Abstract
During the last decade, we saw an explosion of studies investigating the role of lysine methylation/demethylation of histones and non-histone proteins, such as p53, NF-kappaB, and E2F1. These ‘Ying-Yang’ post-translational modifications are important to fine-tuning the activity of these proteins. Lysine methylation and demethylation are catalyzed by protein lysine methyltransferases (PKMTs) and protein lysine demethylases (PKDMs). PKMTs, PKDMs, and their substrates have been shown to play important roles in cancers. Although the underlying mechanisms of tumorigenesis are still largely unknown, growing evidence is starting to link aberrant regulation of methylation to tumorigenesis. This review focuses on summarizing the recent progress in understanding of the function of protein lysine methylation, and in the discovery of small molecule inhibitors for PKMTs and PKDMs. We also discuss the potential and the caveats of targeting protein lysine methylation for the treatment of cancer.
Keywords: methylation, demethylation, methyltransferase, demethylase, cancer, epigenetics
Introduction
Protein lysine methylation has gained tremendous attention since the discovery of SUV39H1 as the first histone lysine methyltransferase in 2000 [1]. Following the discovery, numerous proteins have been found to possess methyltransferase activity, such as G9a/GLP [2,3], MLLs [4], EZH2 [5], SET2 [6], SET7/9 [7], DOT1 [8,9], and PR-SET7 (also known as SETD8) [10]. These enzymes catalyze the transfer of methyl group(s) from the co-factor S-adenosyl-L-methionine (SAM) to the lysine residues of histones. More recently, many non-histone proteins have been identified as substrates for these enzymes, hence the name protein lysine (K) methyltransferases (PKMTs). Apart from lysine methylation, arginine methylation also exists. The history of arginine methylation was recently surveyed [11] and is not the subject of this review. Here we will focus on the implications of PKMTs in cancers and the recent progress in the discovery of selective PKMT inhibitors. It is worth pointing out that PKMTs play important roles in other biological processes including developmental biology and stem cell differentiation. Like other protein modifications, lysine methylation is also subject to its counter modification, demethylation. For histones, the first reported demethylase is lysine-specific demethylase 1 (LSD1, also known as BHC110) [12,13]. However, LSD1 can only demethylate mono- or dimethylated lysines. Shortly after the discovery of LSD1, a second family of enzymes, JmjC domain containing proteins, was found to have demethylation activity for trimethylated, as well as mono- and dimethylated lysines [14]. These enzymes are referred to as protein lysine demethylases (PKDMs). The substrate specificity and potential biological functions of these enzymes have recently been reviewed [11,15]. In this review, we aim to describe the newly discovered inhibitors for PKDMs.
Proteins that are Subject to Lysine Methylation
Lysine methylation has been extensively studied on histones. The potential roles of histone modifications in cancers have been reviewed recently [16]. Besides histone methylation, non-histone methylation is also implicated in tumorigenesis. p53 is arguably the most extensively studied protein for lysine methylation. It is methylated by SET7/9, SMYD2, SETD8, and G9a/GLP at K372, K370, K382, and K373, respectively (Fig. 1) [17–21]. The number of non-histone proteins that are methylated at lysine residues is growing rapidly. DNA methyltransferase 1 (DNMT1) was reported to be methylated by SET7/9 [22] at K142. However, another report concluded that the methylation site of DNMT1 by SET7/9 was at K1094, which was subject to demethylation regulation by LSD1 [23]. It is currently unclear what causes the discrepancy between these two reports. A similar ‘Ying-Yang’ action was also observed on STAT3 and E2F1. STAT3 K140 methylation is carried out by SET7/9 and this methylation is reversed by LSD1 [24]. Similarly, SET7/9 also methylates E2F1 at K185 while LSD1 demethylates at this site [25]. An interesting note is that SET7/9 can carry out dimethylation on STAT3 as well as on p53 in vitro, but has been shown to be a strict mono-methylase for histones according to the X-ray structural data [26]. NF-kappaB subunit, p65, is methylated at K218 and K221 by NSD1 and demethylated by FBXL11 [27]. In addition, another report showed that p65 was methylated by SET7/9 at K37 [28]. Another NF-kappaB subunit, RelA, is methylated by SET7/9 at K314 and K315 [29] and by SETD6 at K310 [30]. Nuclear receptor, estrogen receptor alpha, is methylated by SET7/9 at K302 [31]. Besides the proteins described here, numerous other proteins, for example, Rubisco in plant and cytochrome c in yeast, that are subject to methylation are summarized in a previous review [11]. However, the biological function of the lysine methylation of these proteins is not related to cancers.
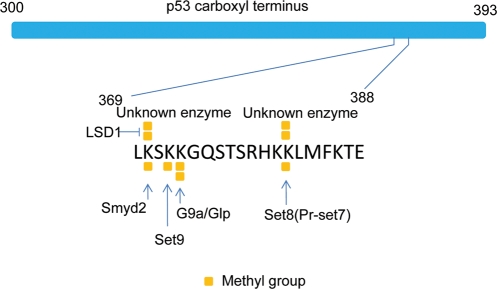
Figure 1.
Lysine methylation sites of p53 as an example of complex protein methylation Scheme showing lysine methylation sites of p53 carboxyl terminus (amino acid residues from 300 to 393). Enzymes that carry out dimethylation at K370 and K382 are unknown. However, dimethylation at these two sites has been detected using mass spectrometry analysis [20]. The existence of a di-methylase of K370 was also predicted based on western blot analysis [18]. Square blocks represent methyl groups.
It is worth noting that most of these studies are based on overexpression of enzymes or substrates in cells. Therefore, the physiological roles of these lysine methylation marks are still elusive. For example, p53 knock-in mice with seven (p537KR: lysines 367, 369, 370, 378, 379, 383 and 384 in mouse) or six (p53K6R: lysines 367, 369, 370, 378, 379 and 383 in mouse) lysine-to-arginine mutations at its carboxyl terminus develop normally and show little defect in p53-mediated damage response [32–34]. This is in drastic contrast to the results observed in the overexpression studies. It is possible that methylation, similar to acetylation, only plays a fine-tuning role in the regulation of the activity of p53. Particularly, two recent studies on SET7/9 knockout cast doubt on the role of K372 methylation in regulating the biological function of p53 [35,36]. Future studies are needed to address these discrepancies.
Potential Biological Functions of PKMTs in Cancers
SUV39H1
SUV39H1 and its homolog SUV39H2 are required for heterochromatin formation. Double knockout of SUV39h1 and SUV39h2 mice are subject to genomic instability [37]. SUV39h1-dependent senescence has been shown to protect mice from Ras-driven invasive T-cell lymphoma [38]. Based on these studies, SUV39H1 appears to play a tumor-suppressive function. Controversially, SUV39H1-mediated H3K9me has been linked to gene silencing of the tumor suppressor genes, such as p15INK4B and E-cadherin, in acute myeloid leukemia (AML) [39]. Therefore, it is highly possible that the default function of SUV39H1 is to maintain genome stability by limiting the acute activation of oncogenes while its dysregulation could cause tumor formation.
EZH2
EZH2 is one of the first PKMTs implicated in human cancers [40,41]. Its expression is highly correlated with the metastasis of various cancers, such as prostate and breast cancers. EZH2 is the enzymatic subunit of polycomb repressive group 2 (PRC2) that methylates histone H3 at K27 [42]. However, the underlying mechanisms of oncogenic effect of EZH2 are not fully understood. It is also unclear whether H3K27 methylation is required for the role of EZH2 in tumorigenesis since EZH2 may have other substrates beyond histone H3 [43]. Nevertheless, ablation of EZH2 in tumor cells using RNA interference technology has demonstrated that EZH2 is a promising drug target to treat cancers [40].
DOT1L
DOT1L performs H3K79 methylation, a modification that is associated with transcription elongation. One of its pathological roles is the mis-regulation of the hox gene expression through interacting with AF9, a fusion partner of mixed lineage leukemia (MLL). The mis-regulation can lead to leukemogenesis [44].
SMYD2
Another promising therapeutic target for cancer is SMYD2. A recent paper has shown that SMYD2 is involved in maintaining an un-differentiated status of MLL-AF9-induced acute myeloid leukemia (AML) [45]. Although the mechanism underlying this leukemia maintenance is unclear, SMYD2 has been shown to methylate p53 [18] and Rb [46], two of the most important tumor suppressors. In addition, SMYD2 is reportedly overexpressed in esophageal squamous cell carcinoma [47]. Knockout mice for SMYD2 have been generated [48]. Future work needs to address whether SMYD2 knockout mice are resistant to tumorigenesis in response to oncogenic insults at various tissues. In addition, the epigenetic role of SMYD2 in cells is largely unknown. One report showed that SMYD2 methylates histone H3K36 [49], while another report suggested that SMYD2 is an H3K4 methyltransferase [50]. These observations merit further studies to elucidate the mechanisms underlying this dual-substrate specificity.
G9a and GLP
G9a and GLP belong to one new group of methyltransferases that methylate p53. They have a wide range of biological and pathological functions. From the cancer perspective, G9a and GLP regulate the apoptotic function of p53. The dimethylation of p53K373 by G9a and GLP decreases the transcriptional activity of p53. Interrogation of Oncomine database reveals that G9a is overexpressed in various tumors, further suggesting its oncogenic effects. Indeed, the overexpression of G9a was shown to increase metastasis and invasion in lung cancer [51]. However, the ultimate outcome of inhibition of G9a/GLP could be complicated by the fact that they methylate other histone and non-histone substrates [52]. G9a and GLP are largely responsible for H3K9 mono- and dimethylation. Recently, a distinguishing feature of cancer cell lines, i.e. the loss of G9a-dependent large block of H3K9me2, was observed [53]. This suggests that the loss of G9a activity or its substrate H3K9me2 confers growth or survival advantage to cancer cells. Therefore, whether the inhibition of G9a/GLP can decrease the growth or increase the apoptosis of cancer cells requires further investigation.
Potential Biological Functions of PKDMs in Cancers
LSD1 and LSD2
LSD1 may serve as a viable target for therapeutic intervention in cancers. It decreases the activity of p53. However, recent studies have uncovered some controversial roles of LSD1. LSD1 has been shown to demethylate p53 and decrease the apoptotic effect of p53, suggesting that it can act as an oncogene. Indeed, the overexpression of LSD1 is observed in prostate cancer [54] and also correlates with poor prognosis of neuroblastoma [55]. Several reports have also shown that LSD1 has a potential role in the repression of E-cadherin, a molecule mediating the cell–cell junction, and cell migration [56–58]. The expression of E-cadherin is inversely correlated with metastasis. All of these studies suggest that LSD1 is a putative onco-protein. However, one report proposes that LSD1 can suppress the metastasis of breast cancer by repressing tumor growth factor (TGF)-beta1 signaling [59]. The exact cause of this controversy is at present unknown. It is possible that the function of LSD1 is regulated by other binding partner(s) and the ultimate effect is context-dependent. LSD2, the homolog of LSD1, was recently shown to have demethylation activity toward H3K4me2 [60]. So far, there is no report to indicate that LSD2 plays a role in tumorigenesis.
JMJD2c
JMJD2c (also known as GASC1) is a member of the JmjC domain-containing protein family. The members of this family, as described above, can demethylate mono-, di- and/or trimethylated lysines. JMJD2c was characterized as an H3K9me3/me2 demethylase. The overexpression of JMJD2c was observed in esophageal squamous carcinoma [61]. It regulates androgen receptor-mediated gene expression [62]. Therefore, it could also play an important role in androgen receptor-dependent prostate cancer, although this hypothesis needs to be tested formally. Very recently, functional interplay between JMJD2c and JAK2, a histone tyrosine kinase [63], has been revealed in B cell lymphoma and Hodgkin's lymphoma [64]. This finding provides a mechanistic rationale for testing the co-inhibition of JAK2 and JMJD2c in cancers. It is worth pointing out that JMJD2c and LSD1 also cooperate in androgen receptor-regulated gene expression [62]. This observation fuels the idea of investigating the effects of LSD1 and JMJD2c inhibition in prostate cancer.
PLU-1
PLU-1 is another member of the JmjC domain-containing protein family. It has H3K4me demethylation activity [65]. Overexpression of PLU-1 has been specifically detected in breast cancer cells and its inhibition by RNA interference decreased the proliferation of breast cancer cells [65,66]. This offers a unique opportunity to treat breast cancer through targeting PLU-1, although it is currently unclear whether PLU-1 plays a role in other types of cancer.
PKMT Inhibitors
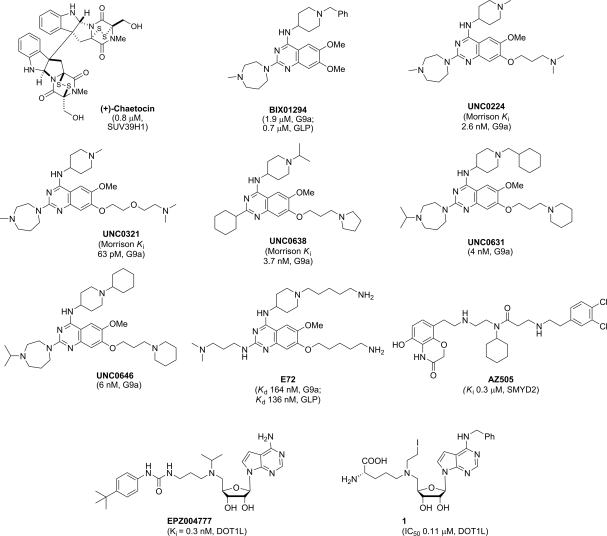
Since Greiner et al. [67] discovered the first selective, non-nucleoside inhibitor of recombinant Drosophila Su(var)3–9, chaetocin (IC50 = 0.6 µM) in 2005, the inhibitor discovery effort has quickly gained momentum and a number of new inhibitors have emerged. Herein, we describe selective PKMT inhibitors with an emphasis on most recently discovered compounds (Fig. 2). PRMT inhibitors were recently surveyed and are not the subject of this review [68]. Chaetocin was found to inhibit SUV39H1 (IC50 = 0.8 µM), the human ortholog of Drosophila Su(var)3–9, as well as other H3K9 PKMTs, including Neurospora DIM5 (IC50 = 3.0 µM) and mouse G9a (IC50 = 2.5 µM). It was selective over non-H3K9 PKMTs, such as H3K27 PKMT dE(z) complex, H3K4 PKMT SET7/9, and H4K20 PKMT SETD8 [67] (IC50 dE(z) complex>90 µM; SET7/9 and SETD8>180 µM). Interestingly, a total synthesis report found natural (+)- and synthetic (−)-chaetocin to be equipotent against G9a (IC50 = 2.4 and 1.7 µM, respectively) [69]. In addition, it was found that chaetocin inhibited thioredoxin reductase [70]. Like other members of the epidithiodiketopiperazine class [71], chaetocin is cytotoxic. Despite its cytotoxicity, it was reported that chaetocin-treated Drosophila SL-2 cells at an inhibitor concentration of 0.5 µM showed marked reduction in cellular levels of di- and trimethylated H3K9 with no apparent changes in cellular levels of methylation of other lysines (H3K27, H3K36, H3K79, and H3K4) [67].
Figure 2.
Structures of select PKMT inhibitors
Discovery of BIX01294, the first selective small molecule inhibitor for G9a and GLP, by Jenuwein and co-workers [72] was an important advance in the PKMT inhibitor discovery field, as this compound was the first PKMT inhibitor that blocks protein–protein interactions [73]. BIX01294 had good in vitro potency against G9a and GLP and was selective over other H3K9 PKMTs (SUV39H1 and SETDB1), H3K4 PKMT SET7/9 [73]. Unfortunately BIX01294 was toxic in cellular assays at concentrations above 4.1 µM. Mechanistically, unlike chaetocin, BIX01294 did not inhibit G9a in a SAM-competitive manner but rather occupied the histone peptide binding pocket, as evidenced by the X-ray crystal structure of BIX01294 and GLP in the presence of SAH (S-adenosyl-l-homocysteine) [73,74]. This crystal structure revealed that BIX01294 did not bind in the SAM-binding site nor did it interact with the lysine binding channel [73].
By elaborating the 7-methoxy moiety of the quinazoline template, it was discovered that a series of new analogs interacted with the lysine-binding channel, including UNC0224, a 7-fold more potent G9a inhibitor (IC50 = 15 nM) in the G9a ThioGlo assay when compared with BIX01294 (IC50 = 106 nM) [75,76]. A high-resolution (1.7 Å) X-ray co-crystal structure of G9a and UNC0224 (PDB: 3K5K) showed that the 7-dimethylamino propoxy side chain of UNC0224 only partially occupied the lysine binding channel of G9a [73,76], and thus space remained to accommodate a longer side chain or larger amino-capping group. The most potent G9a inhibitor to date, UNC0321 was a result of further side chain manipulations [77]. Because UNC0321 likely reached the detection limits of the biochemical assays, Morrison Ki values were determined using an endoproteinase-coupled microfluidic capillary electrophoresis assay [78]. UNC0321 (Morrison Ki = 63 pM) was about 40-fold more potent than UNC0224 (Morrison Ki = 2.6 nM) and 250-fold more potent than BIX01294 (Morrison Ki = 16 nM) [77].
While highly potent in biochemical assays, UNC0321 was less potent in cellular assays in comparison with BIX01294, prompting the development of analogs with higher cellular potency. UNC0638, specifically designed to increase lipophilicity and cell membrane permeability while maintaining high in vitro potency, was indeed found to have excellent in vitro potency (Morrison Ki G9a = 3.7 nM; Ki = 3.0 nM) and was >100-fold selective over a wide range of epigenetic and non-epigenetic targets [79]. Mechanism of action studies revealed that UNC0638 was competitive with the peptide substrate and non-competitive with the co-factor SAM. The MOA findings were confirmed by X-ray crystal structure of the G9a–UNC0638–SAH complex (2.56 Å resolution; PDB: 3RJW). The combination of high potency, excellent selectivity, low cell toxicity and robust on-target activities in cells makes UNC0638 an excellent chemical probe of G9a/GLP for cellular studies. Most recently, UNC0646 and UNC0631, close analogs of UNC0638, were reported to have comparable cellular potency and toxicity and could serve as alternative tool compounds for investigating specific cellular systems [80]. For example, UNC0646 had an outstanding toxicity/function ratio in MCF7 (470), 22RV1 (510), and IMR90 (360) cells, making this compound potentially more useful for studying G9a biology in these specific cell lines [80].
Chang et al. [81] also developed a potent G9a and GLP inhibitor E72 based on the quinazoline template with binding affinities determined by isothermal titration calorimetry (Kd GLP = 136 nM; G9a = 164 nM). A brief selectivity study showed E72 was inactive against SUV39H2 with no inhibition at 5 µM [81]. The X-ray co-crystal structure of the GLP–E72 complex in the presence of SAH (2.19 Å, PDB: 3MO5) showed that E72 analogous to UNC0224 with G9a occupies both the surface of the peptide binding groove and the lysine binding channel [81]. In three separate cell types, E72 was significantly less toxic than BIX01294 at compound concentrations of 10 µM and was able to reactivate K-ras-mediated epigenetic silencing of the Fas gene in NIH 3T3 cells [81].
Most recently, Daigle et al. [82] discovered a highly potent and selective SAM-competitive DOT1L inhibitor EPZ004777 (Ki = 0.3 nM), which is a co-factor product (SAH) mimic. EPZ004777 was>1000-fold selective for DOT1L over CRM1, EHMT2, EZH1, EZH2, PRMT1s, PRMT5, PRMT8, SETD7, and WHSC1, all of which are SAM-utilizing methyltransferases. EPZ004777 was found to selectively kill off cells bearing MLL translocation. In addition, in vivo administrations of EPZ004777 led to extension of survival in a mouse MLL xenograft model [82]. Subsequently, Yao and co-workers showed that protecting the N6 position in SAH does not affect binding to DOT1L, but seems to instill selectivity against CARM1, PRMT1, G9a, and SUV39H2. The best compound in their series, compound 1 had an IC50 of 0.11 µM against DOT1L and was proposed to be capable of covalently binding to the histone [83].
Recently Ferguson et al. [84] described a potent inhibitor of SMYD2, AZ505 (Ki = 0.30 µM) identified by high-throughput screening. As seen from its crystal structure in complex with SMYD2, AZ505 occupies the peptide binding groove and is peptide substrate competitive. Furthermore, this compound was shown to be>83 fold selective for SMYD2 over a panel of PKMTs (SMYD3, DOT1L, EZH2, GLP, G9a, and SET7/9) [84]. Given the broad roles of SMYD2 in cancers, it will be interesting to test the effect of this inhibitor and its analogs in cancer cells.
PKDM Inhibitors
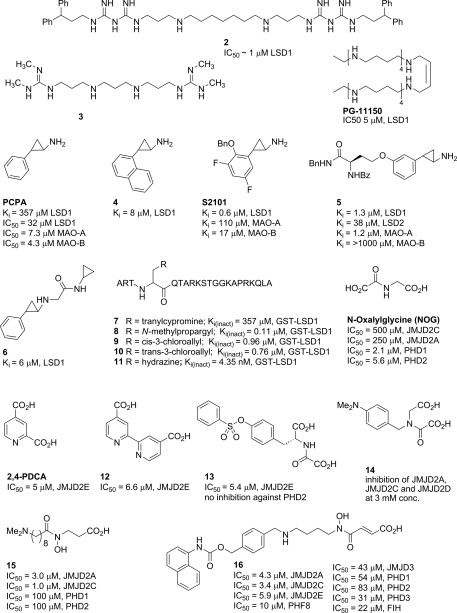
Several groups have investigated polyamine analogs for their ability to inhibit recombinant LSD1 in vitro and in vivo. Among these compounds, the best were polyamines 2 and 3 (Fig. 3), which inhibited LSD1 activity by 85% and 82%, respectively, at 10 µM [85,86]. Subsequently, Huang et al. [87] investigated polyamines, such as PG11150 (IC50 = 5 µM), as inhibitors of LSD1. It was found that colorectal cancer cells treated with polyamine PG11150 display re-expression of multiple aberrantly silenced tumor suppressor genes. In addition, PG-11144 the trans isomer of PG11150, displays a marked decrease in tumor growth and increases H3K4me2 levels in the mouse xenograft model, without significant overall toxicity, when administered in combination treatments alongside a known DNMT inhibitor 5-azacytidine. Interestingly, when used alone PG-11144 displayed antitumor activity, while polyamine 2 did not produce a similar effect without the accompanying DNMT inhibitor. However, selectivity of these inhibitors for LSD1 over monoamine oxidase (MAO)-A, MAO-B and the newly discovered LSD2 is yet to be addressed.
Figure 3.
Structures of select PKDM inhibitors
A high degree of homology exists between the catalytic sites of MAO-A, B, and LSD1; thus, one might reasonably expect that many of the existing monoamine oxidase inhibitors could inhibit LSD1. Recognizing this, researchers tested a known non-selective MAO inhibitor PCPA (trans-2-phenylcyclopyropylamine) and indeed found it to have LSD1 inhibitory activity (Ki = 357 µM; LSD1) [88,89]. MAO and LSD inhibition is fairly insensitive to stereochemistry as has been shown [90,91]; thus (±)-racemates of trans tranylcypromine and its derivatives are commonly used for testing and in the clinic [89]. Inhibition of LSD1 by tranylcypromine has been shown to proceed via ring-opening of the cyclopropyl moiety followed by formation of a covalent adduct with the C(4) of the FAD co-factor. PCPA displays no apparent selectivity for LSD1 over MAO-A, or MAO-B, which prompted a number of groups to investigate LSD1-selective derivatives based on its core structure. Inhibitors based on the tranylcypromine scaffold include compound 4 (Ki = 8 µM) discovered in 2008 [92], S2101 (Ki = 0.6 µM, LSD1; Ki = 110 µM, MAO-A; Ki = 17 µM, MAO-B) in 2010 [93], compound 5 (Ki = 1.9 µM, LSD1; Ki = 290 µM, MAO-A) in 2009 [94], and compound 6 (Ki = 6 µM, LSD1) in 2010 [90]. These inhibitors affect histone H3K9 and H3K4 methylation levels in cells, mediated by the inhibition of LSD1-catalyzed demethylation.
Culhane et al. incorporated tranylcypromine, propargylamine, cis- and trans-chlorovinyl, and hydrazine functionalities as warheads in their peptide scaffold (compounds 7–11) to study LSD1 inhibition by H3 peptides. Through their studies it has been determined that chlorovinyl-H3 (compound 8) is a mechanism-based LSD1 inactivator, while endo-cyclopropylamine-H3 (compound 7) did not show time-dependent inactivation. The best suicide inhibitor hydrazine-H3 (compound 11, Ki(inact) = 4.35 nM, GST-LSD1) was 20-fold more potent than the propargylamine-H3 derivatives [88].
The functional interaction between LSD1 and HDAC has been reported [95]. Lee et al. [95] found that LSD1 and HDAC enhanced the activity of each other. Given the fact that HDAC inhibitors have already been approved for cutaneous T-cell lymphoma treatment, this molecular mechanism raises an interesting hypothesis that LSD1 and HDAC inhibitors could cooperatively inhibit tumorigenesis.
JMJD2 demethylases are inhibited by analogs of the co-factor 2-OG, including N-oxalylglycine (NOG), pyridine dicarboxylate, and the related bipyridyl derivative 12. Other chemotypes that are also presumed to bind to the active site Fe(II) include catechols, hydroxamic acids, and tri-carboxylic acid cycle intermediates, such as succinate and fumarate [61,96,97]. Inhibitors 13 and 14, designed on the basis of the crystal structure of NOG in complex with JMJD2A (PDB ID 2OQ6), are equipped with an appendage intended to engage a large subpocket adjacent to the active site. Hamada et al. [98] have demonstrated inhibition of JMJD2A, 2C and 2D activity by 14 and its analogs in vitro and in vivo. Importantly, only the methyl ester prodrug of 14 was active in cellular assays presumably due to poor cell permeability of the free acid-containing analogs. Rose et al. [99] produced a crystal structure of an analog of their best inhibitor 13 in complex with JMJD2A (PDB ID 2WWJ) confirming the predicted binding mode for their N-oxalyl-d-tyrosinyl derivatives. Compound 13 exhibited some selectivity against prolyl hydroxylase domain-containing protein 2 (PHD2) in biochemical assays, while 14 appeared to inhibit other Fe(II)/α-ketogluterate-dependent oxygenases indiscriminantly [98]. Interestingly, while NOG itself selectively inhibits PHD1/2 over JMJD2C/2A, its analog 15 is selective for JMJD2C and JMJD2A [100]. This selectivity is presumably due to the presence of a methylene group next to the carbonyl of the hydroxamate moiety, and its affinity for JMJD2 is dependent on the length of the linker leading to the tertiary amino group. Another JMJD-selective inhibitor was recently discovered [101]. Compound 16 was designed to incorporate both a substrate mimic and a methyllysine mimic. This inhibitor is more than 9000-fold selective for the Jumonji C domain-containing enzymes over PHDs. Its methyl ester prodrug methylstat is cell active and may have potential for anticancer chemotherapy [101].
A high-throughput assay based on time-resolved fluorescence resonance energy transfer was reported recently to screen the inhibitors for LSD1 and JMJD2c [102]. Numerous inhibitors for these two enzymes have been identified through the assay. Because LSD1 and JMJD2c cooperate in regulating the gene expression in prostate cancer, these inhibitors will be extremely valuable for testing the synergistic effects of co-inhibition of these two enzymes in cancer. Because of the common mechanism underlying the demethylation reaction of JmjC domain-containing proteins, the specificity of these inhibitors needs to be rigorously tested in the future.
Discussion
Given the critical roles of PKMTs and PKDMs in cancers, it is very likely that inhibitors of these enzymes will move forward into clinical trials. However, one must keep in mind the caveat that a single PKMT or PKDM may target multiple substrates as described above, potentially leading to opposite biological effects depending on the context. For example, LSD1 demethylates and inactivates p53 [19]. Further, LSD1 participates in Snail-mediated silencing of E-cadherin [57], whose down-regulation generally correlates with poor prognosis of tumor and metastasis. Both these observations suggest that LSD1 could have an oncogenic effect. However, another report showed that LSD1 could repress breast cancer metastasis by repressing TGF-beta1 expression [59], indicating that LSD1 could also behave as a tumor suppressor. In the same way, LSD1 demethylates and stabilizes E2F1, an apoptosis driver [25]. These results suggest that LSD1 could also have proapoptotic function. Therefore, it is highly possible that the biological effect of inactivation of an enzyme is context dependent. The same is true for G9a, GLP, and SUV39H1 as discussed above. In addition, it is worth noting that some PKMTs or PKDMs are subunits of mega complexes. For example, LSD1, G9a, and GLP are subunits of the CtBP complex [56]. Inhibition of any component of this mega complex may lead to unexpected side-effects. Therefore, caution needs to be taken when extrapolating in vitro data to clinical practice. In addition, many of the enzymes discussed earlier act not only on histone substrates but also on non-histone proteins. However, to our best knowledge, the interplay between histone and non-histone methylations has not been effectively addressed in a well-defined system. Nevertheless, inhibitors of PKMTs or PKDMs, like HDAC and DNMT inhibitors, are bound to eventually become a new generation of epigenetic drugs for cancer treatment.
Funding
This work was supported by the grants from the Intramural Research Program at the Center for Cancer Research (CCR), the National Cancer Institute and the National Institutes of Health, USA for J.H., the University Cancer Research Fund (UCRF) and the Carolina Partnership from the University of North Carolina at Chapel Hill for J.J. and I.K.
References
- 1.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 2.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 5.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 8.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 9.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 10.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 14.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 15.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 16.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 20.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 27.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, et al. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune p53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 34.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campaner S, Spreafico F, Burgold T, Doni M, Rosato U, Amati B, Testa G. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol Cell. 2011;43:681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J, Rossi FM. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell. 2011;43:673–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 38.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 39.Lakshmikuttyamma A, Scott SA, DeCoteau JF, Geyer CR. Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene. 2010;29:576–588. doi: 10.1038/onc.2009.361. [DOI] [PubMed] [Google Scholar]
- 40.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 41.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 44.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 45.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saddic LA, West LE, Aslanian A, Yates JR, 3rd, Rubin SM, Gozani O, Sage J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko SY, et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- 48.Diehl F, Brown MA, van Amerongen MJ, Novoyatleva T, Wietelmann A, Harriss J, Ferrazzi F, et al. Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS One. 2010;5:e9748. doi: 10.1371/journal.pone.0009748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 52.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 55.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 56.Shi Y, Sawada J, Sui G, Affarel B, Whetstine JR, Lan F, Ogawa H, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 57.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial–mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 60.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 62.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 63.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, Wright GW, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 66.Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- 67.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 68.Yost JM, Korboukh I, Liu F, Gao C, Jin J. Targets in epigenetics: inhibiting the methyl writers of the histone code. Curr Chem Genomics. 2011;5(Suppl 1):72–84. doi: 10.2174/1875397301005010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwasa E, Hamashima Y, Fujishiro S, Higuchi E, Ito A, Yoshida M, Sodeoka M. Total synthesis of (+)-chaetocin and its analogues: their histone methyltransferase G9a inhibitory activity. J Am Chem Soc. 2010;132:4078–4079. doi: 10.1021/ja101280p. [DOI] [PubMed] [Google Scholar]
- 70.Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC. The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxid Redox Signal. 2009;11:1097–1106. doi: 10.1089/ars.2008.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gardiner DM, Waring P, Howlett BJ. The epipolythiodioxopiperazine (ETP) class of fungal toxins: distribution, mode of action, functions and biosynthesis. Microbiology (UK) 2005;151:1021–1032. doi: 10.1099/mic.0.27847-0. [DOI] [PubMed] [Google Scholar]
- 72.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 73.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 75.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 76.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, et al. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem. 2009;52:7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem. 2010;53:5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wigle TJ, Provencher LM, Norris JL, Jin J, Brown PJ, Frye SV, Janzen WP. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis. Chem Biol. 2010;17:695–704. doi: 10.1016/j.chembiol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu F, Barsyte-Lovejoy D, Allali-Hassani A, He Y, Herold JM, Chen X, Yates CM, et al. Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J Med Chem. 2011;54:6139–6150. doi: 10.1021/jm200903z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, et al. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J Mol Biol. 2010;400:1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yao Y, Chen P, Diao J, Cheng G, Deng L, Anglin JL, Prasad BV, et al. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. J Am Chem Soc. 2011;133:16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 85.Sharma SK, Wu Y, Steinbergs N, Crowley ML, Hanson AS, Casero RA, Woster PM. (Bis)urea and (bis)thiourea inhibitors of lysine-specific demethylase 1 as epigenetic modulators. J Med Chem. 2010;53:5197–5212. doi: 10.1021/jm100217a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J Am Chem Soc. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 90.Benelkebir H, Hodgkinson C, Duriez PJ, Hayden AL, Bulleid RA, Crabb SJ, Packham G, et al. Enantioselective synthesis of tranylcypromine analogues as lysine demethylase (LSD1) inhibitors. Bioorg Med Chem. 2011;19:3709–3716. doi: 10.1016/j.bmc.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 91.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 92.Gooden DM, Schmidt DM, Pollock JA, Kabadi AM, McCafferty DG. Facile synthesis of substituted trans-2-arylcyclopropylamine inhibitors of the human histone demethylase LSD1 and monoamine oxidases A and B. Bioorg Med Chem Lett. 2008;18:3047–3051. doi: 10.1016/j.bmcl.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010;49:6494–6503. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- 94.Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J Am Chem Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 95.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J Med Chem. 2008;51:7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- 97.Sakurai M, Rose NR, Schultz L, Quinn AM, Jadhav A, Ng SS, Oppermann U, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases. Mol Biosyst. 2010;6:357–364. doi: 10.1039/b912993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, Janknecht R, et al. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett. 2009;19:2852–2855. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- 99.Rose NR, Woon EC, Kingham GL, King ON, Mecinovic J, Clifton IJ, Ng SS, et al. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches. J Med Chem. 2010;53:1810–1818. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, et al. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J Med Chem. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- 101.Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, Che KH, et al. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. J Am Chem Soc. 2011;133:9451–9456. doi: 10.1021/ja201597b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu V, Fisch T, Long AM, Tang J, Lee JH, Hierl M, Chen H, et al. High-throughput TR-FRET assays for identifying inhibitors of LSD1 and JMJD2C histone lysine demethylases. J Biomol Screen. 2011 doi: 10.1177/1087057111418228. in press. [DOI] [PubMed] [Google Scholar]