Abstract
Murine hepatocytes become polyploid, then undergo ploidy reversal and become aneuploid in a dynamic process called the ploidy conveyor. Although polyploidization occurs in some types of human cells, the degree of aneuploidy in human hepatocytes is not known. We isolated hepatocytes derived from healthy human liver samples and determined chromosome number and identity using traditional karyotyping and fluorescence in situ hybridization. Similar to murine hepatocytes, human hepatocytes are highly aneuploid. Moreover, imaging studies revealed multipolar spindles and chromosome segregation defects in dividing human hepatocytes. Aneuploidy therefore does not necessarily predispose liver cells to transformation, but might promote genetic diversity among hepatocytes.
Keywords: chromosomal instability, lagging chromosome, liver regeneration, mitosis
Aneuploidy and polyploidy involve alterations in chromosome number. Whereas aneuploidy refers to the gain or loss of discrete chromosomes, polyploidy indicates numerical change in an entire complement of chromosomes. Aneuploidy is most often associated with malignant transformation 1, but it is also seen in healthy individuals lacking evidence of malignancy in such tissues such as the cytotrophoblast, blood, skin and brain 2–4. The role of aneuploidy in developing and adult tissues is unclear. Polyploidy is similarly found in cancerous 5 and healthy tissues, and it has been extensively described in the mammalian liver 6. Hepatocytes are either mono- or binucleated, and each nucleus is diploid, tetraploid, octaploid or higher 7. Polyploid hepatocytes comprise approximately 50% of adult human hepatocytes (Duncan and Grompe, unpublished observations) and 80–90% of adult murine hepatocytes 8.
We recently identified a relationship between polyploidy and aneuploidy in the murine liver. During normal growth and proliferation polyploid hepatocytes undergo multipolar mitoses, producing an abundance of aneuploid cells as well as daughters with halved chromosome content 8, 9. Approximately 25% of hepatocytes from 3 week old mice are aneuploid, and the frequency increases to 60–70% in 5–12 month old mice. The term “ploidy conveyor” is used to indicate that hepatocytes can sequentially increase and then decrease their ploidy (i.e., diploid hepatocytes become polyploid, and polyploid hepatocytes become diploid again). In addition, chromosome segregation errors by polyploid hepatocytes generate aneuploidy. The extent of aneuploidy in human livers is unknown.
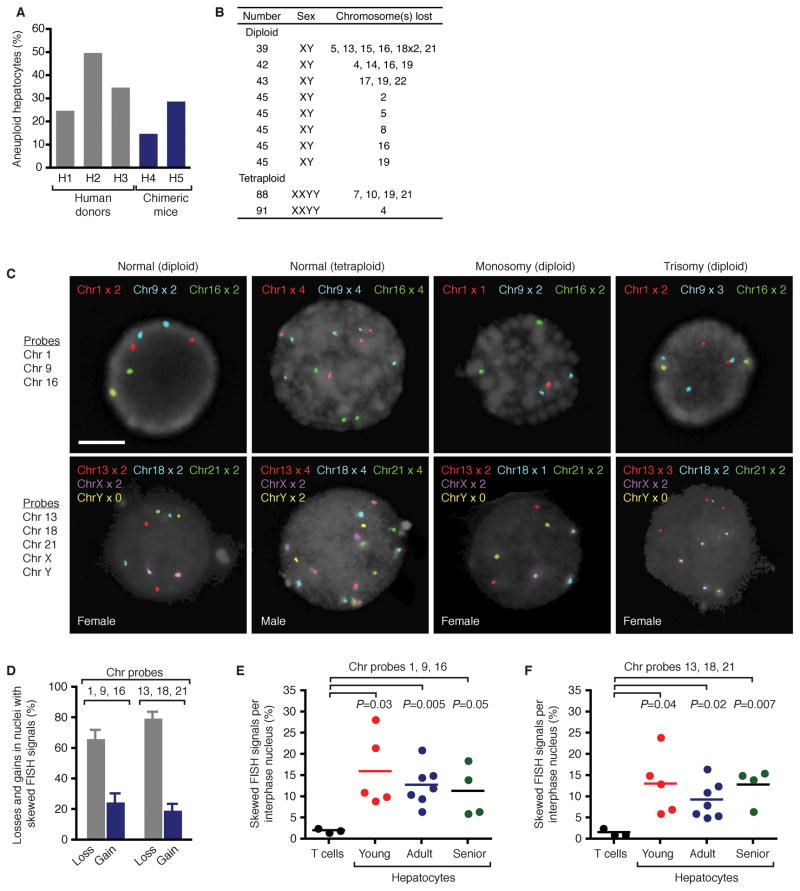
To investigate aneuploidy in human liver, chromosome number and identity was surveyed in hepatocytes from many patients. All samples were derived from tissue devoid of liver cancer. We attempted to karyotype hepatocytes from different aged donors and succeeded in generating high quality metaphases from some young patients. Karyotype analysis of primary hepatocytes revealed numerical aneuploidy in 25–50% of cells (Supplementary Methods, Supplementary Table 1 and Figure 1A). Hepatocyte preparations were highly enriched for hepatocytes, but they also contained low levels of contaminating non-parenchymal cells. To confirm aneuploidy specifically in hepatocytes, liver cells were transplanted into immuno-compromised mice deficient for fumarylacetoacetate hydrolase10. Donor hepatocytes, but not non-parenchymal cells, have a potent selective advantage in this system, repopulating the recipient liver to >75%. Hepatocyte metaphases from 2 independent donors were 15–29% aneuploid, confirming hepatocyte-specific aneuploidy (Figure 1A). Chromosomal losses were seen in both diploid and tetraploid hepatocytes, suggesting that all hepatic ploidy classes are susceptible to aneuploidization (Figure 1B and Supplementary Figure 1). Chromosomal losses were abundant whereas gains and structural rearrangements were not observed. In most cases a single homologue from ≥1 chromosome pairs was lost; however, both homologues of a chromosome pair were lost in 10% of aneuploid hepatocytes. With the exception of chromosome 12, which was never lost, other autosomes were lost at a similar frequency, 3.7% (range 1–7%), suggesting a stochastic mechanism for generating hepatic aneuploidy (Supplementary Figure 2).
Figure 1.
Description of hepatic aneuploidy. A, B, Karyotype analysis. Percentage of human hepatocytes with numerical aneuploidy from primary donors (n=3) and humanized chimeric mice (n=2) (A). Representative aneuploid karyotypes (B). C–F, Hepatic nuclei from young (n=5; 2–11 years), adult (n=7; 23–48 years) and senior (n=4, 66–72 years) donors were hybridized with chromosome-specific probes. Chromosome signals were quantified in 200 nuclei/sample. Representative nuclei are shown; scale bar is 10 μm (C). Chromosomal gains and losses were detected in nuclei with skewed FISH signals (average ± s.em.) (D). Percentage of nuclei (T lymphocytes or hepatocytes) with abnormal FISH signals (E, F).
Metaphase karyotyping has the potential disadvantage of biased selection for rapidly proliferating cells (i.e., potentially restricting analysis to a subset of cells). Therefore, we utilized FISH to enumerate chromosomes in a non-biased manner (Supplementary Methods). Hepatic nuclei from 16 patients, ranging from 2–72 years (Supplementary Table 1) were hybridized with sets of discrete probes, corresponding to chromosomes 1, 9, 16 and 13, 18, 21, X, Y. Patterns of FISH signals consistent with normal diploid and tetraploid nuclei were detected as well as patterns of chromosomal loss and gain (Figure 1C). Approximately 25% of abnormal FISH signals involved chromosome gains (Figure 1D). In contrast to T lymphocytes (Supplementary Table 1), abnormal patterns of FISH signals were frequent in hepatocytes (Figure 1E, F). Although there was variation within each age group, average values between age groups were similar. Bayesian statistics were used to extrapolate the aneuploidy observed with 6 autosomal probes to all 22 autosomes (Supplementary Figure 3). In many cases the calculated aneuploidy was nearly identical for each probe set (e.g., H6); in other instances predicted aneuploidy varied by as much as 2-fold (e.g., H18). To further validate these results obtained by the OHSU Cytogenetics Research Core, hepatocytes were hybridized with additional probes by an independent laboratory. The degree of aneuploidy was similar for each probe set (Supplementary Figure 4). Taken together, the calculated aneuploidy for each age group was 58% (range 31–91%), suggesting the degree of aneuploidy remains constant throughout postnatal life.
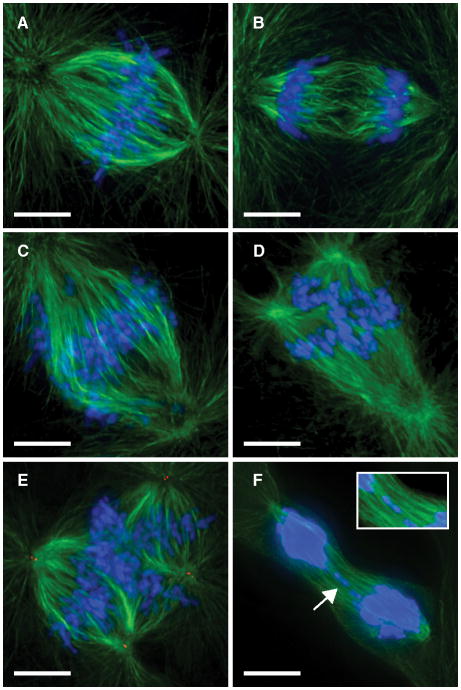
Proliferating mouse polyploid hepatocytes undergo multipolar divisions and chromosome segregation errors to generate aneuploid daughter cells 8. To determine whether similar mechanisms occur in humans, we visualized divisions by hepatocytes from human donors (Supplementary Methods). While most hepatocytes formed bipolar structures (Figures 2A, B), 1–5% of dividing cells had multipolar spindles in prometaphase/metaphase (Figures 2C–E) or lagging chromosomes during nuclear segregation (Figure 2F). Thus, the data strongly indicate that numerical aneuploidy in the human liver also occurs through segregation defects and multipolar divisions.
Figure 2.
Diversity of cell divisions by human hepatocytes. A–F, Mitotic structures were identified in cultured hepatocytes by visualizing DNA (blue) and microtubules (green). Centrioles (red) were detected in E. n=5 experiments from independent donors. Dividing hepatocytes contained bipolar spindles in metaphase (A) and anaphase (B). Multipolar spindles were detected in prometaphase/metaphase (C–E). Lagging chromosomes (arrow; inset) were also detected (F). Scale bars are 10 μm.
Human hepatocytes have the capacity to polyploidize, undergo multipolar divisions and aneuploidize, demonstrating that the ploidy conveyor is active in the human liver. Hepatic aneuploidy was independent of patient age and gender, and it was characterized in tissue without evidence of malignant transformation. Strikingly, livers from every patient we analyzed displayed aneuploidy, and the incidence of hepatic aneuploidy was very high (~50%). Since polyploid hepatocytes undergo chromosome segregation errors during mitosis (i.e., multipolar spindles, lagging chromosomes, etc.) to generate aneuploidy, aneuploid daughters are likely found specifically in pericentral regions of the liver, an area enriched for polyploid hepatocytes11.
It is unclear how hepatocytes tolerate aneuploidy. While polyploid hepatocytes should be minimally affected by the gain/loss of one or more chromosomes, aneuploid diploid hepatocytes could have altered cellular function. For example, loss of a single homologue has the potential to generate tremendous genetic diversity. Approximately 5–10% of genes are monoallelically expressed, therefore chromosomal monosomies can lead to complete functional loss of a large number of genes 12. We suggest that aneuploidy is not necessarily a predisposition for transformation in the liver, and it may actually play a beneficial role 13. We hypothesize that aneuploid hepatocytes serve as a reservoir of genetic diversity. Hence, chronic liver injury could result in selective expansion of injury-resistant hepatocytes with a favorable karyotype. Indeed, clonal cirrhotic liver nodules have already been identified in patients with hepatitis C 14. Considering the potential role of aneuploidy in adaptation to chronic liver injury, caution is advised in the proposed use of anti-cancer therapies that specifically target aneuploid cells 15.
Supplementary Material
Acknowledgments
Support
This work was supported by grants from the National Institute of Health to M.G. (R01DK051592) and A.W.D. (F32DK076232). Human hepatocytes were obtained from Pittsburgh, PA (S.C.S) through the Liver Tissue and Cell Distribution system (N01-DK-7-0004 / HHSN267200700004C). We thank A. Snyder (Advanced Light Microscopy Core at OHSU, Core grant S10-RR023432) for microscopy assistance. We are grateful to K. Dorko for distribution of human hepatocytes and to W. Fleming and D. Goldman for providing mobilized peripheral blood. Finally, thanks to E. Lagasse for advice on hepatocyte expansion in vitro.
Abbreviations
- FISH
Fluorescence in situ hybridization
Footnotes
Disclosure
M.G., E.M.W. and S.C.S. have financial interests in Yecuris Corporation.
Transcript profiling
N/A
Writing assistance
N/A
Author contributions
A.W.D designed the experiments, performed immunocytochemistry, analyzed data and wrote the paper. Karyotypes were generated by A.E.H.N. and S.B.O. FISH experiments were performed by A.E.H.N., S.B.O., L.S. and M.J.T. E.M.W. generated chimeric mice with humanized livers. S.C.S. supervised isolation of all primary human hepatocyte specimens. M.G. supervised all aspects of this work. All authors discussed the results and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gasparini P, et al. J Cell Biochem. 2007;102:320–31. doi: 10.1002/jcb.21481. [DOI] [PubMed] [Google Scholar]
- 2.Conlin LK, et al. Hum Mol Genet. 2010;19:1263–75. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weier JF, et al. Dev Biol. 2005;279:420–32. doi: 10.1016/j.ydbio.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Westra JW, et al. J Comp Neurol. 2008;507:1944–51. doi: 10.1002/cne.21648. [DOI] [PubMed] [Google Scholar]
- 5.Ganem NJ, et al. Curr Opin Genet Dev. 2007;17:157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Gentric G, et al. Clin Res Hepatol Gastroenterol. doi: 10.1016/j.clinre.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Guidotti JE, et al. J Biol Chem. 2003;278:19095–101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 8.Duncan AW, et al. Nature. 2010;467:707–10. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan AW, et al. PLoS Genet. 2009;5:e1000385. doi: 10.1371/journal.pgen.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azuma H, et al. Nat Biotechnol. 2007;25:903–10. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmucker DL. J Electron Microsc Tech. 1990;14:106–25. doi: 10.1002/jemt.1060140205. [DOI] [PubMed] [Google Scholar]
- 12.Gimelbrant A, et al. Science. 2007;318:1136–40. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 13.Rancati G, et al. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paradis V, et al. Lab Invest. 2000;80:1553–9. doi: 10.1038/labinvest.3780165. [DOI] [PubMed] [Google Scholar]
- 15.Tang YC, et al. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.