Abstract
In animal cells, small RNA molecules, called piRNAs, defend the genome against selfish DNA elements such as transposons. In this issue, Klattenhoff et al. (2009) report that an HP1 family protein, Rhino, is required for piRNA generation and transposon silencing in Drosophila germline cells. The results provide a link between heterochromatin and piRNA-mediated genome defense.
Transposons are selfish DNA elements that exploit the genome and replicative machinery of host cells in order to survive and proliferate (O’Donnell and Boeke, 2007). These elements occupy nearly one-third to one-half of the genomes of the fruit fly Drosophila and human, respectively. Movement of transposons within the genome (transposition) induces mutations at their excision and insertion sites, which can result in genomic instability. Therefore, transposons and genomes are in a constant evolutionary arms race. Although transposons have evolved to proliferate in the host genome, organisms have evolved multiple mechanisms to regulate the mobilization of transposable elements and to maintain genome integrity. In this issue of Cell, Klattenhoff et al. (2009) provide evidence for the convergence of two major transposon defense pathways involving heterochromatin and RNA silencing mechanisms.
It is well established that one of the cellular defense mechanisms at the center of the arms race between transposons and the host genome involves the packaging of transposon-rich regions into heterochromatin. These regions are characterized by specific modifications on histones, such as methylation of histone H3 lysine 9 (H3K9). These modifications recruit proteins, such as heterochromatin protein 1 (HP1), which promote transcriptional and transposon silencing. A spectacular series of discoveries have uncovered roles for RNA-based silencing mechanisms in defending cells against transposons as well as viruses. Cells of the germline are particularly sensitive to transposition, an event that can be passed on to the next generation, thus endangering genetic stability. To provide an additional level of fidelity in transmitting the genetic material to the next generation, some multi-cellular eukaryotes have evolved additional RNA-based pathways to silence transposable elements in the germline. One such pathway involves the generation of piRNAs, a class of small RNAs that are associated with Piwi proteins of the Argonaute (Ago) family. In Drosophila, distinct heterochromatic loci are the source of primary antisense piRNAs, which then target a large number of transposons that are dispersed throughout the genome and are active in the germline. Primary piRNAs are amplified by a ping-pong mechanism where antisense piRNAs, associated with the Piwi clade Aubergine (Aub) protein, target sense transcripts and generate sense piRNAs, which then associate with Ago3 and target antisense transcripts (Brennecke et al., 2007; Gunawardane et al., 2007).
The mechanism responsible for the generation of primary piRNAs and how piRNAs silence transposons is not well understood. Piwi proteins loaded with piRNAs can target and cleave RNA molecules and degrade them posttranscriptionally. Although previous data suggested that piRNAs could also silence target transposons by promoting chromatin modifications, a clear relationship between heterochromatin and piRNAs has been lacking. In their new work, Klattenhoff et al. (2009) establish a role for Rhino (HP1d), one of five HP1-like proteins in Drosophila, in the generation of piRNAs. Unlike HP1a, b, and c, which are expressed ubiquitously, Rhino is expressed predominantly in the fly ovaries, and mutations in the rhi gene lead to the activation of transposons and female sterility (Vermaak et al., 2005; Volpe et al., 2001).
Klattenhoff and colleagues show that Rhino is required for transposon silencing, production of piRNAs by dual-strand heterochromatic clusters, and efficient amplification via the ping-pong mechanism. Genome-wide transcriptional mapping revealed that mutations in the rhi gene and in the armi gene (which encodes a putative helicase also required for piRNA generation) increased the expression of a subset of transposons. However, rhi mutations did not significantly affect the expression of protein coding genes carrying transposons inserted in their introns. The authors suggest that piRNA-dependent silencing occurs after RNA splicing, which removes intronic transposon elements from protein coding genes. Furthermore, they find that rhi mutations disrupt localization of Aub and Ago3 to a perinuclear structure implicated in RNA processing called nuage. This suggests that Rhino might act upstream of Ago3 and Aub and that the nuage body and piRNA-mediated silencing are interdependent. These results suggest that piR-NAs scan transcripts in the nuage body, after splicing and export of precursor RNAs. Nuage may have a conserved role in RNA surveillance, eliminating dangerous transposon RNAs before they can be exported to the cytoplasm. This would prevent translation of transposon transcripts and synthesis of proteins required for the reverse transcription of transposons and their insertion back into the genome.
HP1 proteins are required for heterochromatin assembly and transcriptional gene silencing within heterochromatic domains. Rhino, however, appears to have an unusual role in the generation of piRNAs. It is required for the generation of precursor RNAs from dual-cluster piRNA loci, including the 42AB region on chromosome 2, a major piRNA cluster in Drosophila. Using chromatin immunoprecipitation, the authors show that Rhino associates with the 42AB cluster, thus directly linking an HP1 protein to a major piRNA cluster. Like most heterochromatic piRNA clusters, the 42AB cluster produces piRNAs from both genomic strands. A key finding is that expression of long precursor RNAs from the 42AB cluster in rhi homozygous mutant flies is dramatically reduced, suggesting that Rhino promotes the production of long precursor RNAs from the 42AB cluster and possibly from other dual-strand clusters in the germline. This result provides a possible explanation for an unexpected observation by Yin and Lin, who found that the Piwi protein is required for the generation of specific piRNAs from another dual-cluster piRNA locus in flies called TAS. Surprisingly, loss of Piwi increased HP1a at the TAS locus as well as methylation of H3K9, suggesting that Piwi is normally required for maintaining a transcriptionally open chromatin conformation at this piRNA cluster (Yin and Lin, 2007). Although additional information on the chromatin structure of the 42AB cluster in wild-type and mutant fly ovaries is required before any clear conclusions can be drawn, an intriguing possibility is that Piwi proteins regulate chromatin structure and transcription at some piRNA clusters by governing the recruitment of different HP1 proteins. Rhino may prevent heterochromatin-dependent silencing at the 42AB cluster in the germline by competing with and excluding other HP1 proteins from associating with the cluster (Figure 1). HP1 proteins recruit downstream effectors that mediate either transcriptional gene silencing or cotranscriptional RNA processing. The germline-specific expression of Rhino reported by Klattenhoff et al. therefore may shift the balance toward expression from dual-strand clusters to allow piRNA production in the germline. It is also possible that the accumulation of long precursor RNAs is due to a change in RNA stability rather than increased transcription. In the latter model, Rhino would interact with the long RNAs and prevent their degradation in the nucleus so that they could be exported to the nuage. In fission yeast, which do not have piRNAs, small interfering RNAs (siRNAs) target the RNA-induced transcriptional silencing (RITS) complex containing Argonaute and the chromodomain protein Chp1 to noncoding RNAs, which are transcribed from centromeric repeat regions. Subsequently, RITS promotes amplification of siRNAs, the degradation of transcripts, and recruitment of HP1 proteins and H3K9 methylation. The HP1 proteins of fission yeast have roles in controlling RNA synthesis and processing within centromeric regions. Such roles include facilitating the association of RNAi complexes with noncoding RNAs, recruiting deacetylase complexes to mediate transcriptional gene silencing, and recruiting Epe1, a protein that antagonizes H3K9 methylation to allow transcription of centromeric RNAs (Motamedi et al., 2008; Zofall and Grewal, 2006). These centromeric RNAs are the precursors of repeat-associated siRNAs in fission yeast. That Rhino is required for the accumulation of 42AB dual-cluster RNAs may reflect a similar, evolutionarily conserved, division of labor among multiple HP1 proteins.
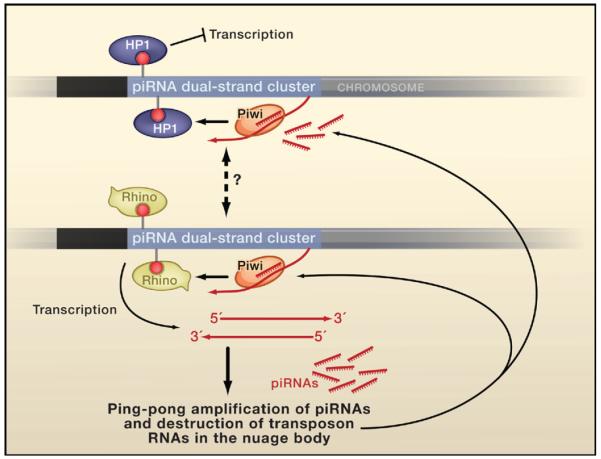
Figure 1. Rhino, HP1, and piRNA Generation.
The heterochromatin protein Rhino is a key player in the generation of piRNAs in fly ovaries, which help to protect the germline genome by silencing transposons. At heterochromatic dual-strand piRNA clusters, the structure of heterochromatin and transcription are regulated by the competing activities of Rhino and another heterochromatin protein, HP1. Rhino is preferentially expressed in the germline and may possibly dislodge other HP1 proteins from the cluster to allow transcription of long precursor RNAs, which are required for the generation of primary piRNAs and their amplification in the nuage body. (Black, heterochromatin; red lollipops, H3K9 methylation.)
The exciting findings of Klattenhoff and colleagues have set the stage for addressing several important questions about piRNA biology and its relationship to heterochromatin. For example, how is Rhino targeted to specific piRNA clusters? Ago3 and Aub localize preferentially to nuage, where they amplify piRNAs by the ping-pong mechanism. However, Piwi is present in the germline and is localized preferentially to the nucleus. One possibility is that existing piRNAs associated with Piwi target Rhino to transposon clusters and thus promote the production of additional piRNAs from the cluster. It would be interesting to know whether Piwi, Ago3, or Aub localize to the 42AB and other dual-strand clusters. A physical interaction between Piwi and HP1a has been observed in somatic cells of Drosophila (Brower-Toland et al., 2007), where Piwi might recruit HP1a to transcriptionally silence these regions. In the fly ovaries, Rhino may take the place of HP1 in a similar kind of interaction to promote the expression of dual-cluster RNAs and the generation of piRNAs to silence transposons. An intriguing possibility is that another HP1 protein, HP1e, which is preferentially expressed in the male germline (Vermaak et al., 2005), promotes dual-cluster RNA expression in sperm. Future studies on the interplay between Rhino, other HP1 proteins, and piRNA generation should provide further insights into this fascinating area of genome biology.
REFERENCES
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. Cell. 2009;(this issue) doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. Mol. Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Boeke JD. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. Genetics. 2001;159:1117–1134. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin H. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. Mol. Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]