Abstract
Three recent papers (Gu et al., 2009; Claycomb et al., 2009; van Wolfswinkel et al., 2009) provide evidence that links a new class of small RNAs and Argonaute-associated complexes to centromere function and genome surveillance.
RNA interference plays a conserved role in gene regulation and in maintaining the stability of repetitive DNA sequences associated with transposons and retro-elements. Small RNAs interact with target RNAs by base pairing interactions and promote translational inhibition, degradation of complementary RNAs, or chromatin modifications (Malone and Hannon, 2009). Several classes of small RNAs are described in plants, flies, yeast, and mammals. Small RNAs often target repetitive sequences and transposable elements as well as pseudogenes and other aberrant transcripts. In many organisms, repetitive sequences flank centromeric DNA regions. For example, in the fission yeast Schizosaccharomyces pombe, small RNAs have been implicated in establishment of pericentric heterochromatin, which is essential for proper centromere function and chromosome segregation (Moazed, 2009). In this issue of Molecular Cell, Gu et al. (2009) describe a new class of Caenorhabditis elegans small RNAs (22G-RNAs), which associate with two distinct Argonaute proteins, a worm-specific clade of Argonaute proteins (WAGOs) and CSR-1, and have been implicated in silencing of transposons and repetitive elements as well as chromosome segregation. Two related papers published in Cell (Claycomb et al., 2009; van Wolfswinkel et al., 2009) provide evidence for the involvement of 22G-RNA/CSR-1 complexes in the assembly and function of holocentric centromeres.
Transposons are selfish DNA elements that exploit the genome in order to survive and proliferate. Movements of transposons within the genome can result in genetic instability. Cells have therefore evolved several mechanisms to silence transposons and prevent their proliferation. In Drosophila and other multicellular eukaryotes, piRNA-based mechanism defends the germline against transposons. Previous studies in C. elegans have implicated 21U-RNAs associated with PRG-1 in transposon silencing in an analogous manner to piRNAs. However, silencing of most transposons in C. elegans depends on WAGOs, which associate with the newly described 22G class of small RNAs, as reported by Gu and colleagues (Gu et al., 2009). The 22G-RNAs are abundantly expressed in the germline and are maternally deposited in embryos. Germline cells are particularly sensitive to transposition, which can be passed to the next generation, thus endangering genetic stability, and it seems that WAGO-associated 22G-RNAs are an important part of a germline surveillance system that silences transposons and aberrant transcripts (Figure 1).
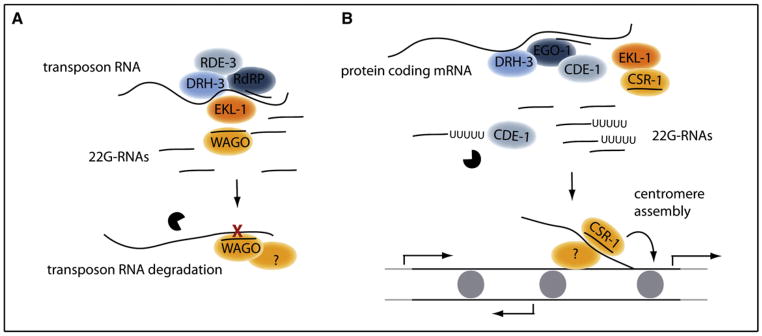
Figure 1. Transposon Silencing and Centromere Assembly Mediated by 22G-RNAs.
(A) 22G-RNAs generated from transposons and repetitive sequences are loaded onto WAGOs and promote RNA degradation.
(B) CSR-1, loaded with 22G-RNAs generated from protein-coding mRNAs, targets chromatin and promotes centromere assembly. Gray circles indicate centromere-specific histone H3 (cenH3). Arrows indicate protein-coding RNA transcription. See text for details.
In S. pombe, centromeric transcripts become targets of Dicer and RNA-dependent RNA polymerase (RdRP). The fission yeast RdRP, Rdp1, is associated with two conserved proteins, RNA helicase Hrr1 and nucleotidyltransferase Cid12, in a functional RdRP complex (RDRC) (Motamedi et al., 2004). siRNAs are generated by Dicer from double-stranded precursor and loaded onto Argonaute in the RNA-induced transcriptional silencing (RITS) complex, which targets centromeric transcripts to establish heterochromatin and stabilize centromeres (Moazed, 2009; Verdel et al., 2004). Although 22G-RNAs are produced independently of Dicer, their biogenesis displays remarkable similarity to the biogenesis of S. pombe siRNAs. 22G-RNA generation involves two C. elegans RdRPs, RRF-1 and EGO-1, and an RNA helicase, DRH-3, which most likely helps to unwind the template RNA and thus increases the RdRP processivity. Furthermore, nucleotidyltransferase RDE-3, a homolog of Cid12, is required for the generation of 22G-RNAs. RDE-3 and Cid12 may have a similar role in the RDRC that involves controlling RdRP activity or recognizing aberrant transcripts to initiate silencing. However, RDE-3 might also be involved in sorting of 22G-RNAs into distinct RNAi pathways in C. elegans. The exact role of RDE-3 in the generation of 22G-RNAs remains unclear.
Several nucleotidyltransferases have been implicated in RNAi in C. elegans (Chen et al., 2005). Besides RDE-3, biogenesis of 22G-RNAs involves another nucleotidyltransferase (CDE-1), which is associated with RdRP EGO-1 and, contrary to RDE-3, promotes the degradation of a subclass of 22G-RNAs by uridinylation. This subclass of 22G-RNAs is EGO-1-dependent and is derived exclusively from protein-coding loci from all chromosomes, as reported by van Wolfswinkel, Claycomb, and colleagues (Claycomb et al., 2009; van Wolfswinkel et al., 2009). The subclass of small RNAs is loaded onto the Argonaute protein CSR-1 and does not downregulate its protein-coding targets. In CDE-1 mutants, CSR-1 22G-RNAs accumulate and may be loaded onto other Argonaute proteins like WAGOs, which promote target degradation. Therefore, it is critical that small RNAs are loaded only onto their specific Argonaute proteins, and CDE-1may provide a separation of different RNAi pathways by helping sort small RNAs into different Argonaute proteins. CDE-1 homologs are conserved throughout evolution and might play a crucial role in target recognition and small RNA sorting in other eukaryotes in various RNAi pathways.
What is the function of CSR-1 22G-RNAs in targeting protein-coding genes? Claycomb and colleagues (2009) observe that CSR-1 is essential for proper chromosome segregation, and remarkably, it associates directly with the chromatin in a 22G-RNA-dependent manner. In S. pombe, small RNAs guide the RITS complex to noncoding RNAs that are transcribed from pericentromeric repeats to mediate heterochromatin formation and stabilize centromeres during mitosis. In contrast to the S. pombe Argonaute, C. elegans CSR-1 targets protein-coding euchromatic regions distributed throughout the genome. Despite the difference, both Argonaute proteins may use similar mechanisms to target chromosomes and may perform similar functions in defining centromeres (Figure 1). C. elegans chromosomes are holocentric, and centromeres are formed over the entire chromosome. CSR-1 targeting of protein-coding genes might define the centromeric domains on holocentric chromosomes in a similar way to how the S. pombe Argonaute defines centromeres by establishing pericentromeric heterochromatin, which can in turn promote de novo centromere assembly (Folco et al., 2008).
However, it remains unclear whether and how CSR-1 modifies chromatin structure to promote centromere establishment. One possibility is that CSR-1 establishes transient heterochromatin, which serves as a signal for deposition of centromere-specific histone CENP-A. Alternatively, CSR-1 may directly promote CENP-A deposition and kinetochore assembly without changing the underlying chromatin structure. Most eukaryotes have monocentric chromosomes in which the centromere is surrounded by repetitive sequences that assemble into heterochromatin. Although once assembled, the centromere is epigenetically inherited (Ekwall, 2007), heterochromatin can promote de novo centromere assembly in S. pombe, and this role may be conserved in multicellular eukaryotes (Folco et al., 2008). The exciting studies described above define an alternative pathway of kinetochore assembly. Future studies should provide new insights into this emerging area of chromosome biology. In particular, we can look forward to new discoveries in this area that will elucidate how the 22G-RNA pathway is linked to the recruitment of CENP-A and other centromere-specific factors and the extent to which RNA-dependent pathways contribute to centromere assembly in other multicellular eukaryotes.
References
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC. Curr Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. Cell. 2009;139:123–234. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman E, Keys J, Stoltz MJ, et al. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PM, Mello CC, Berezikov E, Ketting RF. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]