Abstract
G-quadruplex formation in the sequences 5′-(TTAGGG)n and 5′(TTAGGG)nTT (n=4,8,12) was studied using circular dichroism, sedimentation velocity, differential scanning calorimetry and molecular dynamics simulations. Sequences containing 8 and 12 repeats formed higher-order structures with two and three contiguous quadruplexes, respectively. Plausible structures for these sequences were determined by molecular dynamics simulations followed by experimental testing of predicted hydrodynamic properties by sedimentation velocity. These structures featured folding of the strand into contiguous quadruplexes with mixed hybrid conformations. Thermodynamic studies showed the strands folded spontaneous to contain the maximum number contiguous quadruplexes. For the sequence 5′(TTAGGG)12TT, more than 90% of the strands contained completely folded structures with three quadruplexes. Statistical mechanical-based deconvolution of thermograms for three quadruplex structures showed that each quadruplex melted independently with unique thermodynamic parmameters. Thermodynamic analysis revealed further that quadruplexes in higher-ordered structures were destabilized relative to their monomeric counterparts, with unfavorable coupling free energies. Quadruplex stability thus depends critically on the sequence and structural context.
Introduction
Telomeres are regions at the ends of chromosomes that contain highly repetitive DNA sequences. In humans, the sequence 5′-TTAGGG is repeated within the telomere. Several kilobases of this sequence are paired with a complementary strand to form duplex DNA, but approximately 200 bp are unpaired as a single-stranded overhang. These repetitive sequences protect the chromosome from damage, and prevent chromosome fusion 1,2. There is evidence that supports the existence of quadruplex structures in vivo 3-9, along with evidence suggesting that quadruplexes form in telomeric DNA at specific times during the cell cycle 5,6.
Oligonucleotides containing approximately four repeats of the 5′-TTAGGG sequence readily fold into unimolecular quadruplex structures 10-15. The exact structure of the folded form depends critically on the cation composition of the solution. In the presence of sodium, an antiparallel “basket” structure forms that features one diagonal and two lateral loops16. In potassium solution, two types of antiparallel “hybrid” structures form that feature one “side” (chain-reversal) loop and two lateral loops 17-22. The location of the side loop differentiates the two hybrid forms. An unusual parallel-stranded “propeller” structure with three side loops was observed by x-ray crystallography23, but that form is not the major conformation in solution 24. The “propeller” structure was recently seen in NMR studies under extreme solution conditions with high concentrations of cosolutes and greatly diminished water activity 25. Additional conformations, based on 125I radiocleavage experiments26, have been reported. A unique “basket” form containing only two stacked quartets in potassium solution was recently reported 27. Recent reviews concisely summarize the structures characterized to date for human telomeric quadruplexes 15,28
Folding of telomeric quadruplex sequences is spontaneous and thermodynamically favored 11,29,30. Folded quadruplexes are stable, but not extraordinarily so in comparison to duplex DNA 31. The folding is rapid in both sodium and potassium, and occurs through pathways that include intermediate states 32. Conversion of the potassium “hybrid” forms to the sodium “basket” form can occur readily and rapidly, and is characterized by a surprisingly small energy barrier of only about 2 kcal mol−1 33.
While the structure and stability of monomeric telomere quadruplexes are now reasonably well characterized, little is known about possible higher-order quadruplex forms. The ~200nt single-strand overhang may fold into structures containing multiple contiguous quadruplexes. Understanding such structures is critically important for understanding the interactions of telomerase and other telomeric proteins with telomeric DNA, and how these interactions may regulate changes in telomere structure throughout the cell cycle. There are scattered reports of selected properties of such quadruplex multimers. Yu and coworkers34 reported that Oxytricha or human telomeric sequences containing 1 to 3 folded quadruplex units melted in a two-state manner and proposed a “beads-on-a-string” model in which contiguous quadruplex did not interact. Circular dichroism and gel electrophoresis studies35 indicated that intermolecular quadruplex structures are less likely to form in long telomeric repeat sequences, which instead preferentially fold into intramolecular structures with contiguous quadruplex units. Dai and coworkers14 proposed a model of the long telomeric overhang that featured a compact structure composed of stacked, contiguous hybrid quadruplex units. Mass spectrometry and Taq polymerase stop assay were used to study the binding of sanguinarine to quadruplex monomers and to a longer sequence that formed tandem quadruplexes arranged as “beads-on-a-string”, and it was proposed that an additional ligand bound to the interface between the quadruplex units 36. Circular dichroism and electrophoresis were used to investigate long telomeric sequences of the type G3(TTAG3)n, with n=1-1637. A variety of both inter- and intramolecular antiparallel and parallel forms was observed, and quadruplex thermal stability was found to decrease with oligonucleotide length. A subsequent study from the same laboratory 38 proposed that a variety of intramolecular multimeric quadruplexes can form in long telomeric sequences. Atomic force microscopy (AFM) was used to visualize global structures of single-stranded telomere repeat sequences that form compact contiguous quadruplex structures 39, although the resolution of the method could not definitively assign the conformations of the individual quadruplex units. Sannohe and coworkers40 prepared end-extended and (Br)G-substituted oligonucleotides of the human telomere repeat sequence and showed by several biophysical methods that the ends of stable G-quadruplex structures point in opposite directions. Their results indicate that the human telomere DNA is likely to form rod-like higher-order structures, and they proposed a model with interacting quadruplex units, although the model was not tested by additional experiments with longer repeat sequences. A more recent attempt was made to characterize higher-order quadruplex structures by AFM41 . Wang and coworkers41 claimed that “physiologic” tails rarely form the maximum potential number of quadruplex units, and that single-stranded gaps separated the quadruplexes that did form. This claim is contradicted by an earlier AFM study 39 that showed that four contiguous quadruplexes, separated by only a short single-stranded TTA linker, readily formed in a 96 nt telomere repeat sequence. The same study used FRET to show that two contiguous quadruplexes formed a 46 nt repeat sequence, in full accord with the detailed model proposed by Petraccone and coworkers 42,43. Circular dichroism and thermal-gradient electrophoresis were recently used to study telomeric G-quadruplex motifs arranged in tandem44. Structures with two and three contiguous quadruplexes were observed, and quadruplex thermal stability was diminished upon formation of the higher-ordered structures.
The exact structure of the single-strand telomeric overhang is not known, nor is it likely that current x-ray crystallographic or NMR methods will be able to obtain high-resolution structures because of the inherent difficulty of coping with longer DNA sequences by those techniques. There have been attempts to simulate plausible higher-order structures based on the high-resolution structures of monomeric quadruplexes. Haider and coworkers reported a molecular dynamics study based on the parallel-stranded propeller quadruplex structure observed in crystals45. A compact cylindrical structure was observed in which contiguous quadruplexes stacked upon one another. The quadruplex core was very stable, with the TTA side loops being the most flexible part of the structure. The proposed model was not verified by any direct experimental data, and the proposed parallel structure is inconsistent with the CD spectra in solution reported for longer telomere repeat sequences as described above. The all-parallel tandem repeat structure was subsequently used in a molecular modeling study that explored possible drug binding in the longer telomeric overhang46. Again there was no experimental validation of the proposed drug binding modes.
In our opinion, molecular dynamics simulations are most valuable and informative when tightly coupled to rigorous experimental validation. Accordingly, we devised and implemented a strategy for exploring higher-order structures in the telomeric overhang 43 in which several plausible models are constructed using known monomeric quadruplex structures. These models are then subjected to molecular dynamics simulations to arrive at the most stable structures, which are then used to predict testable experimental properties such as sedimentation coefficients or the solvent accessibility of specific nucleotide bases within loop structures. The results from our initial exploration of contiguous dimer structures43 showed that the most plausible structure in solution was one with two quadruplexes in an alternating hybrid 1 - hybrid 2 arrangement. That structure featured a unique interface structure that was stabilized by interactions involving loop residues from both quadruplex units. The hybrid 1-hybrid 2 model predicted biophysical properties that were most consistent with experimental measurements in solution. Sedimentation velocity, circular dichroism and fluorescence studies using strategically substituted 2-aminopurine residues validated the hybrid 1-hybrid 2 structure, and clearly eliminated the very compact all parallel-stranded, propeller quadruplex model, as well as several other models with a variety of combinations and arrangements of hybrid and propeller structures43.
Described here are studies that extend our investigations to include longer telomeric sequences that might fold into structures containing three quadruplex units, and which use differential scanning calorimetry to better evaluate the stability of monomer, dimer and trimer structures.
Materials and Methods
Preparation of the samples
The DNA oligonucleotides were synthesized by IDT (Integrated DNA Technologies, Inc.) and used without further purification. All studies were done in a buffer consisting of 10 mM potassium phosphate, 100 mM KCl, 0.1 mM EDTA at pH 7.0. The oligonucleotides were dissolved in the buffer and then slowly heated in a water bath until the temperature reached 95°C. The oligonucleotide solutions were allowed to equilibrate for 10 minutes at 95°C and then were cooled overnight in the water bath. The samples were placed in a 4°C refrigerator for 48 hours before dialysis was performed. Pierce Slide-A-Lyzer® Dialysis Cassettes or Slide-A-Lyzer® MINI Dialysis Units (MWCO 3.5K) were used to dialyze the samples at 4°C. Dialysis was allowed to proceed for 24 hrs using four buffer changes within that period; after the last buffer change, dialysis was allowed to equilibrate overnight. Oligonucleotides were then transferred into microfuge tubes and kept at 4°C until needed for experiments. Oligonucleotide concentrations were determined by their absorbance at 260 nm measured at 90°C using the following molar extinction coefficients at 260 nm calculated from the nearest neighbor model for the unfolded forms: (TTAGGG)4 = 244600 M−1cm−1, (TTAGGG)4TT = 261200 M−1cm−1, (TTAGGG)8 = 489000 M−1cm−1, (TTAGGG)8TT = 505600 M−1cm−1, (TTAGGG)12 = 733400 M−1cm−1 and (TTAGGG)12TT = 750000 M−1cm−1 for the (TTAGGG)12TT.
Sedimentation velocity experiments
Sedimentation velocity experiments were performed at a temperature of 20 °C and a rotor speed of 50,000 rpm using a Beckman Optima XL-A analytical ultracentrifuge. Following loading and before data collection, samples were allowed to equilibrate for one hour after vacuum and temperature had been established. Data were collected at 260 nm as a function of radial position. Each centrifuge cell was scanned sequentially with zero time delay between scans until no further sedimentation was observed. For each sample, data were collected at three loading concentrations of A260(1.2 cm) ~ 0.25, 0.5 and 1. Primary sedimentation data were transferred to the program Sedfit for analysis47,48 . Data were analyzed using a continuous c(s) model using a range of 0.5 to 10 S and a confidence level of 0.68 (1 standard deviation). Solution density and viscosity were calculated from buffer composition as 1.00419 g/mL and 1.0030 cP, respectively, using the program Sednterp49. A value of 0.55 mL/g was assumed for the partial specific volume50. Fitting was performed using alternating rounds of the simplex and Marquardt-Levenberg algorithms until there was no further decrease in rmsd. Data in the form of c(s) distributions were exported to Origin v7.0 (OriginLab Corporation, Northampton, MA) for graphing.
CD Experiments
CD spectra of the quadruplexes were recorded on a Jasco 810 circular dichroism spectrophotometer equipped with a Peltier heating/cooling device and nitrogen purging capabilities. The spectra were recorded in the range 220-320 nm with a response of 1 s, at 2.0 nm bandwidth and corrected by subtraction of the background scan with buffer. The oligonucleotides concentrations were in the range 2-5 M and a 1 cm path length cuvette was used. For the CD melting experiments a scan rate of 1 °C/min was used for the melting and the annealing curves, CD spectra were recorded at 1 °C steps in the range 10-95°C and the melting curves were obtained by reporting the molar ellipticity at 290 nm versus the temperature.
Singular Value Decomposition Analysis
The CD spectra versus temperature were analyzed by singular value decomposition (SVD) to determine the number of significant spectral species involved in the CD melting experiments51,52. All SVD calculations were performed using routines in Matlab 7.1 software (Mathworks). The matrix of the CD spectra A is decomposed by the SVD method into the product of three matrices: The matrix U contains the basis spectra, S is a diagonal matrix that contains the singular values, and V is a matrix containing the amplitude vectors. Examination of the autocorrelation functions of the basis spectra and of the amplitude vectors permits us to determine the minimum number of component spectra required to describe the data within the random noise. The value of the autocorrelation function is a measure of the smoothness between adjacent row elements. Values near 1 indicate slow variation from row-to-row or “signal”, a value of 0.8 corresponds to the signal/noise ratio of 1. A value of the autocorrelation function higher than 0.8 for both the U and V matrices was selected as a cutoff criterion for accepting a significant spectral species.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) measurements were carried out using a VP-DSC Microcalorimeter (Microcal, Northampton, MA). The experiments were performed at single strand concentrations in the range 40-100 M. Scans were performed at 1.0 °C/min in the 5-105 °C temperature range. Reversibility and repeatability were established for each sample by multiple (3-4) up scans after cooling. A buffer-buffer scan was subtracted from the buffer-sample scans and linear-polynomial baselines were drawn for each scan. Baseline corrected thermograms were then normalized with respect to the single strand molar concentration to obtain the corresponding molar heat capacity curves. Model-free enthalpy estimates for the overall unfolding of quadruplex structures were obtained by integrating the area under the heat capacity versus temperature curves. Tm values were estimated as the temperatures corresponding to the maximum of each thermogram peak. Entropy values were obtained by integrating the curve Cp/T versus T (where Cp is the molar heat capacity and T is the temperature in Kelvin) and the free energy values were computed by the equation G = H-T S. The thermodynamic parameters in Table 1 represent averages of heating curves from three to five experiments. The reported errors for thermodynamic parameters are the standard deviations of the mean from the multiple determinations.
Table1.
Thermodynamic parameters obtained from the analysis of the CD and DSC melting curves
| Sequence | Tm (°C) (± 1)a |
Tm (°C) (± 0.2)b |
ΔHcal (kJ/mol) |
|---|---|---|---|
| (TTAGGG)4 | 61 | 62.7 | 235 ± 5 |
| (TTAGGG)4TT | 58 | 59.7 | 213 ± 4 |
| (TTAGGG)8 | 57 | 57.1 | 475 ± 8 |
| (TTAGGG)8TT | 57 | 57.3 | 455 ± 7 |
| (TTAGGG)12 | 57 | 57.3 | 585 ± 10 |
| (TTAGGG)12TT | 57 | 57.1 | 605 ± 11 |
Tm values evaluated by CD melting
Tm values evaluated by DSC melting
A more ambitious analysis and deconvolution of DSC thermograms to identify possible intermediate states was attempted using the statistical mechanical approach of Freire and Biltonen 53,54 as recently implemented in Origin™ Software 55.
Molecular Modeling
To build the trimer models, we started from coordinate files of dimer models previously developed by some of us43. All of these dimer models corresponded to the 50-mer (TTAGGG)8TT sequence. In building the hybrid-121 trimer we started from the coordinate file of the Hybrid-12 model consisting of 5′-hybrid-1 and hybrid-2 quadruplex units with optimal stacking interactions at the quadruplex-quadruplex interface. The hybrid-121 trimer was then built by adding to the 3′-end of the dimer structure an hybrid-1 quadruplex unit corresponding to the 22-mer oligonucleotide AG3(TTAG3)3. In building the hybrid-2-hybrid-1 interface we tried to maximize the inter-quadruplex interactions. The coordinates for the hybrid-2 quadruplex were taken from the reported NMR structure (PDB code 2jpz). The final trimer structure contained 72 oligonucleotides corresponding to the (TTAGGG)12 telomeric sequence. In each quadruplex unit a K+ ion was placed between the adjacent G-tetrads.
To build the all-propeller model we started from the coordinates of the all-propeller dimer model of sequence (TTAGGG)8TT previously optimized through MD simulation43 and then added to the 3′-end of this structure an additional 22-mer propeller quadruplex unit generated from the coordinates of the reported X-ray structure (PDB code 1kf1). In the all-propeller model the G-tetrad planes of the two quadruplex units were allowed to stack on each other and one additional K+ ion was placed between the two stacked quartets. The all-propeller and hybrid-121 models were built also for the (TTAGGG)12TT sequence by adding two additional thymine residues at the 3′-end of the corresponding models built for the (TTAGGG)12 sequence.
To build the dt12 model we added to the Hybrid-12 dimer model, corresponding to the (TTAGGG)8TT sequence, a 12-mer single strand overhang at the 5′ end and a 10-mer single strand overhang at the 3′end to reach the total 72-mer sequence of (TTAGGG)12. The models were solvated in a 10 Å box of TIP3P water using standard Amber 9.0 leap rules to hydrate the systems and potassium counterions were added for overall charge neutrality. Molecular dynamics (MD) calculations were carried out with the AMBER 9.0 version of sander and parm99.dat parametrization56. The force field was modified using the frcmod.parmbsc0 parameter file57. The systems were heated slowly and equilibrated for 250 ps with gradual removal of positional restraints on the DNA following this protocol: (i) minimize water, (ii) 50 ps MD (T=100K) holding DNA fixed (100 kcal/mol Å−1), (iii) minimize water with DNA fixed (100 kcal/mol Å−1), (iv) minimize total system, (v) 50 ps MD (T=100 K) holding DNA fixed (100 kcal/mol Å−1), (vi) 50 ps MD (T=300 K) holding DNA fixed (100 kcal/mol Å−1), (vii) 50 ps MD (T=300 K) holding DNA fixed (50 kcal/mol Å−1), (viii) 50 ps MD (T=300 K) holding DNA fixed (10 kcal/mol Å−1),(ix) 50 ps MD (T=300 K) holding DNA fixed (1 kcal/mol Å−1). Simulations were performed in the isothermic isobaric ensemble (P=1 atm, T=300 K). Periodic boundary conditions and the Particle-Mesh-Ewald algorithm were used. A 2.0 fs time step was used with bonds involving hydrogen atoms frozen using SHAKE. The sedimentation coefficients were computed from the MD trajectories using the Hydropro558 program on snapshots extracted each 10ps from the last 5 ns of each MD run. Calculations were performed at the J. G. Brown Cancer Center Molecular Modeling Facility.
Results and Analysis
Sedimentation Velocity
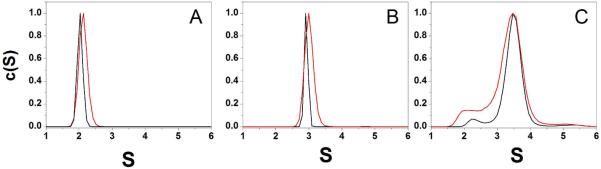
To explore the structure of the human telomeric DNA formed by 4, 8 and 12 TTAGGG repeats and the effect of the 3′-flanking bases we carried out analytical ultracentrifugation (AUC) experiments on each sequence at three different strand concentrations (Figures 1 and S1). Sedimentation velocity experiments were done to determine the distribution of sedimentation coefficients (c(s)). The S-values were found to be independent of the loading concentration indicating that these sequences form only intramolecular complexes (Figure S1). For the (TTAGGG)4 and (TTAGGG)4TT sequences S values of 2.03S and 2.13S were measured. Previous NMR studies have shown that (TTAGGG)4 and (TTAGGG)4TT sequences form hybrid-1 and hybrid-2 quadruplex structures, respectively, in K+ buffer14. These structures are very similar and they are predicted to have S values of ~2.0S which is in accord with the experimental observations (Fig. 1A). The observed S values for the (TTAGGG)8 and (TTAGGG)8TT sequences are 2.92S and 2.99S, respectively (Fig. 1 B). These values are in good agreement with predictions from molecular modeling studies for structures formed by two consecutive quadruplex units with alternating hybrid 1 - hybrid 2 structures43. The slight difference between these two values is very close to the experimental error and could also result from the slight difference in molecular weight of the two sequences.
Figure 1.
Sedimentation coefficient distributions of (TTAGGG)4 and (TTAGGG)4TT (A), of (TTAGGG)8 and (TTAGGG)8TT (B), (TTAGGG)12 and (TTAGGG)12TT (C). The sequence with the two thymine residues at 3′end is shown in red.. Each distribution was normalized with respect to the highest c(s) value.
The (TTAGGG)12 and (TTAGGG)12TT sequences show more complex sedimentation coefficient distributions (Fig. 1C). The distributions show a slight but pronounced shoulder at lower sedimentation coefficient (~ 2.3S), and a prominent peak at 3.49S for both sequences. These data suggest that (TTAGGG)12 and (TTAGGG)12TT adopt the same major conformation in solution, but that there is slight heterogeniety that most probably arises from different folded forms.
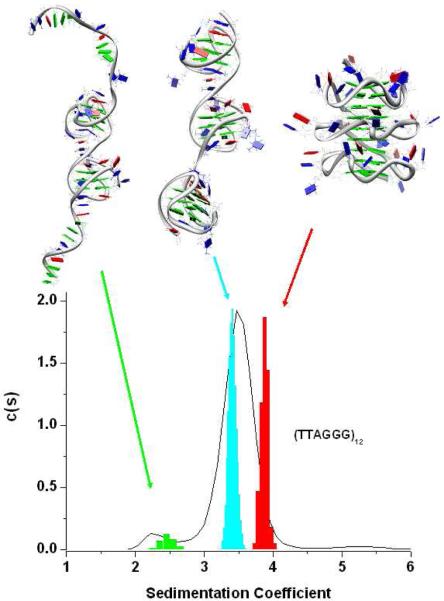
To extract more information from these AUC profiles we compared experimental c(s) distributions with calculated sedimentation coefficient distributions obtained from different possible models of structures containing three contiguous quadruplex units following a procedure already tested 24. We built two possible structures (Figure S2), one containing three all-parallel, propeller quadruplex units (all-parallel trimer) and another one containing an hybrid-2 quadruplex in the middle and two hybrid-1 quadruplexes at the extremities (hybrid-121 trimer). These constructs, after a 250 ps equilibration period, were subjected to 10 ns of free molecular dynamics simulations to obtain stable structures which were then used to predict the c(s) distributions (see material and methods for details). For both of the initial models, the integrity of the individual quadruplex units was preserved throughout the simulation. Further, in the hybrid-121 model the stacking interactions between loops residues at the hybrid-1-hybdrid-2 interface were retained during all the simulation time whereas the interaction between the hybrid-1 quadruplex at the 3′ and the central hybrid-2 quadruplex were lost after 2 ns. As expected, greater flexibility between the individual quadruplex units was observed in the hybrid-121 model compared to the all-propeller model in which the presence of the stacking interactions between the G-tetrads of the adjacent quadruplexes makes the system more rigid.
In Figure 2 the average structures from the MD simulations are shown and the corresponding predicted sedimentation coefficient distributions are superimposed on the experimental c(s) distribution of (TTAGGG)12 (a similar result was obtained with (TTAGGG)12TT)). We found that the predicted S value for the hybrid-121 (3.40S) is very close (within 3%) to the experimental value whereas the value predicted by the all-propeller model (3.87S) is inconsistent (≈ 15% larger) than the experimental value. The all-parallel trimer is predicted to be a hydrodynamically more compact structure than is actually observed in solution. As seen in Figure 2, the experimental c(s) distribution slightly overlaps the predicted distribution for the all-propeller structure. What is the probability that the experimental and computed distributions are different? From the c(s) distribution, a weight-average sedimentation coefficient of 3.48 ± 0.25 (95% confidence interval 3.445;3.515) is found for the major component. From the distribution calculated for the all-propeller structure, the mean is 3.87 ± 0.05 (95% confidence interval 3.863;3.877). Comparison of these means by an unpaired two-tailed t test shows that they differ significantly (P<0.0001) and that the observed difference in the distributions is greater than expected by chance. Similarly, the difference between the calculated distributions for the propeller and hybrid (mean=3.41±0.06; 95%CI 3.402;3.418) models is highly significant (P<0.0001) and greater than expected by chance. Although these results do not allow us to establish unequivocally the trimer structure, it clearly suggests that a major fraction of (TTAGGG)12 and (TTAGGG)12TT can fold in three contiguous quadruplex units and that these units are likely a mixture of hybrid conformations.
Figure 2.
The average structure of the Hybrid-121, all-propeller and dt12 models are shown on the (top). The colors of the residues are: green for dG, blue for dT and red for dA residues. On the bottom of the figure, the sedimentation coefficient distributions obtained from the MD trajectories of the model (indicated by the colored arrows) are superimposed on the experimental distribution (black line) obtained for the (TTAGGG)12 telomeric sequence in K+ solution.
Figure 2 shows the power of sedimentation velocity experiments in revealing even slight heterogeneity. The slight shoulder near 2.3S represents no more than 7% of the total sedimenting material, and is hydrodynamically distinct from the folded two-quadruplex structures that sediment near 2.9S (Fig. 1B). To account for this slower sedimenting material in the c(s) profiles, we explored the hypothesis that a fraction of the total (TTAGGG)12 and (TTAGGG)12TT samples formed structures with only two quadruplex units, with unfolded single-strand regions. A variety of such models are possible, ones with single-strand tails, or ones with quadruplex units at the ends with a single-strand linker in between. While we did not exhaustively explore all possible models, one such model is portrayed in Fig. 2, along with its predicted c(s) distribution. This structural model (named dt12) is formed by two consecutive quadruplex units (hybrid-1 and hybrid-2) in the middle of the (TTAGGG)12 sequence leaving two single-strand overhangs on both the 5′ and 3′ ends (Fig. 2). After an equilibration period of 250 ps the initial model was subjected to 8 ns of free molecular dynamics. It can be seen from Figure 2 that the predicted c(s) distribution is very broad as expected for a structure with flexible single-strand overhangs and it is close to the value of the experimentally observed shoulder in the (TTAGGG)12 and (TTAGGG)12TT AUC profiles. Although the dt12 modeling is not an exhaustive search of all the possible dimers conformations that the (TTAGGG)12 and (TTAGGG)12TT sequences can adopt in solution, it shows that the models containing only two quadruplex units can explain the shoulder observed at lower sedimentation coefficient. In the next sections we will present other experimental data consistent with this structural hypothesis. It is notable that only 7% of the strands are incompletely folded to contain two quadruplex units, and that the trimer is the thermodynamically favored species.
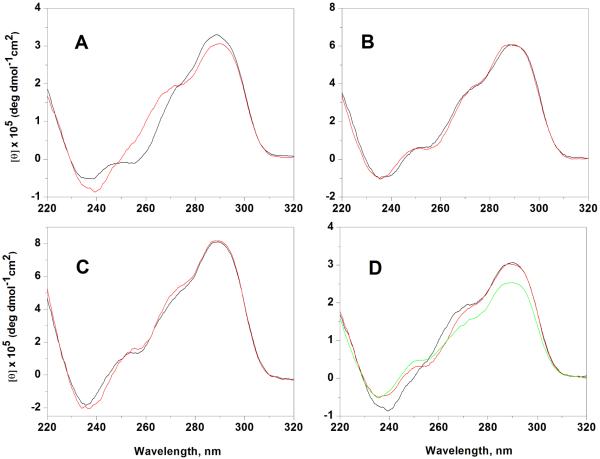
Circular dichroism studies
Figure 3 shows the CD spectra of the six oligonucleotides at T= 20°C and in the same buffer conditions. The CD spectra of all telomeric sequences show a peak at 290 nm, a major shoulder at 270 nm and a smaller one at 250 nm with a small negative band around 240 nm. The amplitudes of the CD spectra, when normalized with respect to strand concentration, steadily increase with oligonucleotide length. The spectral shapes seen in Fig. 3 are consistent with the reported spectra of hybrid-type structures 14, and clearly indicate that none of these strands fold into an all-parallel stranded structure, which would be expected to show a positive maximum near 260 nm 59-61. Slight but significant differences are observed in the spectra of the monomer sequences (TTAGGG)4 and (TTAGGG)4TT, with the spectrum of the latter differing over the shoulder region 250-270 nm and with the negative band at 240 nm having slightly greater amplitude. These results are consistent with previous NMR studies showing that the predominant conformation adopted by the monomeric sequences in solution is critically affected by the 3′-end flanking bases14. Particularly, the presence of the 3′-flanking bases promotes the hybrid-2 conformation over the hybrid-1. On the other hand, the CD spectra of the dimer ((TTAGGG)8 and (TTAGGG)8TT) and trimer ((TTAGGG)12 and (TTAGGG)12TT) sequences are very similar and are not as perturbed by the presence of the 3′-end flanking bases. These findings suggest that the 3′-flanking bases are less important in determining the quadruplex units folding in the longer DNA telomeric sequences. To further check for the presence of very slow conformational changes, we followed the CD spectra of all the sequences at 20°C over a period of several months after the first annealing procedure and no significant changes were observed (data not shown).
Figure 3.
CD spectra of the (TTAGGG)4 and (TTAGGG)4TT (A), (TTAGGG)8 and (TTAGGG)8TT (B), (TTAGGG)12 and (TTAGGG)12TT C).The sequence with the two thymine residues at 3′end are shown in red. In the panel D the CD spectrum of the (TTAGGG)4TT (black line) is compared to the CD spectrum of (TTAGGG)8TT (red line) and (TTAGGG)12TT (green line). The CD spectrum of (TTAGGG)8TT and (TTAGGG)12TT were divided by factors of two and three , respectively.
In a previous report43 we showed that the (TTAGGG)8TT spectra is best represented by a combination of the (TTAGGG)4 and (TTAGGG)4TT spectra rather than by only one of these spectra. The same components can be recognized in the (TTAGGG)12 and (TTAGGG)12TT spectra. These findings suggest the presence of both the hybrid-1 and hybrid-2 quadruplexes in the multimer structure. In Figure 3d the CD spectra of the monomer, dimer and trimer sequences with the 3′-end flanking bases are compared, with each of the spectra in Fig. 3a-3c now normalized with respect to the expected number of quadruplexes present in each folded structure. That is to say dividing by one for (TTAGGG)4TT, two for (TTAGGG)8TT and three for (TTAGGG)12TT. The normalized intensity at 290 nm (Figure 3d) for the sequence (TTAGGG)8TT is fully consistent with a completely folded strand containing two quadruplex units whereas the CD intensity of the (TTAGGG)12TT sequence is slightly less than expected for a fully folded stand with three quadruplex units. Similar results were obtained for the sequences (TTAGGG)4, (TTAGGG)8 and (TTAGGG)12. The slightly diminished CD intensity of the trimer sequence is consistent with the hypothesis that some small fraction of the (TTAGGG)12 and (TTAGGG)12TT oligonucleotides is incompletely folded and contains only two quadruplex units. This suggestion is consistent with our interpretation of the observed ~2.3S shoulder seen in c(s) distributions (Fig. 2). By comparing the CD intensity at 290 nm and assuming that the (TTAGGG)4 and (TTAGGG)4TT sequences fold completely in one quadruplex unit we can estimate that at least 87% of the (TTAGGG)12 and (TTAGGG)12TT sequences are composed of three quadruplex units and the remaining 13% contains only two quadruplex units, in fair agreement with the 7% estimated by sedimentation.
Thermodynamic characterization CD melting
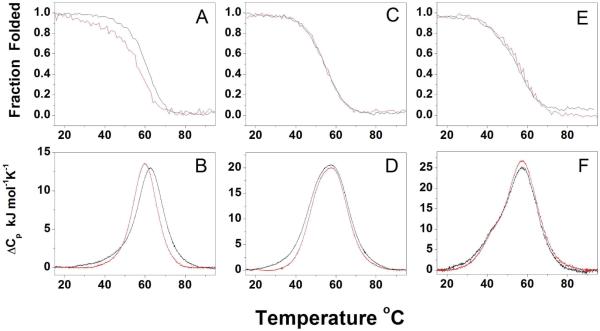
In Figure 4 (top) the CD melting profiles monitored at 295 nm for the sequences studied are shown. These melting curves are fully reversible, with no significant hysteresis observed between heating or cooling scans (Figure S3). The (TTAGGG)8, (TTAGGG)8TT, (TTAGGG)12 and (TTAGGG)12TT sequences have very similar melting behavior with a Tm of about 57 °C whereas (TTAGGG)4 has Tm ~ 62 °C and (TTAGGG)4TT of about 3 °C lower ( ~59 °C). These results indicate that the 3-flanking bases do not affect the thermal stability of the longer DNA sequences as much as they do for the shorter sequences ((TTAGGG)4 and (TTAGGG)4TT) and further strengthen the hypothesis that the same major conformation is formed by the longer DNA telomeric sequences independent of the presence of the 3-flanking bases.
Figure 4.
CD (top) and DSC (bottom) melting profiles of (TTAGGG)4 and (TTAGGG)4TT (A,B), (TTAGGG)8 and (TTAGGG)8TT (C,D), (TTAGGG)12 and (TTAGGG)12TT (E,F). The sequences with the two thymine residues at 3′end are shown in red.
To enumerate the number of significant spectral species involved in the melting processes, we performed singular value decomposition analysis (SVD) on the data matrices obtained by monitoring CD spectra as a function of temperature 51,62. The magnitudes of the singular values provide the first indication of the number of significant spectral species. We found that, for all the sequences studied, at least 3 singular values appear to significantly deviate from the linear behavior of the remaining S values (Figure S4) suggesting that the melting processes are not a simple two-state processes, and involves intermediate species. Analysis of the first-order autocorrelation of the columns of the V and U matrices (Table 2S) supports this conclusion
DSC measurements
DSC provides a method for the model-independent analysis of denaturation thermodynamics. In Figure 4 (bottom) the DSC thermograms for denaturation of the various quadruplex structures are shown. The reversibility of DSC scans was tested by repeat scans of the same sample after cooling, with 3-4 such repeat scans yielding the same thermogram within experimental error. DSC thermograms obtained over a strand concentration range of 40 – 100 μM were identical within error, suggesting that there were no complicating intermolecular reactions. The thermodynamic parameters for the melting transitions are reported in Table 1. As for the CD melting, a slight effect of the 3-end bases on thermal stability was observed only for the shorter sequences ((TTAGGG)4 and (TTAGGG)4TT) whereas the (TTAGGG)8, (TTAGGG)8TT, (TTAGGG)12 and (TTAGGG)12TT sequences show similar Tm values of about 57°C. There is a good agreement between the DSC and CD derived Tm values considering that the single strand concentrations used in the DSC experiments are approximately 20 times higher than those used in CD melting experiments, again suggesting no complications from competing intermolecular interactions. This agreement is further confirmation of the unimolecular nature of the telomeric DNA structures present in solution under our experimental conditions.
The DSC peak of the (TTAGGG)4 sequence shows slight asymmetry at lower temperatures, suggesting the presence of more than one species in the melting process. In contrast, the (TTAGGG)4TT sequence has a more symmetrical peak. The thermodynamic parameters for these two monomeric quadruplexes (Table 1) are slightly different, with the introduction of the 3′-end flanking bases resulting in a decrease of both the Tm and the enthalpy.
The calorimetric enthalpy values for the (TTAGGG)8 and (TTAGGG)8TT sequences are 475 and 455 kJ/mol, respectively. These enthalpic values corresponds to an average enthalpy value per quadruplex unit of about 237 kJ/mol for (TTAGGG)8 and 227 kJ/mol for (TTAGGG)8TT, similar to the values reported for the single quadruplexes. The total enthalpy changes for (TTAGGG)8 and (TTAGGG)8TT unfolding are slightly higher than the sum of the (TTAGGG)4 and (TTAGGG)4TT enthalpies perhaps indicating the presence of additional stabilizing interfacial interactions between contiguous quadruplex units.
The (TTAGGG)8 and (TTAGGG)8TT sequences have the same Tm values, CD spectra and similar thermograms, thus suggesting that they adopt the same main conformation in solution. Closer inspection of their thermograms (Figure 4, bottom), however, reveals that the peak for the (TTAGGG)8 sequence is broader than the one for the (TTAGGG)8TT sequence, and particularly that the former displays a broad tail at lower temperature ( T < 35 °C) that is completely absent in the thermogram of the latter sequence.
The (TTAGGG)12 and (TTAGGG)12TT sequences have nearly identical DSC thermograms, indicating similar melting pathways for these two sequences. This result indicates that with these longer sequences the effect of the 3′-flanking bases on the folding is diminished. Both the DSC thermograms are asymmetric and display a major peak at about 57 °C and a shoulder at lower temperature (~45 °C). These results are consistent with the SVD analysis that shows that multiple species are involved in the melting process. The two sequences have very close thermodynamic parameters with an average enthalpy value of 590 kJ/mol This enthalpy value is slightly less than expected from the denaturation of three quadruplex units and could result from a small percentage of strands that form only two quadruplex units as suggested by the corresponding AUC and CD data. Alternatively, the lower than expected total enthalpy could reflect destabilizing interactions among quadruplex units.
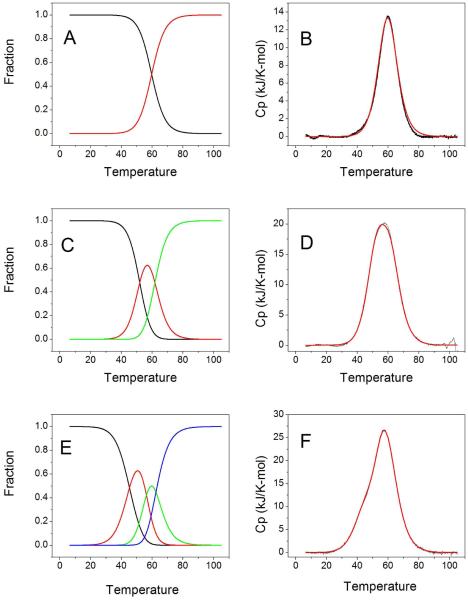
A more ambitious analysis of the DSC data was attempted using the statistical mechanical model and deconvolution procedure developed by Freire and Biltonen 53,54,63. Their procedure exploits the fact that DSC thermograms are unique in that they can be transformed directly to yield the partition function for the unfolding process without any assumptions or recourse to any specific mechanistic model. The partition function may then be used to enumerate the fractions of folded, unfolded and intermediate species at any temperature, and thus can define the complexity of the unfolding reaction process. With such information in hand, DSC thermograms may then be fit to rationally chosen reaction models to provide thermodynamic characterization of the steps in the process. Spink has provided a detailed protocol for the deconvolution procedure and a discussion of the process55. A description of the deconvolution and fitting of all of the thermograms shown in Figure 4 may be found as Supplementary Material. Figure 5 shows results obtained for three sequences, (TTAGGG)nTT with n = 4, 8 and 12, and Table 2 shows the thermodynamic parameters obtained from fits to the data.
Figure 5.
Deconvolution of DSC profiles for the (TTAGGG)nTT (n=4,8,12) series. Panels A, C and E show the species plots for the monomer, dimer and trimer structures, respectively. Panels B, D, and F show the corresponding best fits to experimental thermograms.
Table 2.
Deconvolution of DSC thermograms
| Sequence | Tm | ΔH | ΔS | ΔGfolding (20°C) |
|---|---|---|---|---|
| °C | kJ/mol | J/K-mole | kJ/mol | |
| A. (TTAGGG)4 | 63.1 | 228 | 681 | −25.8 |
| B. (TTAGGG)4TT | 59.8 | 213 | 639 | −25.8 |
| C. (TTAGGG)8 | ||||
| Transition 1 | 53.4 | 263 | 806 | −26.8 |
| Transition 2 | 63.3 | 249 | 739 | −32.5 |
| D. (TTAGGG)8TT | ||||
| Transition 1 | 52.0 | 219 | 676 | −20.9 |
| Transition 2 | 61.8 | 222 | 662 | −28.0 |
| E. (TTAGGG)12 | ||||
| Transition 1 | 43.1 | 159 | 503 | −11.6 |
| Transition 2 | 54.9 | 203 | 620 | −21.3 |
| Transition 3 | 61.2 | 216 | 646 | −26.7 |
| F. (TTAGGG)12TT | ||||
| Transition 1 | 45.4 | 176 | 553 | −14.0 |
| Transition 2 | 56.3 | 221 | 672 | −24.1 |
| Transition 3 | 62.6 | 204 | 606 | −26.4 |
Figure 5 shows an interesting progression for quadruplex-forming sequences of increasing repeat length. DSC thermograms become increasingly complicated with an increasing number of intermediate states. For (TTAGGG)4TT, deconvolution revealed negligible intermediate species, and the thermogram can be fit to a simple two-state model (Figure 5A, B) with Tm= 59.8 °C and an enthalpy of 213 kJ/mol (Table 2). (TTAGGG)8TT denaturation shows a distinct intermediate species (Figure 5C), requiring a fit to a sequential model characterized by the two melting transitions with the parameters shown in Table 2. Finally, deconvolution of the (TTAGGG)12TT thermogram reveals two intermediate species (Figure 5E), and a best fit to a sequential model with three melting transitions (Table 2). This progression suggests that melting of each of the quadruplex units in the higher-order structures is not identical, and is different from the melting of monomeric quadruplexes. For melting of the dimer quadruplex structure, the enthalpy of both units is greater than for the monomer, perhaps reflecting stabilizing interactions at the quadruplex-quadruplex interface. For melting of the three-quadruplex structure, one quadruplex unit seems to be greatly destabilized relative to the monomer structure. A more detailed discussion of the deconvolution and analysis of DSC thermograms is included in Supplementary Material. The free energies for each transition, obtained by the standard Gibbs equation ΔG = ΔH –TΔS, are shown in Table 2. The signs of the free energies were changed to refer to the folding reaction.
Discussion
These results show that the deoxyoligonucleotides (TTAGGG)n and (TTAGGG)nTT (with n = 4, 8 and 12) spontaneously fold into stable quadruplex structures. For the longer sequences (n = 8, 12), higher-order quadruplex structures form spontaneously in which the maximum number of contiguous quadruplex units form. The favorable folding free energies for these quadruplex structures result from large favorable enthalpy contributions, and folding is opposed by an unfavorable entropy contribution.
Our studies provide a detailed molecular model, with experimental validation, for the longest quadruplex assembly reported to date. Detailed molecular dynamics simulations and sedimentation studies 42,43 previously showed that the sequences (TTAGGG)8 and (TTAGGG)8TT both formed stable structures containing two contiguous quadruplexes with alternating hybrid 1 and hybrid 2 conformations. A unique interface formed between the quadruplex units that featured stabilizing interactions between bases within particular loops of each quadruplex. The new computational, CD and sedimentation studies reported here show that the sequences (TTAGGG)12 and (TTAGGG)12TT form structures with three contiguous quadruplex units in hybrid conformations. As seen in Figure 2, a structure containing all parallel (“propeller”) quadruplex units cannot account for the experimentally observed sedimentation coefficient distribution, and is predicted to be a hydrodynamically more compact structure than is observed in sedimentation velocity experiments. Exhaustive comparison of all possible three-quadruplex structures constructed from hybrid 1 and 2 conformations is restricted by computational demands. Other hybrid combinations than the 1-2-1 arrangement shown in Figure 2 would likely predict similar sedimentation coefficients and may be difficult to distinguish. A variety of “beads-on-a-string” models for higher-order quadruplex structures at telomere tails have been proposed 34,38,64. Our model differs from these models in several respects. First, our atomic-level model was used to predict experimental properties that were then measured in order to validate and test the proposed structure. Second, the models that are most consistent with experimental measurements are those that feature mixed quadruplex conformations with interactions between quadruplex units. Simpler “bead-on-a-string” models generally assume identical, independent conformational units with no interactions between the “beads”. We caution that rigid body hydrodynamic calculations involve a number of assumptions (chief among them the effects of hydration) that influence their comparison to experimental hydrodynamic properties. Our previously published studies24,42,43 have discussed these caveats and have provided additional validation for our approach.
A recent AFM study suggested that human telomeric tails rarely formed the maximum number of possible quadruplex units, and that single-stranded gaps separated those quadruplex that did form 41. That is not the case in our hands, and also stands in disagreement with a previous AFM study 39. Our data (Figures 2 and S1) show that over 90% of the (TTAGGG)12 and (TTAGGG)12TT sequences fold into structures containing the maximum number of three contiguous quadruplex units, with the remaining ≈10% forming two-quadruplex structures. No structures with only one quadruplex were evident from sensitive sedimentation velocity measurements that could easily detect a few percent of such species.
Several laboratories have reported that higher-order quadruplex assemblies are destabilized, with decreased melting temperatures, relative to their monomeric counterparts 34,37,44. Our results (Table 1) are consistent with these reports, although we find that the melting of higher-order structures is not a simple two-state process as was assumed in these earlier studies. Our integrated CD and DSC denaturation studies provide data for a detailed thermodynamic analysis that reveals subtleties and complexities in the unfolding mechanism not previously recognized.
Table 1 shows that melting temperatures decrease for higher-order structures relative to single quadruplexes, and that model-independent enthalpy values obtained by DSC are lower than expected for the longer trimer sequences. Singular value decomposition of 3D CD melting data (Table 2S) suggests that all quadruplex structures deviate from simple two-state melting, and that intermediate states along the melting pathway may need to be considered. Such complexity was explored by analysis of DSC thermograms. Freire and Biltonen provided a statistical mechanical method for deconvoluting thermograms to enumerate the number of intermediate species and their fractional contribution at each temperature along the melting curve. Their method exploits the fact that the double integral of the DSC thermogram provides the partition function of the transition. Figure 5 shows the deconvolution of thermograms for the (TTAGGG)nTT (n=4,8,12) series. What is notable is that the complexity of the species plots (Figure 5A, C, E) increase with the number of quadruplex units within the sequence. The DSC data can be adequately represented by two species (folded and unfolded) for the single quadruplex sequence, but there are clearly three species for the two quadruplex assembly and four species for the three quadruplex assembly. This suggests that each quadruplex unit in the higher-order structures is not independent and identical, but is thermodynamically unique and is influenced by its neighbors. To account for this, DSC thermograms were fit to sequential melting models (figures 5B,D,F) to yield the thermodynamic parameters shown in Table 2.
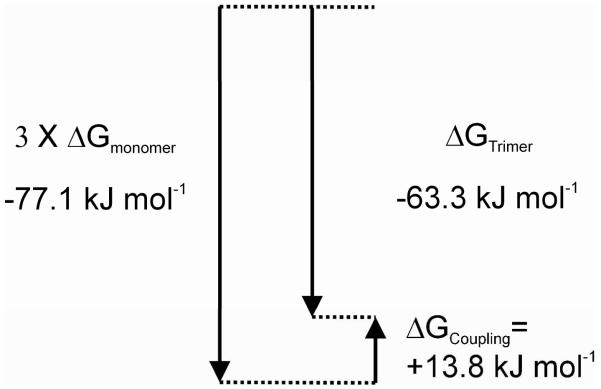
The thermodynamic data in Table 2 enables us to quantify the apparent coupling free energy65 for multimeric quadruplex assembly as shown in Figure 6. The difference between the total free energy of the folding of three contiguous quadruplexes (−63.3 kJ/mol) and three times the folding free energy of a single quadruplex (−77.1 kJ/mol) defines the coupling free energy ∆GCoupling = +13.8 kJ/ mol. The positive sign indicates an unfavorable coupling free energy in the multimeric assembly, arising from unfavorable interaction of an unknown nature between the quadruplex units. Inspection of the data in Table 2 suggests that ∆GCoupling contains contributions from both enthalpy and entropy components. A similar analysis for the dimer sequences shows small coupling free energies of only 1-2 kJ/mol, indicating lesser destabilizing interactions between two contiguous quadruplexes compared to three found in the trimer. Detailed analysis of the origin of the differences in coupling free energies is beyond the scope of this work, but may arise from destabilization of interfacial interactions in longer assemblies. It is possible that coupling free energies may become increasingly unfavorable in longer quadruplex assemblies. The ≈200 nt telomeric overhang could potentially fold into a structure with about 8 contiguous quadruplex units. Larger unfavorable coupling free energies could, however, limit complete folding of the overhang, leaving single-stranded regions that might facilitate or nucleate protein binding.
Figure 6.
Free energy diagram65 illustrating the coupling free energy for folding of three contiguous quadruplexes compared to folding of three individual quadruplex structures.
Our data show that folding of the single-stranded telomeric overhang is energetically favorable with a substantial free energy change. It is important to recognize, then, that any process that requires unfolding of the overhang must somehow overcome this folding free energy. For example, POT1 is thought to bind to the single-stranded overhang as part of the shelterin complex, and its binding is coupled to quadruplex unfolding. Its binding free energy must therefore be large enough to both drive quadruplex unfolding and to stabilize its complex with DNA.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants CA35635 (J.B.C.), GM077422 (J.B.C & J.O.T).) and a MFAG grant of the “Associazione Italiana per la Ricerca sul Cancro”, A.I.R.C. project n° 6255 (L. P.).
References
- (1).McEachern MJ, Krauskopf A, Blackburn EH. Annu Rev Genet. 2000;34:331. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- (2).Rhodes D, Giraldo R. Curr Opin Struct Biol. 1995;5:311. doi: 10.1016/0959-440x(95)80092-1. [DOI] [PubMed] [Google Scholar]
- (3).Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Anal Chem. 2004;76:4490. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- (4).Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, Mailliet P, Dutrillaux B, Boussin FD. Nucleic Acids Res. 2005;33:4182. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Paeschke K, Juranek S, Rhodes D, Lipps HJ. Chromosome Res. 2008;16:721. doi: 10.1007/s10577-008-1222-x. [DOI] [PubMed] [Google Scholar]
- (6).Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Nat Struct Mol Biol. 2005;12:847. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- (7).Lipps HJ, Rhodes D. Trends in cell biology. 2009;19:414. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- (8).Degtyareva NN, Wallace BD, Bryant AR, Loo KM, Petty JT. Biophys J. 2007;92:959. doi: 10.1529/biophysj.106.097451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Oganesian L, Bryan TM. Bioessays. 2007;29:155. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- (10).Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Nucleic Acids Res. 2006;34:5402. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lane AN, Chaires JB, Gray RD, Trent JO. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Neidle S, Parkinson GN. Curr Opin Struct Biol. 2003;13:275. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- (13).Patel DJ, Phan AT, Kuryavyi V. Nucleic Acids Res. 2007;35:7429. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dai J, Carver M, Yang D. Biochimie. 2008;90:1172. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yang D, Okamoto K. Future medicinal chemistry. 2010;2:619. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang Y, Patel DJ. Structure. 1993;1:263. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- (17).Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Nucleic Acids Res. 2007;35:4927. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Nucleic Acids Res. 2007;35:2440. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Phan AT, Kuryavyi V, Luu KN, Patel DJ. Nucleic Acids Res. 2007;35:6517. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xu Y, Noguchi Y, Sugiyama H. Bioorg Med Chem. 2006;14:5584. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- (21).Okamoto K, Sannohe Y, Mashimo T, Sugiyama H, Terazima M. Bioorg Med Chem. 2008;16:6873. doi: 10.1016/j.bmc.2008.05.053. [DOI] [PubMed] [Google Scholar]
- (22).Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. The FEBS journal. 2007;274:3545. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- (23).Parkinson GN, Lee MP, Neidle S. Nature. 2002;417:876. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- (24).Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Nucleic Acids Res. 2005;33:4649. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Heddi B, Phan A. T. n. J Am Chem Soc. 2011;133:9824. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- (26).He Y, Neumann RD, Panyutin IG. Nucleic Acids Res. 2004;32:5359. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. J Am Chem Soc. 2009;131:4301. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Phan AT. The FEBS journal. 2010;277:1107. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- (29).Antonacci C, Chaires JB, Sheardy RD. Biochemistry. 2007;46:4654. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- (30).Chaires J,B. FEBS Journal. 2009;9999 [Google Scholar]
- (31).Ida R, Wu G. J Am Chem Soc. 2008;130:3590. doi: 10.1021/ja709975z. [DOI] [PubMed] [Google Scholar]
- (32).Gray RD, Chaires JB. Nucleic Acids Res. 2008;36:4191. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gray RD, Li J, Chaires JB. J Phys Chem B. 2009;113:2676. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Yu HQ, Miyoshi D, Sugimoto N. J Am Chem Soc. 2006;128:15461. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- (35).Pedroso IM, Duarte LF, Yanez G, Burkewitz K, Fletcher TM. Biopolymers. 2007;87:74. doi: 10.1002/bip.20790. [DOI] [PubMed] [Google Scholar]
- (36).Bai L-P, Hagihara M, Jiang Z-H, Nakatani K. ChemBioChem. 2008;9:2583. doi: 10.1002/cbic.200800256. [DOI] [PubMed] [Google Scholar]
- (37).Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Nucleic Acids Res. 2005;33:5851. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Renciuk D, Kejnovska I, Skolakova P, Bednarova K, Motlova J, Vorlickova M. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Xu Y, Ishizuka T, Kurabayashi K, Komiyama M. Angew Chem. 2009;48:7833. doi: 10.1002/anie.200903858. [DOI] [PubMed] [Google Scholar]
- (40).Sannohe Y, Sato K, Matsugami A, Shinohara K, Mashimo T, Katahira M, Sugiyama H. Bioorg Med Chem. 2009;17:1870. doi: 10.1016/j.bmc.2009.01.051. [DOI] [PubMed] [Google Scholar]
- (41).Wang H, Nora GJ, Ghodke H, Opresko PL. J Biol Chem. 2011;286:7479. doi: 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Petraccone L, Garbett NC, Chaires JB, Trent JO. Biopolymers. 2010;93:533. doi: 10.1002/bip.21392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Petraccone L, Trent JO, Chaires JB. J Am Chem Soc. 2008;130:16530. doi: 10.1021/ja8075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Bauer L, Tluckova K, Tothova P, Viglasky V. Biochemistry. 2011;50:7484. doi: 10.1021/bi2003235. [DOI] [PubMed] [Google Scholar]
- (45).Haider S, Parkinson GN, Neidle S. Biophys J. 2008;95:296. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Haider SM, Neidle S. Biochem Soc Trans. 2009;37:583. doi: 10.1042/BST0370583. [DOI] [PubMed] [Google Scholar]
- (47).Brown PH, Schuck P. Comput Phys Commun. 2008;178:105. doi: 10.1016/j.cpc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Schuck P. SEDFIT (v. 11.3b) National Institutes of Health; 2008. Available from: http://www.analyticalultracentrifugation.com/download.htm. [Google Scholar]
- (49).Hayes DB, Laue T, Philo J. Sedimentation Interpretation Program (version 1.09) University of New Hampshire; Durham, NH: 2006. Available from http://www.jphilo.mailway.com/download.htm. [Google Scholar]
- (50).Hellman LM, Rodgers DW, Fried MG. Eur Biophys J. 2010;39:389. doi: 10.1007/s00249-009-0411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Haq I, Chowdhry BZ, Chaires JB. Eur Biophys J. 1997;26:419. doi: 10.1007/s002490050096. [DOI] [PubMed] [Google Scholar]
- (52).Henry RW, Hofrichter J. In: Methods in Enzymology. Brand L, Johnson ML, editors. Vol. 210. Academic Press; New York: 1992. p. 129. [Google Scholar]
- (53).Freire E, Biltonen R,L. Biopolymers. 1978;17:463. doi: 10.1002/bip.1978.360170512. [DOI] [PubMed] [Google Scholar]
- (54).Freire E;, Biltonen R,L. Biopolymers. 1978;17:481. doi: 10.1002/bip.1978.360170512. [DOI] [PubMed] [Google Scholar]
- (55).Spink CH. Methods in cell biology. 2008;84:115. doi: 10.1016/S0091-679X(07)84005-2. [DOI] [PubMed] [Google Scholar]
- (56).Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179. [Google Scholar]
- (57).Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Biophys J. 2007;92:3817. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Garcia De La Torre J, Huertas ML, Carrasco B. Biophys J. 2000;78:719. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Chirality. 2008;20:431. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- (60).Gottarelli G, Lena S, Masiero S, Pieraccini S, Spada GP. Chirality. 2008;20:471. doi: 10.1002/chir.20459. [DOI] [PubMed] [Google Scholar]
- (61).Karsisiotis AI, Hessari NM, Novellino E, Spada GP, Randazzo A, Webba da Silva M. Angew Chem. 2011 doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- (62).Gray RD, Chaires JB. In: Current protocols in nucleic acid chemistry / Serge L. Beaucage, et al., editors. 2011. Chapter 17, Unit17 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Freire E, Biltonen RL. Biopolymers. 1978;17:497. doi: 10.1002/bip.1978.360170512. [DOI] [PubMed] [Google Scholar]
- (64).Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Nucleic Acids Res. 2006;34:2723. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Weber G. Advances in protein chemistry. 1975;29:1. doi: 10.1016/s0065-3233(08)60410-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.