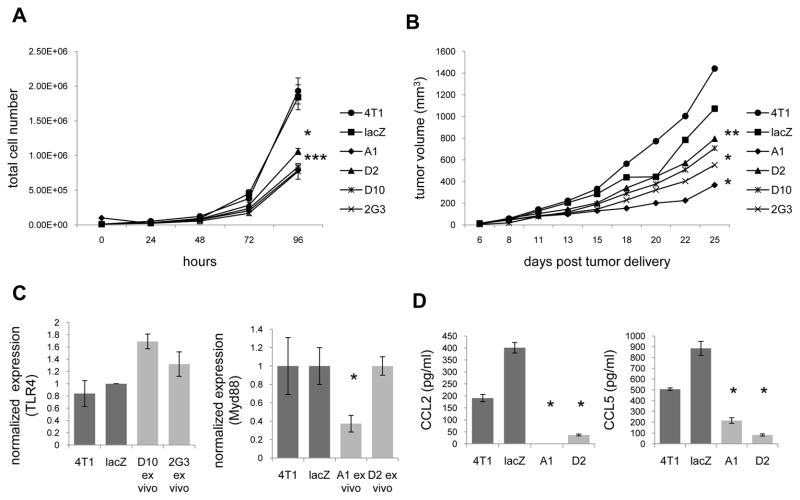
While monitoring responsiveness to LPS it became apparent that reducing TLR4 or Myd88 expression was sufficient to influence tumor growth rates. With regard to in vitro growth rates, all four clones exhibited significantly slower growth than either 4T1 or lacZ controls (Fig. 4A). Following 96 hours in culture there were 1.9e6 4T1 cells and 1.8e6 lacZ control cells, while clones with reduced levels of TLR4 reached 7.7e5 cells (D10) and 8.3e5 cells (2G3), and clones with reduced levels of Myd88 reached 7.8e5 cells (A1) and 1.0e6 cells (D2, Fig. 4A). A similar pattern was evident with the in vivo growth rates (Fig. 4B). Following 25 days of growth 4T1 tumors averaged 1442 mm3 and lacZ controls averaged 1072 mm3, while clones with reduced levels of TLR4 averaged 707 mm3 (D10) and 552 mm3 (2G3), and clones with reduced levels of Myd88 averaged 367 mm3 (A1) and 795 mm3 (D2, Fig. 4B).
Fig. 4.
Inhibiting Myd88 expression impacts tumor growth and chemokine expression. (A) In vitro growth was evaluated by culturing 1×104 cells, and counting at the specified time points. (B) In vivo growth was evaluated by delivering 5×104 cells/mouse and measuring tumors at the specified time points. The data represent the average of 3–4 experiments with 5–6 mice/experiment for each tumor. (C) After 25 days of growth the tumors were harvested, digested, and cultured in G418 for 2 weeks. TLR4 and Myd88 expression were then assessed by QRT-PCR using gapdh as the reference gene, and cells transfected with a lacZ specific shRNA expression vector as the control. (D) Tumors were cultured for 24 hours and the supernatants were then harvested and assessed for CCL2 and CCL5 levels by ELISA. All data represent at least 3 separate experiments. Where indicated (*) p < 0.05 using Student’s t-Test relative to the lacZ control.
Since we confirmed stability of the clones with regard to reduced levels of TLR4 and Myd88 expression in vitro (Fig. 2), we were interested in determining whether the clones maintained reduced levels of TLR4 and Myd88 expression following in vivo growth. For this purpose we analyzed TLR4 and Myd88 mRNA levels from 3 separate D10, 2G3, A1, and D2 tumors ex vivo by QRT-PCR. Even though the ex vivo tumors were cultured in, and resistant to G418, both of the TLR4 clones, and one Myd88 clone (D2) regained TLR4 and Myd88 expression (Fig. 4C). Interestingly, the clone with reduced levels of Myd88 that was stable in vitro and ex vivo (A1) exhibited the slowest growth rate. Following 25 days of growth 4T1 tumors averaged 1442 mm3, while A1 tumors averaged 367 mm3. These data suggest that D10, 2G3, and D2 may have grown faster in vivo because they regained expression of TLR4 or Myd88. Since the greatest effect was seen by inhibiting Myd88 expression we subsequently focused our efforts on Myd88.
In addition to evaluating growth rates, we noticed that tumor-derived CCL2 and CCL5 secretion were reduced by the A1 and D2 clones (Fig. 4D). 4T1, lacZ, A1, and D2 expressed 191, 401, 0, and 37 pg/ml of CCL2, and 507, 885, 216, and 82 pg/ml of CCL5 respectively. Although Myd88 levels rose in D2 in vivo, we have not looked at whether expression of these chemokines were stable after ex vivo culturing. Regardless, the data reveal that reduced expression of Myd88 correlates with reduced growth, CCL2, and CCL5 secretion.
3.4 Impact of Myd88 expression on metastasis
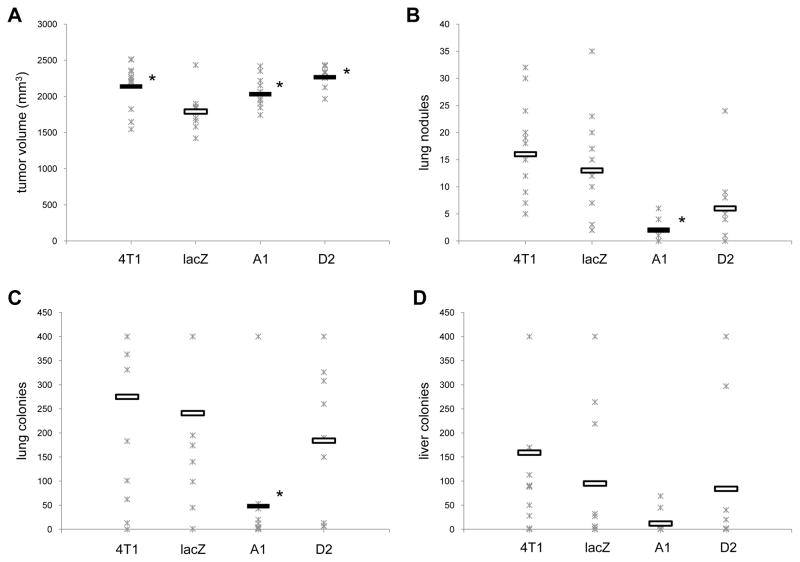
To determine whether expression of Myd88 influenced metastasis the lungs and livers were harvested from tumor-bearing mice. Because the tumors grew at different rates (Fig. 4B), and because A1 tumor-bearing mice appeared healthy even when the tumors reached 2000 mm3, a size at which the lacZ tumor-bearing mice did not often reach, we compared metastasis just among mice with tumors that ranged from 1500 mm3 to 2500 mm3. Using these parameters the average size of the 4T1, A1, and D2 tumors were slightly, yet significantly larger than the lacZ tumors (Fig. 5A). Despite having larger tumors, mice bearing A1 tumors had significantly fewer metastatic nodules visible on their lungs, and significantly fewer colonies grew from the lung digests compared to the lungs from lacZ tumor-bearing mice (Figs. 5B, 5C). Although D2 tumor-bearing mice exhibited fewer lung nodules and colonies than lacZ tumor-bearing mice, the results were not significant. Lung metastasis were similar among 4T1 and lacZ tumor-bearing mice. Despite fewer colonies from the digested livers of A1 tumor-bearing mice the results were not significantly different than the colonies from the digested livers of lacZ tumor-bearing mice (Fig. 5D). These data reveal that reduced expression of Myd88 correlates with reduced lung metastasis.
Fig. 5.
Inhibiting Myd88 expression impacts metastasis. Mice were sacrificed when their tumors reached 1500–2500mm3 and their lungs and livers were assessed for metastasis. Data from each mouse (Ж) and the average (ˉ) from 2–3 separate experiments using 5–6 mice/experiment are shown. Where indicated (*) p < 0.05 using Student’s t-Test relative to the lacZ control.
3.5 Inhibiting Myd88 function also decreases tumor growth and chemokine expression
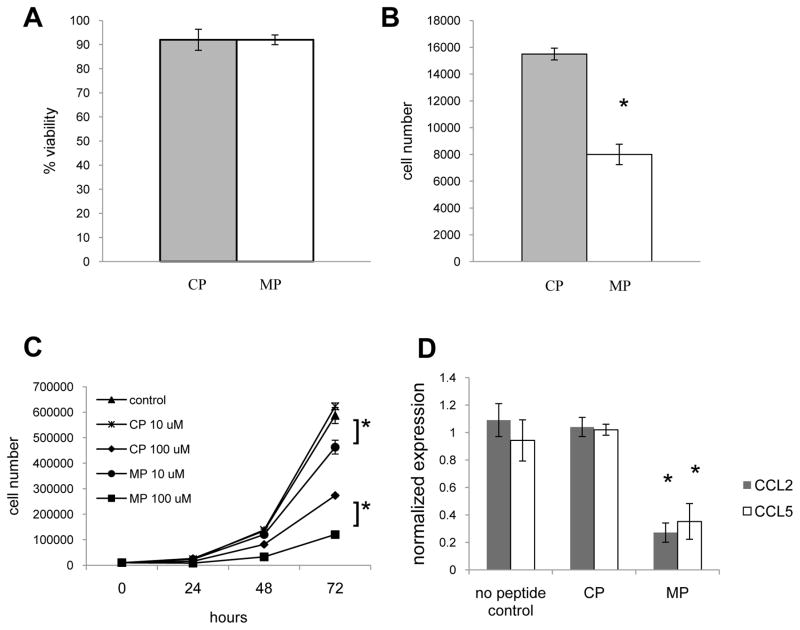
To determine whether inhibiting Myd88 function rather than expression would also inhibit tumor growth and CCL2 and CCL5 expression we used a Myd88 homodimerization inhibitory peptide [13]. Strikingly, viability was unaffected by neither the control peptide nor the Myd88 inhibitory peptide (Fig. 6A), while 24 hours of treatment with the Myd88 inhibitor prevented growth of 4T1, whereas cells treated with the control peptide proliferated (Fig. 6B). A dose titration and time course evaluation revealed that treatment with 100uM of control peptide, as well as 10uM and 100uM of Myd88 inhibitory peptide significantly influenced growth relative to untreated cells. More importantly, there were significantly fewer cells following treatment with 10uM of the Myd88 inhibitory peptide compared to treatment with 10uM of the control peptide, and significantly fewer cells following treatment with 100uM of the Myd88 inhibitory peptide compared to treatment with 100uM of the control peptide (Fig. 6C). Treatment with 1uM of the peptides had no effect on cell number (data not shown). Analysis of gene expression revealed that treatment with the Myd88 inhibitory peptide also significantly decreased CCL2 and CCL5 expression (Fig. 6D).
Fig. 6.
Inhibiting Myd88 function impacts tumor growth and chemokine expression. 4T1 were treated with 100uM control peptide (CP) or Myd88 inhibitory peptide (MP) for 24 hours and the viability (A) and cell number (B) were determined using trypan blue staining. (C) 4T1 were treated with HBSS as a control, the CP, or MP for 24–72 hours and then the cell number was determined. (D) After 72 hours of treatment with HBSS as a control, 100uM CP or MP, mRNA was isolated and QRT-PCR was used to determine CCL2 and CCL5 expression levels using gapdh as the reference gene and CP treated cells as the control. All data represent the average and standard deviation of 3 separate experiments. Where indicated (*) p < 0.05 using Student’s t-Test relative to the same dose of CP treated cells.
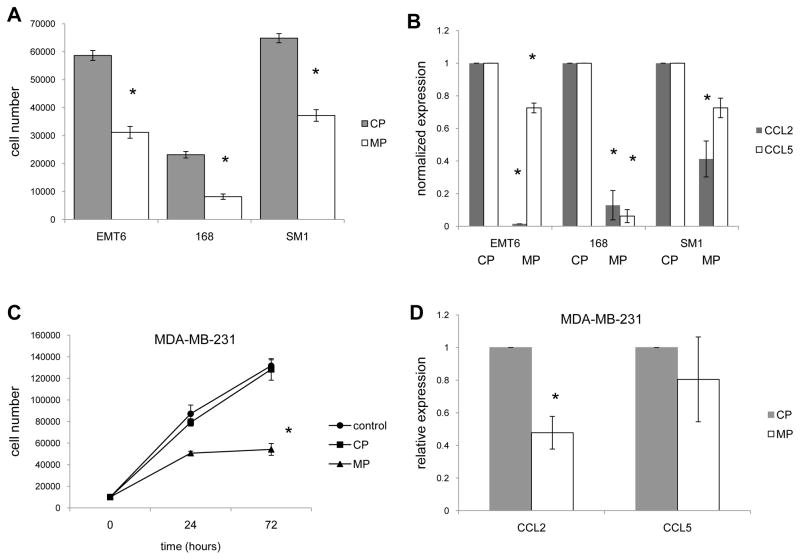
To determine whether these findings were relevant to additional murine mammary carcinomas we screened the EMT6, 168, and SM1 lines for growth and chemokine expression following treatment with the Myd88 inhibitory or control peptides. Similar to 4T1, the EMT6, 168, and SM1 tumors exhibited decreased growth as well as CCL2 and CCL5 expression following treatment with the Myd88 inhibitor (Figs. 7A, 7B). Also similar to 4T1, viability was unaffected by neither the control peptide nor the Myd88 inhibitor (data not shown). Finally, to determine whether the results had potential relevance to humans we examined the effect of the inhibitory peptide on the human breast cancer cell line MDA-MB-231. Similar to the murine mammary carcinomas, MDA-MB-231 exhibited decreased growth as well as CCL2 and CCL5 expression following treatment with the Myd88 inhibitor (Figs. 7C, 7D). Collectively, these data suggest that Myd88 is important for growth of breast cancer and expression of at least two proinflammatory chemokines.
Fig. 7.
Inhibiting Myd88 function impacts growth and chemokine expression by additional breast cancer models. (A) Three murine mammary carcinoma were treated with 100uM control peptide (CP) or Myd88 inhibitory peptide (MP) for 24 hours and then the cell number was determined using trypan blue staining. (B) After 72 hours of treatment with 100uM CP or MP, mRNA was isolated from the murine mammary carcinomas and QRT-PCR was used to determine CCL2 and CCL5 expression levels using gapdh as the reference gene and CP treated cells as the control. (C) The human breast cancer cell line MDA-MB-231 was treated with 100uM control peptide (CP) or Myd88 inhibitory peptide (MP) for 24–72 hours and then the cell number was determined using trypan blue staining. (D) After 72 hours of treatment with 100uM CP or MP, mRNA was isolated from MDA-MB-231 and QRT-PCR was used to determine CCL2 and CCL5 expression levels using gapdh as the reference gene and CP treated cells as the control. All data represent the average and standard deviation of 3 separate experiments. Where indicated (*) p < 0.05 using Student’s t-Test relative to CP treated cells.
4. Discussion
While many studies have explored how treatments with TLR agonists can lead to tumor progression or regression in different tumor models, we have been focusing our efforts on the implications of treating murine mammary carcinomas with LPS. Initially we reported that the 4T1, 168, and SM1 tumors treated with LPS grew slower in vivo, but not in vitro, and that this was due to the presence of CD8+ T cells [12]. Subsequently, we found that altering the dose, length, or frequency of LPS treatments could result in either enhanced or delayed tumor growth [11]. These data imply that utilizing TLR agonists for the treatment of cancer requires an enhanced understanding of the signal transduction systems so that a response could be better predicted. Consequently, to start deciphering the mechanism(s) through which 4T1 respond to LPS, we focused on the beginning of the signaling cascade. Our results show that 4T1 respond to LPS in a TLR4 and Myd88 dependent manner, and reveal another important role for Myd88 in several different breast cancer models; Myd88 is important for tumor cell growth as well as CCL2 and CCL5 expression.
The involvement of Myd88 in tumor-derived CCL2 and CCL5 expression is of particular interest since expression of these chemokines has been correlated with disease progression in patients with breast cancer [14–18]. As a result, targeting Myd88 may serve as a novel way to control expression of these tumor-derived proinflammatory chemokines. Although inhibiting TLR4 expression was sufficient to inhibit growth of the breast cancer cell line MDA-MB-231 [19], there is a paucity of information on the involvement of Myd88 in breast cancer growth. The contribution of Myd88 in growth of other types of tumors however has been established. Using a murine intestinal cancer model Rakoff-Nahoum and Medzhitov [20] reported that Myd88 played a role in tumor development in mice with mutations in the adenomatous polyposis coli (APC) gene. Similar findings were obtained using a murine hepatocellular carcinoma (HCC) model, as well as T and B cell malignancies [21–23]. While some these findings may be attributed to Myd88 dependent proinflammatory gene expression, there is growing evidence that Myd88 can play a direct role in cell cycle progression. For instance, using a Ras-dependent skin carcinogenesis model Myd88 was shown to contribute to Ras signaling, cell-cycle control, and cell transformation [24]. Given these findings it is not surprising that activating mutations of Myd88 could cause it to function as an oncogene [25]. Based upon these reports it is interesting to speculate that our inability to inhibit TLR4 or Myd88 expression using bulk transfected tumor cells may in part be attributed to the importance of these proteins for cell growth, and may also explain why 3 out of the 4 clones we generated regained TLR4 or Myd88 expression in vivo. In addition to its role in growth and chemokine expression, Myd88 may also contribute to the immunogenicity of the tumors as well. Following 25 days of tumor growth, 4T1 tumors averaged 1442 mm3, while the A1 tumors (the only tumor to retain reduced levels of Myd88 in vivo) averaged 367 mm3, and in 4/16 mice the A1 tumors regressed following a brief period of tumor growth (data not shown). Moreover, ¾ of these mice rejected a rechallenge with the parental 4T1 tumor supporting the necessity to investigate immunogenicity of tumors with altered Myd88 levels.
These studies do not however explain how signaling through Myd88 under some conditions may lead to tumor progression while other conditions can slow tumor growth. A recent study by Ahtiainen et al., [26] may shed light on the issue by revealing that cancer stem cells/initiating cells (CIC) and non-CIC cells have different capacities to signal through Myd88. Thus, the outcome and extent of Myd88 signaling should be taken in context of other critical proteins such as STAT3 or SOCS expression. Additionally, it should not be forgotten that Myd88 is important for IL-1β signaling which has a range of implications such as HIF-1α transcription which also contributes to tumorigenesis [27]. Undoubtedly, studying Myd88 in context of several other signaling cascades may help unravel the complexity and consequences of Myd88 signaling in cancer, and may shed light upon when Myd88 signaling leads to tumor progression or regression.
In addition to unraveling the role of Myd88 in tumor growth, identifying what is triggering Myd88 dependent signaling in the absence of activation mutations would also be helpful in developing ways to target this cascade. Since there are numerous reports that tumors constitutively express some damage associated molecular patterns (DAMPs) which have been shown to mediate signaling through TLR [28–30], it may be worthwhile to determine whether tumor-derived DAMPs mediate autocrine signaling in a Myd88 dependent manner. Towards this aim, we recently found that inhibiting several DAMPs expressed by the 4T1 line can recapitulate some of the findings of inhibiting Myd88 expression in 4T1 (data not shown). Nevertheless, regardless of the initiator of the signaling cascade, our data, in addition to what has been reported in several different tumor systems, supports the contention that signaling through Myd88 is important for breast cancer progression.
Highlights.
This is the first paper showing the importance of Myd88 for breast cancer
Targeting Myd88 can significantly slow growth and metastasis of breast cancer
CCL2 and CCL5 expression (which are correlated with progression of disease in patients with breast cancer) are Myd88 dependent
Acknowledgments
This project was supported by award number R15CA137858 from the National Cancer Institute (to R.A.K). The Department of Biology and the Excel Scholars Program at Lafayette College also supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lakomy D, Janikashvili N, Fraszczak J, Trad M, Audia S, Samson M, Ciudad M, Vinit J, Vergely C, Caillot D, Foucher P, Lagrost L, Chouaib S, Katsanis E, Larmonier N, Bonnotte B. Cytotoxic dendritic cells generated from cancer patients. J Immunol. 2011;187:2775–2782. doi: 10.4049/jimmunol.1004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogunovic D, Manches O, Godefroy E, Yewdall A, Gallois A, Salazar AM, Marie I, Levy DE, Bhardwaj N. TLR4 engagement during TLR3-induced proinflammatory signaling in dendritic cells promotes IL-10-mediated suppression of antitumor immunity. Cancer Res. 2011;71:5467–5476. doi: 10.1158/0008-5472.CAN-10-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Luo F, Cai Y, Liu N, Wang L, Xu D, Chu Y. TLR1/TLR2 agonists induces tumor regression by reciprocal modulation of effector and regulatory T cells. J Immunol. 2011;186:1963–1969. doi: 10.4049/jimmunol.1002320. [DOI] [PubMed] [Google Scholar]
- 4.So EY, Ouchi T. The application of Toll-like receptors for cancer therapy. Int J Biol Sci. 2010;6:675–681. doi: 10.7150/ijbs.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostuni R, Zanoni I, Granucci F. Deciphering the complexity of Toll-like receptor signaling. Cell Mol Life Sci. 2010;67:4109–4134. doi: 10.1007/s00018-010-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirabayashi K, Yano J, Inoue T, Yamaguchi T, Tanigawara K, Smyth GE, Ishiyama K, Ohgi T, Kimura K, Irimura T. Inhibition of cancer cell growth by polyinosinic-polycytidylic acid/cationic liposome complex: a new biological activity. Cancer Res. 1999;59:4325–4333. [PubMed] [Google Scholar]
- 7.Tormo DA, Ferrer A, Bosch P, Gaffal E, Basner-Tschakarjan E, Wenzel J, Tuting T. Therapeutic efficacy of antigen-specific vaccination and toll-like receptor stimulation against established transplanted and autochthonous melanoma in mice. Cancer Res. 2006;66:5427–5435. doi: 10.1158/0008-5472.CAN-06-0399. [DOI] [PubMed] [Google Scholar]
- 8.Huang B, Zhao J, Li H, He K, Chen Y, Chen SH, Mayer L, Unkeless JC, Xiong H. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005;65:5009–5014. doi: 10.1158/0008-5472.CAN-05-0784. [DOI] [PubMed] [Google Scholar]
- 9.Huang B, Zhao J, Shen S, Li H, He K, Shen G, Mayer L, Unkeless J, Li D, Yuan Y, Zhang GM, Xiong H, Feng ZH. Listeria monocytogenes promotes tumor growth via tumor cell toll-like receptor 2 signaling. Cancer Res. 2007;67:4346–4352. doi: 10.1158/0008-5472.CAN-06-4067. [DOI] [PubMed] [Google Scholar]
- 10.Sfondrini L, Rossini A, Besusso D, Merlo A, Tagliabue E, Menard S, Balsari A. Antitumor activity of the TLR-5 ligand flagellin in mouse models of cancer. J Immunol. 2007;176:6624–6630. doi: 10.4049/jimmunol.176.11.6624. [DOI] [PubMed] [Google Scholar]
- 11.Palha De Sousa C, Blum CM, Sgroe EP, Crespo AM, Kurt RA. Murine mammary carcinoma cells and CD11c+ dendritic cells elicit distinct responses to lipopolysaccharide and exhibit differential expression of genes required for TLR4 signaling. Cell Immunol. 2010;266:67–75. doi: 10.1016/j.cellimm.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair P, O’Donnell CM, Janasek K, Sajduk MK, Smith EA, Golden JM, Vasta CA, Huggins AB, Kurt RA. Lipopolysaccharide-treated mammary carcinomas secrete proinflammatory chemokines and exhibit reduced growth rates in vivo, but not in vitro. Immunol Invest. 2009;38:730–748. doi: 10.3109/08820130903177810. [DOI] [PubMed] [Google Scholar]
- 13.Loiarro M, Capolunghi F, Fanto N, Gallo G, Campo S, Arseni B, Carsetti R, Carminati P, De Santis R, Ruggiero V, Sette C. Inhibition of Myd88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J Leuk Biol. 2007;82:801–810. doi: 10.1189/jlb.1206746. [DOI] [PubMed] [Google Scholar]
- 14.Niwa Y, Akamatsu H, Niwa H, Sumi H, Ozamki Y, Abe A. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clin Cancer Res. 2001;7:285–289. [PubMed] [Google Scholar]
- 15.Luboshits G, Shina S, Kaplan O, Engelberg S, Nass D, Lifshitz-Mercer B, Chaitchik S, Keydar I, Ben-Baruch A. Elevated expression of the CC chemokine regulated on activation, normal T cell expressed and secreted (RANTES) in advanced breast carcinoma. Cancer Res. 1999;59:4681–4687. [PubMed] [Google Scholar]
- 16.Saji H, Koike M, Yamori T, Saji S, Seiki M, Matsushima K, Toi M. Significant correlation of monocyte chemoattractant protein-1 expression with neovascularization and progression of breast carcinoma. Cancer. 2001;92:1085–1091. doi: 10.1002/1097-0142(20010901)92:5<1085::aid-cncr1424>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 17.Lebrecht A, Grimm C, Lantzsch T, Ludwig E, Hefler L, Ulbrich E, Koelbl H. Monocyte chemoattractant protein-1 serum levels in patients with breast cancer. Tumour Biol. 2004;25:14–17. doi: 10.1159/000077718. [DOI] [PubMed] [Google Scholar]
- 18.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, Koike M, Inadera H, Matsushima K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–3289. [PubMed] [Google Scholar]
- 19.Yang H, Zhou H, Feng P, Zhou X, Wen H, Xie X, Shen H, Zhu X. Reduced expression of Toll-like receptor 4 inhibits human breast cancer cells proliferation and inflammatory cytokine secretion. J Exp Clin Cancer Res. 2010;29:92–99. doi: 10.1186/1756-9966-29-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through adaptor protein Myd88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 21.Naugler WE, Sakurai T, Kim S, Maeda S, Kim KH, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in Myd88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 22.Bao H, Lu P, Li Y, Wang L, Li H, He D, Yang Y, Zhao Y, Yang L, Wang M, Yi Q, Cai Z. Triggering of toll-like receptor-4 in human multiple myeloma cells promotes proliferation and alters cell responses to immune and chemotherapy drug attack. Cancer Biol Ther. 2011;11:58–67. doi: 10.4161/cbt.11.1.13878. [DOI] [PubMed] [Google Scholar]
- 23.Morrison C, Baer MR, Zandberg DP, Kimball A, Davila E. Effects of toll-like receptors signals in T-cell neoplasms. Future Oncol. 2011;7:309–320. doi: 10.2217/fon.10.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coste I, Le Corf K, Kfoury A, Hmitou I, Druillennec S, Hainaut P, Eychene A, Lebecque S, Renno T. Dual function of Myd88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest. 2010;120:3663–3667. doi: 10.1172/JCI42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim K, Kohlhammer H, Xu W, Tang Y, Zhao H, Shaffer AL, Romesser P, Wright G, Powell J, Rosenwald A, et al. Oncogenically active Myd88 mutations in human lymphoma. Nature. 2011;470:115–122. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahtiainen L, Mirantes C, Jahkola T, Escutenaire S, Diaconu I, Osterlund P, Kanerva A, Cerullo V, Hemminki A. Defects in innate immunity render breast cancer initiating cells permissive to oncolytic adenovirus. PLoSOne. 2010;5:1–15. doi: 10.1371/journal.pone.0013859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma V, Dixit D, Koul N, Mehta VS, Sen E. Ras regulates interleukin-1β-induced HIF-1α transcriptional activity in glioblastoma. J Mol Med. 2011;89:123–136. doi: 10.1007/s00109-010-0683-5. [DOI] [PubMed] [Google Scholar]
- 28.Apetoh L, Tesniere A, Ghiringhelli F, Kroemer G, Zitvogel L. Molecular interactions between dying tumor cells and the innate immune system determine the efficacy of conventional anticancer therapies. Cancer Res. 2008;68:4026–4030. doi: 10.1158/0008-5472.CAN-08-0427. [DOI] [PubMed] [Google Scholar]
- 29.Yu M, Wang H, Ding A, Golenbock DT, Latz E, Czura CJ, Fenton MJ, Tracey KJ, Yang H. HMGBI signals through toll-like receptor (TLR) 4 and TLR2. Shock. 2006;26:174–179. doi: 10.1097/01.shk.0000225404.51320.82. [DOI] [PubMed] [Google Scholar]
- 30.Desmetz C, Bibeau F, Boissiere F, Bellet V, Rouanet P, Maudelonde T, Mange A, Solassol J. Proteomics-based identification of HSP60 as a tumor-associated antigen in early stage breast cancer and ductal carcinoma in situ. J Proteome Res. 2008;7:3830–3837. doi: 10.1021/pr800130d. [DOI] [PubMed] [Google Scholar]