Abstract
Motivation: Genome-wide mRNA profiling provides a snapshot of the global state of cells under different conditions. However, mRNA levels do not provide direct understanding of upstream regulatory mechanisms. Here, we present a new approach called Expression2Kinases (X2K) to identify upstream regulators likely responsible for observed patterns in genome-wide gene expression. By integrating chromatin immuno-precipitation (ChIP)-seq/chip and position weight matrices (PWMs) data, protein–protein interactions and kinase–substrate phosphorylation reactions, we can better identify regulatory mechanisms upstream of genome-wide differences in gene expression. We validated X2K by applying it to recover drug targets of food and drug administration (FDA)-approved drugs from drug perturbations followed by mRNA expression profiling; to map the regulatory landscape of 44 stem cells and their differentiating progeny; to profile upstream regulatory mechanisms of 327 breast cancer tumors; and to detect pathways from profiled hepatic stellate cells and hippocampal neurons. The X2K approach can advance our understanding of cell signaling and unravel drugs mechanisms of action.
Availability: The software and source code are freely available at: http://www.maayanlab.net/X2K.
Contact: avi.maayan@mssm.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
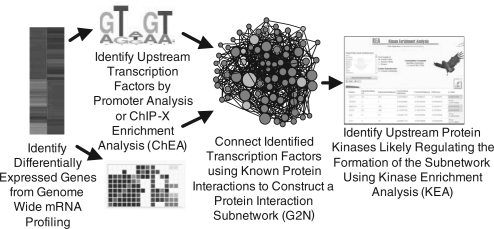
Although genome-wide proteomic approaches are rapidly improving, the most widely available and cost-effective genome-wide expression data is still collected at the mRNA level. These experiments are carried out using either microarrays or more recently RNA sequencing (RNA-seq) (Wang et al., 2009). Commonly, studies examine cells under different experimental conditions such as control versus drug treated, disease versus normal states or as a time-series, for example, during cell differentiation. Since quantitative changes in mRNA levels do not directly explain how cell signaling mechanisms are altered to induce changes in gene expression, and in turn lead to changes in cellular phenotype, identification of such upstream regulatory mechanisms has been the focus of many computational systems biology studies. Such understanding will enable us, among other things, to better control cell behavior with small molecules, and in turn translate such ability to therapeutics. Most popular approaches for data interpretation of changes in genome-wide gene expression include promoter analysis (Matys et al., 2006; Portales-Casamar et al., 2010), gene ontology (The Gene Ontology Consortium) or pathway enrichment analyses (Kanehisa et al., 2010), as well as reverse engineering of networks from mRNA expression data (Margolin et al., 2006). The ultimate goal of many of these approaches is to identify and rank potential target genes/proteins that if knocked down or overexpressed would explain the observed changes by, for example, reversing them. Such proteins may ultimately become drug targets. Here, we present a rational approach called Expression2Kinases (X2K) to identify and rank putative transcription factors, protein complexes and protein kinase that are likely responsible for the observed changes in genome-wide mRNA expression. By combining data from chromatin immuno-precipitation (ChIP)-seq/chip experiments and/or position weight matrices (PWMs), protein–protein interactions and kinase–substrate protein phosphorylation reactions, we demonstrate how we can better identify regulatory mechanisms responsible for genome-wide differences in gene expression. The idea is to first infer the most likely transcription factors that regulate the differences in gene expression, then use protein–protein interactions to connect the identified transcription factors using additional proteins to build transcriptional regulatory subnetworks centered on these factors and finally use kinase–substrate protein phosphorylation reactions to identify and rank candidate protein kinases that most likely regulate the formation of the identified transcriptional complexes (Fig. 1).
Fig. 1.
The X2Ks workflow.
We show how transcription factors, protein complexes and protein kinase candidate identification and ranking are inferred robustly by cross-validating the method with additional data such as those from drug perturbations followed by genome-wide mRNA expression profiling. Furthermore, we demonstrate the application of the method to in several case studies, where we developed several visualization methods that present a global view of cell-fate trajectories at different layers of regulation. All together, X2K can rapidly advance our understanding of cell signaling networks' regulation of gene expression by utilizing different modalities of prior knowledge. The X2K approach can assist in drug target discovery and help in unraveling drug mechanisms of action.
2 METHODS
2.1 Identifying differentially expressed genes
The first step of the X2K computational approach is a standard procedure where differentially expressed genes (mRNAs) are identified. Such sets of genes can originate from experiments that profiled cells under different conditions, during different stages of differentiation, from tissues of different patients or different cell-lines. The identified sets of differentially expressed genes can then be grouped into up or down subgroups, clusters of genes that behave similarly across different perturbations, or gene modules that behave similarly over a time course. The outputs from such analyses produce sets of unranked lists of genes. For microarray analysis we performed here, MAS5.0-processed data from the gene expression omnibus (GEO) database was used. Quantile normalization was then required for cross-assay comparisons. Following normalization, differentially expressed genes were identified via the R statistical package LIMMA (Smyth, 2004).
2.2 Identifying upstream transcription factors
Once sets of differentially expressed genes are identified, these gene lists can be fed into the transcription factor inference module of X2K using the tool and database ChIP-seq/chip Enrichment Analysis (ChEA) (Lachmann et al., 2010) or PWMs to obtain a list of transcription factors that are the likely upstream regulators of the identified differentially expressed gene set. The ChEA database and software contains manually extracted results of transcription factor/target-gene interactions from ChIP-seq/chip experiments applied to human or mouse cells. This database currently contains a network of 361 299 interactions, manually extracted from 157 publications, describing the binding of 159 transcription factors to their putative targets covering almost all annotated human or mouse target genes. On average, each transcription factor/experiment entry lists ~ 1300 target genes. Most interactions were extracted based on authors' selection of target genes for each factor, but in few cases, we directly processed the raw fastq files from sources such as the National Center for Biotechnology Information's sequence read archive (SRA) database or processed the WIG or BED files provided by the authors to identify the top peaks near genes, while setting an arbitrary cut-off to obtain the top ~ 1000–2000 target genes for each experiment based on distance to the start site and peak height. We used BowTie (Langmead et al., 2009) for reads alignment and MACS (Zhang et al., 2008) for peak calling. With the ChEA database, we compute enrichment for overlap between the input set of differentially expressed genes and entries in the ChEA database using either the Fisher's exact test, an alternative method that computes the deviation from expected rank for random input gene-set, or a combination of these two scoring schemes. As an alternative to ChEA, we used TRANSFAC (Matys et al., 2006) and JASPAR (Portales-Casamar et al., 2010), which are two state-of-art databases for PWMs. From TRANSFAC and JASPAR, we generated a Gene Matrix Transposed (GMT) file (Subramanian et al., 2007) listing putative target genes for each transcription factor for human or mouse by scanning the promoters (−2000 to +500 from the transcription factor start site) for all annotated genes for these two organisms. The program Patch, provided by TRANSFAC, was used to scan promoter sequences. We kept all individual entries from both databases even though for some transcription factors there are more than one PWM. For JASPAR we used the JASPAR Core.
The ChEA or the GMT file created from TRANSFAC and JASPAR were used to analyze lists from mRNA expression profiling by performing gene-list enrichment analysis with the Fisher's exact test using the ChEA or the PWMs dataset as the prior biological knowledge gene-list library. ChEA and PWMs, each have their own advantages and disadvantages. ChEA is created from empirical observations in different cell types and conditions. On one hand, ChEA considers the chromatin state of the cell under a specific condition, which is not done by PWMs and may produce more specific overlapping genes with fewer false positives. However, the ChEA approach may miss hits for transcription factors if the examined expression is derived from completely different cell types or the transcription factor is missing from ChEA. Another advantage of the PWM GMT library is that it provides more coverage for factors. For example, TRANSFAC contains 830 mouse and 1113 human matrices for about ~ 300 transcription factors, whereas the ChEA database currently only has 159 factors.
2.3 Connecting transcription factors with protein–protein interactions
Most analyses that attempt to link gene expression changes to upstream regulators stop at the step of promoter analysis, or attempt to infer pathways directly from differentially expressed genes. However, X2K further ‘connects’ the identified transcription factors using networks of experimentally reported protein–protein interactions or protein complexes. Genes2Networks (G2N) is command-line and web-based software that we developed in the past to connect lists of mammalian genes/proteins in the context of background mammalian signalome and interactome protein networks (Berger et al., 2007). The background protein–protein interactions network we use in X2K is made of experimentally determined mammalian interactions collected from 18 databases/datasets and currently contains 24 036 proteins connected through 389 959 interactions. The input to the program is a list of human Entrez gene symbols and background protein interaction networks, while the output is a subnetwork made of ‘intermediate’ proteins that ‘connect’ the ‘seed’ list of genes/proteins. This is achieved by finding all shortest paths between all pairs of seed nodes with a specified maximum path length and then adding additional interactions between intermediates. Different settings allow for filtering interactions from background networks by limiting the number of interactions from a specific paper, limiting the selection of background databases or only including interactions that are reported more than once. Once transcription factor-centered complexes upstream of differentially expressed gene modules are identified, using the G2N module of X2K, we identify the protein kinases that are most likely responsible for the transcription factor complexes' formation and functional regulation.
2.4 Identifying protein kinases upstream of transcriptional complexes
Once we build a subnetwork/protein complex that connects the identified transcription factors to each other, we convert this subnetwork to a list of proteins and feed it as input to the Kinase Enrichment Analysis (KEA) (Lachmann and Ma'ayan, 2009) module of X2K. KEA is web-based and command-line software with an underlying database that provides users with the ability to link lists of mammalian proteins with the protein kinases that likely phosphorylate them. The system draws from several available kinase–substrate databases to compute kinase enrichment probability based on the distribution of kinase–substrate proportions in the background kinase–substrate database compared with the protein kinases found to be associated with an input list of proteins using the Fisher's exact test. Using information available in the public domain, we reconstructed a mammalian kinase–substrate network. The kinase–substrate interactions are from the human protein reference database (HPRD) (Keshava Prasad et al., 2009), PhosphoSite (Hornbeck et al., 2004), phospho.ELM (Diella et al., 2004), NetworKIN (Linding et al., 2008) and Kinexus (www.kinexus.ca). In total, the consolidated dataset contains 14 374 interactions from 3469 publications involving 436 kinases.
2.5 The X2K software
All together, starting from a set of differentially expressed genes, we end up with protein kinases, transcription factors and protein complexes that are putative regulators of the inputted differentially expressed genes. The X2K system was developed as an open source Java desktop application and is available at http://www.maayanlab.net/X2K with documentation. The underlying code for X2K was developed using the Java 6 SDK under the Eclipse IDE. Using an Apache Maven build process, command-line and Swing GUI versions are packed into an executable JAR with all the necessary background files included. User can unpack the JAR to access the background databases used. Since the code does not use any operating-system-specific methods, the application is inherently multiplatform. After entering a list of differentially expressed genes, the program outputs Excel spreadsheets, text files and network files in different formats, including networks that can be visualized with Cytoscape (Shannon et al., 2003), SNAVI (Ma'ayan et al., 2009), Pajek, or yEd. User manual is available as supporting materials.
3 RESULTS
3.1 Application of X2K to recover drug-targeted pathways from gene expression signatures
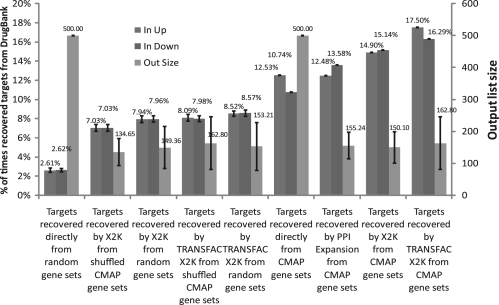
To demonstrate how X2K can be used to infer upstream regulators given gene expression changes, we first applied the tool to analyze expression data from the Connectivity Map (CMAP) (Lamb et al., 2006). The CMAP database developed by the Broad Institute is a large dataset of mRNA microarray gene expression profiles made from experiments where four different types of human cancer cell lines were treated with many single FDA-approved drugs and then gene expression was measured after 6 hours. CMAP contains 6100 perturbations with 1309 single drugs, where compounds were applied in different concentrations, to different cell types, or other variable experimental conditions. Using CMAP, we examine if the known drug target proteins fall within the subnetworks created by the intermediate steps of X2K. We omitted G-protein coupled receptors (GPCR) targeting drugs because X2K is not designed to recover those. First, we extracted the top 500 upregulated and bottom 500 downregulated genes from CMAP for each drug perturbation experiment based on the ranked gene lists provided for download from the CMAP website. We then entered these lists as input into the X2K pipeline. Once we collected all the transcription factors, protein complexes and protein kinases based on gene expression changes induced by the different drug perturbations, using the default settings of X2K, we asked whether the genes/proteins appearing in these pathways are enriched in known drug targets reported in DrugBank (Wishart et al., 2008) (Fig. 2).
Fig. 2.
Validation of X2K with CMAP and DrugBank. X2K recovered ~15–17% of the times at least one known non-GPCR primary drug target in pathways created upstream from gene expression profiles from CMAP using the 500 up and down lists from individual experiments in CMAP (first two sets of bar graphs from right). The X2K software was used with the default settings: 10 top transcription factors based on ChEA or TRANSFAC/JASPAR (ChEA is used as default, otherwise TRANSFAC is labeled), G2N to expand the initial list with all protein–protein interactions datasets, and 10 top kinases from KEA. These results were compared with the percent of recovered drug targets in randomly generated gene lists, randomly generated lists piped into X2K, shuffled lists from CMAP, targets recovered directly from the top 500 up and down lists from CMAP or targets found in protein–protein interaction networks created from the top 500 up and down lists from CMAP (left to right bar graphs).
We show that ~15–17% of the time we can recover the drug target in pathways created by X2K using ChEA or TRANSFAC/JASPAR. The TRANSFAC/JASPAR option is slightly better in recovering targets as compared with ChEA. Interestingly, targets can be recovered directly within the differentially expressed genes better than by chance but with much less recall and specificity as compared with X2K. Having targets appearing in differentially expressed genes more than by chance was previously reported by Iskar et al. (2010), which is consistent with our findings. In addition, targets can be found in pathways constructed directly from the differentially expressed genes, but this procedure too has less recall and specificity as compared with X2K. Other statistical controls show that targets can be found in randomly generated gene lists of 500 human genes ~2–3% of the time, and in pathways created from randomly generated lists of genes ~7% of the time. Hence, X2K is capable of recovering drug targets from gene expression better than other methods. More parameter tuning, as well as expansion and improvement of the databases quality and coverage used by X2K are expected to improve performance. This is reserved to future studies.
3.2 Application of X2K to obtain a global view of cellular differentiation
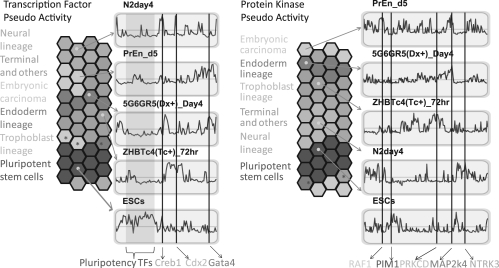
The X2K method can be applied globally to map the putative upstream ‘regulatory state’ of mammalian cells by comparing and contrasting the subnetworks generated by the program across different cell types and cell states. Our hypothesis is that given a set of samples from genome-wide expression data across many cell types and experimental conditions, we can correctly infer the activity patterns of the upstream transcription factors and protein kinases across samples to obtain a global picture of cell regulation across multiple regulatory layers (Supplementary Fig. S1). Such activity patterns can be approximated by enrichment analyses applied to the weighted expression of differentially expressed gene modules. This approach can also be used to validate whether X2K is identifying a set of transcription factors and protein kinases that are unique to specific cell types and experimental conditions. Developing an initial approach to achieve this goal, we first analyzed 44 samples from genome-wide expression data collected from embryonic stem cells induced to differentiate toward different lineages as well as several other terminal cell types all collected and previously analyzed by other studies (Aiba et al., 2009). The gene expression data matrix was subjected to an iterative consensus agglomerative clustering algorithm with within-module-coherence threshold of 0.7 and merging threshold set to 0.8 (Qiu et al., 2011). As a result, 300 expression modules were identified, but only 49 modules had a hundred or more genes, and these modules were retained for further analysis. Upstream transcription factors enriched for each module were computed using ChEA. An enrichment significance matrix M was then generated with entries mjk representing the -log(p-value) of the enriched transcription factor j for module k. A pseudo activity matrix P was then generated with pij representing pseudo activity for transcription factor j in sample i calculated as follows: pij=max(mjk)×akj×eij×iij where akj is the mean expression of module k in sample i, eij is the expression level of the transcription factor and iijis a Boolean indicator function that checks if the transcription factor is expressed above average in the sample. Hence, the pseudo activity pij is composed of the binding score for the transcription factor, the average mRNA expression of the regulated module and the expression level of the transcription factor in the sample (Supplementary Fig. S1). To visualize the preservation of ordering of the samples across regulatory layers for the 44 cell types and conditions, we implemented four data visualization methods: (i) Principle Component Analysis (PCA) (Supplementary Fig. S2); (ii) Minimum Spanning Trees (MST), implemented with a modified script based on the recently published sample progression discovery (SPD) package (Qiu et al., 2011) (Supplementary Fig. S3); (iii) hierarchical clustering (Supplementary Figs S4 and S5); and (iv) our Grid Analysis of Time-series Expression (GATE) software (MacArthur et al., 2010), repurposed to have each hexagon representing a cell type (Fig. 3).
Fig. 3.
GATE visualization of transcription factor and protein kinase pseudo-activity scores applied to 44 cell types. Each hexagon represents a different cell type, color-coded based on the different lineage groups, but ordered based on pseudo-activity scores correlation with other cell types. Next to each hexagonal heatmap there are representative (left) transcription factors and (right) protein kinases, pseudo-activity score profiles for all transcription factors and kinases for selected samples. Red represents up-regulation and green represents down-regulation of the transcription factors or kinases regulating co-expressed modules across the 44 samples for a specific sample. Specific transcription factors and protein kinases are highlighted with straight vertical lines and are annotated at the bottom. Transcription factors and kinases are ordered along the line corresponding to the order determined by hierarchical clustering. The hexagonal grid folds on itself to form a torus such that hexagons at the edges are close to hexagons from the opposite side.
The GATE software takes as input a data table, where rows are variables and columns are measurements. The software uses simulated annealing to arrange variables on a hexagonal grid based on correlations between variables across all measurements. In our case, the variables are cell types or tumor samples, and the columns, representing measurements, are inferred pseudo-activity levels of transcription factors and protein kinases. Similar to the way we compute pseudo-activity for transcription factors, we can identify the upstream protein kinases enriched for each module using the command-line version of X2K and the same steps performed for the transcription factors. Consequently, by using the upstream regulatory transcription factors and protein kinases activity patterns, the landscape of samples can be correctly time-ordered and samples of the same subtype are closer to each other than to other subtypes. To test whether the preserved ordering of samples across regulatory layers arises by chance, we applied the same procedure to shuffled data (Supplementary Fig. S6). We quantified the preservation of the ordering by an objective error function, counting the times neighbors of each node are preserved, and clearly saw that the ordering is far from random (P-value <10−10, two-tailed t-test for both transcription factors and protein kinases). Enriched transcription factors with relatively high predicted activity in the pluripotent stem cells are generally known factors such as Oct4/Pou5f1, Nanog and Sox2. In comparison, enriched lineage commitment regulators are predicted to be active in more differentiated cell types. For example, Gata4 is a known master regulator for the endoderm lineage and correspondingly displays high pseudo activity scores in late-stage endoderm cells. These results can be used to characterize the upstream regulatory profile of the 44 different cell types. This approach can be used to tune the parameters and datasets used by X2K to validate the approach by setting the thresholds that best preserve the ordering of samples and recovering the already known transcription factors and protein kinases for cell types.
3.3 Application of X2K to unravel regulatory mechanisms of subtypes of breast cancer
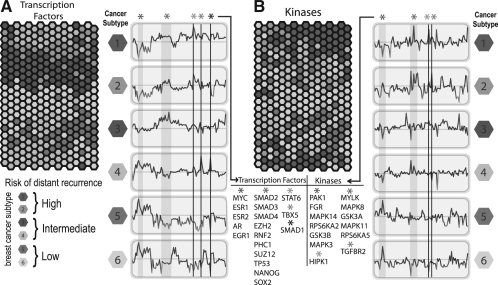
The inherent inter-patient heterogeneity of breast cancer motivates the identification of unique molecular signatures of the disease at the individual patient level. The ability to identify molecular regulatory differences at the genome, transcriptome, gene-regulome and kinome levels for particular cancer subtypes may enable us to better tailor and optimize therapeutics for individual patients. To achieve this, we illustrate the utility of X2K to uncover putative upstream regulatory mechanisms from previously published gene expression data collected from a large cohort of breast cancer tumors. We show that subtype similarities in gene expression can be grouped and visualized based on the pseudo-activity scores of upstream transcription factors and protein kinases that likely regulate differentially expressed genes in the breast cancer subtypes. Specifically, X2K was applied to analyze a publicly available breast cancer gene expression dataset from fresh frozen tissues of 327 patients that were randomly selected from a group of diagnosed individuals between 1991 and 2004 at the Koo Foundation Sun-Yat-Sen Cancer Center (Kao et al., 2011). In the original study, the cancer tissues were categorized into six subtypes based on differential gene expression signatures. Based on OncotypeDX (Paik et al., 2004) and MammaPrint (van 't Veer et al., 2002) signatures, the risk for distant cancer recurrence and metastasis was assessed such that subtypes 1 and 2 had high risk, subtypes 3 and 4 had intermediate risk and subtypes 5 and 6 had low risk. The normalized gene expression data and corresponding patient subtype designations were obtained from Gene Expression Omnibus (accession GSE20685), subjected to probe-set consolidation, hierarchical clustering, and gene co-expression module identification. X2K was applied to co-expressed modules, where each module consisted of a list of genes whose expression profiles correlated across all patients. The detailed procedure and parameters are similar to those applied above for analyzing the 44 stem cells and their differentiated progeny. The correlation of pseudo activity contribution scores of the transcription factors and protein kinases that were enriched in the maximum-score gene modules were used to cluster and visualize the 327 patients using GATE (MacArthur et al., 2010) (Fig. 4).
Fig. 4.
GATE visualization of transcription factor and protein kinase pseudo-activity scores applied to 327 breast cancer tumor samples. Each hexagon represents a different profiled tumor from an individual patient. Each hexagon is color-coded based on previous classification of tumor invasiveness and recurrence. Next to each hexagonal heatmap there are representative (left) transcription factor and (right) protein kinase, pseudo activity score profiles for all transcription factors and kinases for selected samples. Red represents up-regulation and green represents down-regulation for a specific sample. Specific transcription factors and protein kinases are highlighted with straight vertical lines and are annotated at the bottom. Transcription factors and kinases are ordered along the line profile that corresponds to ordering determined by hierarchical clustering. The hexagonal grid folds on itself to form a torus such that hexagons at the edges are close to hexagons from the opposite side.
As shown by the list of identified key transcription factors in the subtypes' representative profiles, our analysis confirmed the downregulation of the nuclear receptors ESR1, ESR2 and AR in the high risk tumors. This is consistent with the original expression-based analysis that reported that subtypes 1 and 2 (high risk group) were estrogen receptor (ESR) negative. On the other hand, the high risk subtypes display upregulation of target genes regulated by members of the Polycomb group (EZH2, RNF2, PHC1, SUZ12), which are known to be downregulated in many cancers (Raaphorst, 2005). Together with identified enrichment for NANOG and SOX2, the high risk tumors suggest a stem cell-like regulatory signature. Moreover, the tumor suppressor TP53, as well as the receptor-mediated SMADs (SMAD1, SMAD2, SMAD3) and their associated common mediator (SMAD4), are predicted to be upregulated in the high-risk subtypes. Indeed, the TGF-β pathway is commonly implicated in distant metastasis of breast cancer (Kang et al., 2005), and reduction of SMAD2/3 signaling in breast cancer has been shown to suppress distant metastasis (Tian et al., 2003). Furthermore, we assessed the differential pseudo activity of the putative protein kinases predicted by X2K for the different breast cancer subtypes. We predict that MAPK14, RPS6KA2, GSK3B and MAPK3 are downregulated in the high- and intermediate metastasis-risk subtypes relative to the low-risk subtypes, while other isoforms of the same kinases, namely MAPK8, RPS6KA5, GSK3A and MAP11, exhibit the opposite pattern. Furthermore, we predict that the protein kinase HIPK1 and the TGF-β receptor TGFBR2 are more active in the high-risk group relative to the low-risk group. These predictions mostly agree with already known pathways, for example, HIPKs were reported to phosphorylate p53 (Arai et al., 2007), while the SMAD family members and TGF-β signaling are connected to TGFBR2. However, many novel candidates were also identified. Subsequent experimental verification of these is necessary but is beyond our expertise.
3.4 Application of X2K for reanalyzing expression data collected from hippocampal neurons and activated hepatic stellate cells in liver fibrosis
Lastly, in two additional case studies, we applied the X2K approach to analyze data from prior studies that applied microarray genome-wide gene expression analyses to investigate two commonly studied mammalian systems: (i) investigating differences in genome-wide expression profiles collected from hepatic stellate cells (HSCs) in liver fibrosis; and (ii) detecting upstream regulatory pathways by reanalyzing microarray data collected from hippocampal neurons treated with bicuculline during development. For the HSCs case study, we reanalyzed gene expression profiles to investigate regulatory mechanisms of hepatic fibrosis. Hepatic fibrosis is a scarring response to liver damage often due to chronic liver disease. In fibrogenesis, HSCs become activated and differentiate into extra-cellular-matrix-producing myofibroblasts. To better understand the gene expression changes that occur during such a process, De Minicis et al. (2007) conducted a microarray study to examine differences in gene expression profiles between cultured and in vivo-activated HSCs. Using the study as a source of microarray data for X2K, we identified the putative upstream transcription factors, intermediate proteins and protein kinases that may regulate the fibrosis response of HSCs. The transcription factor Tcf3 was predicted as a top candidate and this is supported by studies investigating the anti-adipogenic role of Wnt signaling in the pro-fibrogenic response, as a loss of adipogenic transcriptional regulation has been shown to be important for HSC activation (She et al., 2005). Among the predicted kinases are members of the ribosomal s6 kinase (RSK) family of serine/threonine kinases that can phosphorylate C/EBPbeta, an adipogenic transcription factor known to regulate collagen type I expression. RSK-mediated phosphorylation of C/EBPbeta at Thr217 appears to be crucial for the progression of fibrosis; Rsk inhibition led to regression of fibrosis in CCl4-treated mice, and increased activation of RSK and phosphorylated C/EBPbeta both were found in activated HSCs of liver fibrosis patients (Buck and Chojkier, 2007). Hence, it appears that X2K was able to correctly identify known upstream regulators based on the differentially expressed gene alone.
For the next case study, we reanalyze data relevant to long-term potentiation (LTP). Zhang et al. (2007) examined gene expression changes in neonatal mouse hippocampal neurons undergoing induction of rhythmic network activity. Reanalysis of this data using X2K has recaptured the transcription factor CREB and the protein CaMK4 as important upstream regulators. Activation of CREB is heavily implicated in the literature (Hardingham et al., 1999). In addition, the calmodulin-dependent kinase (CamK4) was also recovered from the X2K analysis. The link between CamK4 and CREB dependent transcription is well established (Matthews et al., 1994; Sun et al., 1994). Following this link to N-Methyl-D-aspartic acid (NMDA) receptor activation is clearly through calcium signaling. It was shown that CamK4 activation is important for different forms of LTP that depend on NMDA receptor activation (Kang et al., 2001). Activity-dependent increases in intracellular calcium, likely through voltage-gated calcium channels, affect increases in nuclear calcium where CamK4 is preferentially localized. Hence, the X2K pipeline is capable of recovering known pathways and likely predicting pathways not known to be involved before. More details about the two case studies from this section are available as Supplementary Material and as part of the X2K online documentation.
4 DISCUSSION
The X2K pipeline presents a new rational approach to identify and rank upstream regulators that are responsible for observed changes in gene expression collected at the genome-wide scale from mammalian cells. The approach, applied to datasets such as CMAP, has the potential to rapidly advance drug target discovery and help in unraveling drug mechanisms of action. The application to mapping transcription factor profiles and kinome profiles of many individual cell types, i.e. different cells during lineage commitment or tumors from patients, can be useful to obtain a global view of the axis of cell signaling networks across many cell types or to compare individual patients for suggesting appropriate pharmacological interventions. In addition, specific applications to common studies that examine genome-wide gene expression under two conditions, such as the two case studies we presented for HSCs and hippocampal neurons, can benefit from X2K analysis for generating hypotheses for further functional experiments following the global expression profiling.
While currently the X2K method uses only protein/DNA interactions, protein–protein interactions and kinase–substrate reactions, other types of data could be added. For example, histone modifications, microRNAs and other types of post-translational modifications could be incorporated into the pipeline. While more sophisticated enrichment analyses tests could be implemented, i.e. gene set enrichment analysis (GSEA) (Subramanian et al., 2005), and better parameter tuning can be achieved by cross-validation, the initial application of the approach shows great promise. The X2K approach is useful for data integration across layers and the reuse of prior knowledge within newly acquired expression datasets linking expression changes to upstream regulation. Another limitation of the method is the assumption of independence between regulators and targets when applying the ChEA or KEA steps. It is known that the kinome and transcriptional regulome networks are made of tightly coupled protein kinases regulating other kinases and transcription factors regulating other transcription factors. Several recent studies considered such interactions for transcription factors (Asif and Sanguinetti, 2011; Novershtern et al., 2011). Such interdependencies could be added to the X2K analysis where these two regulatory networks could be dynamically modeled. Regardless of these limitations and future directions, the current application of X2K presents an advancement toward our ultimate goal of understanding mammalian cell signaling networks from a global perspective at a molecular level of resolution.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Cijiang John He for useful discussions.
Funding: The project was partially funded by NIH grants (P50GM071558-03, R01DK088541-01A1, RC2LM010994-01, P01DK056492-10, RC4DK090860-01 and KL2RR029885-0109).
Conflict of Interest: none declared.
Footnotes
†The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First authors.
REFERENCES
- Aiba K., et al. Defining developmental potency and cell lineage trajectories by expression profiling of differentiating mouse embryonic stem cells. DNA Res. 2009;16:73–80. doi: 10.1093/dnares/dsn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S., et al. Novel homeodomain-interacting protein kinase family member, HIPK4, phosphorylates human p53 at serine 9. FEBS Lett. 2007;581:5649–5657. doi: 10.1016/j.febslet.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Asif H.M., Sanguinetti G. Large-scale learning of combinatorial transcriptional dynamics from gene expression. Bioinformatics. 2011;27:1277–1283. doi: 10.1093/bioinformatics/btr113. [DOI] [PubMed] [Google Scholar]
- Berger S., et al. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Chojkier M. A ribosomal S-6 Kinase-mediated signal to C/EBP-beta is critical for the development of liver fibrosis. PLoS One. 2007;2:e1372. doi: 10.1371/journal.pone.0001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.H., et al. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am. J. Physiol.– Gastrointest. Liver Physiol. 2008;294:G39–G49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- De Minicis S., et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- Diella F., et al. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham G.E., et al. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- Hornbeck P.V., et al. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- Iskar M., et al. Drug-induced regulation of target expression. PLoS Comput. Biol. 2010;6:e1000925. doi: 10.1371/journal.pcbi.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., et al. An important role of neural activity-dependent CaMKIV signaling in the consolidation of long-term memory. Cell. 2001;106:771–783. doi: 10.1016/s0092-8674(01)00497-4. [DOI] [PubMed] [Google Scholar]
- Kang Y., et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl Acad. Sci. USA. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K.J., et al. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad,T.S., et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A., Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A., et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, Genes, and Disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Langmead B., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R., et al. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A., et al. SNAVI: desktop application for analysis and visualization of large-scale signaling networks. BMC Syst. Biol. 2009;3:10. doi: 10.1186/1752-0509-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur B.D., et al. GATE: software for the analysis and visualization of high-dimensional time series expression data. Bioinformatics. 2010;26:143–144. doi: 10.1093/bioinformatics/btp628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin A., et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews R.P., et al. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol. Cell. Biol. 1994;14:6107–6116. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., et al. TRANSFAC(R) and its module TRANSCompel(R): transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Portales-Casamar E., et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N., et al. Physical Module Networks: an integrative approach for reconstructing transcription regulation. Bioinformatics. 2011;27:i177–i185. doi: 10.1093/bioinformatics/btr222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., et al. Discovering biological progression underlying microarray samples. PLoS Comput. Biol. 2011;7:e1001123. doi: 10.1371/journal.pcbi.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst F.M. Deregulated expression of Polycomb-group oncogenes in human malignant lymphomas and epithelial tumors. Hum. Mol. Genet. 2005;14(1):R93–R100. doi: 10.1093/hmg/ddi111. [DOI] [PubMed] [Google Scholar]
- Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She H., et al. Adipogenic transcriptional regulation of hepatic stellate cells. J. Biol. Chem. 2005;280:4959–4967. doi: 10.1074/jbc.M410078200. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Sun P., et al. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., et al. Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 2003;63:8284–8292. [PubMed] [Google Scholar]
- van 't Veer L.J., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.-J., et al. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.