Abstract
Mitochondrial DNA (mtDNA) is replicated by the DNA polymerase γ in concert with accessory proteins such as the mitochondrial DNA helicase, single stranded DNA binding protein, topoisomerase, and initiating factors. Nucleotide precursors for mtDNA replication arise from the mitochondrial salvage pathway originating from transport of nucleosides, or alternatively from cytoplasmic reduction of ribonucleotides. Defects in mtDNA replication or nucleotide metabolism can cause mitochondrial genetic diseases due to mtDNA deletions, point mutations, or depletion which ultimately cause loss of oxidative phosphorylation. These genetic diseases include mtDNA depletion syndromes (MDS) such as Alpers or early infantile hepatocerebral syndromes, and mtDNA deletion disorders, such as progressive external ophthalmoplegia (PEO), ataxia-neuropathy, or mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). This review focuses on our current knowledge of genetic defects of mtDNA replication (POLG, POLG2, C10orf2) and nucleotide metabolism (TYMP, TK2, DGOUK, and RRM2B) that cause instability of mtDNA and mitochondrial disease.
Keywords: DNA polymerase γ, mitochondrial DNA replication, nucleotide pools, mitochondrial DNA depletion syndrome, progressive external ophthalmoplegia, ataxia-neuropathy
Introduction
The mitochondrial genome is a closed circular genome of 16,569 bp that codes for 37 genes, all of which are directly or indirectly involved in the production of ATP. Thirteen of these genes encode protein subunits involved in electron transport to carry out oxidative phosphorylation. The remaining 24 genes encode the transfer RNAs (22 genes) and ribosomal RNAs (2 genes) required for mitochondrial protein synthesis of the 13 polypeptides. Animal tissue cells can contain several thousand copies of mtDNA spread out over hundreds of mitochondria (Miller et al., 2003). The mtDNA is located in discrete nucleods in the inner mitochondrial matrix of the mitochondrion and contain between 1–2 copies of mtDNA (Kukat et al., 2011). MtDNA is replicated by the concerted action of DNA polymerase γ (pol γ) encoded by POLG or POLG1(alias), its accessory subunit, p55 encoded by POLG2, and replication factors such as the mitochondrial single-stranded DNA binding protein and the mitochondrial DNA helicase (C10orf2 or Twinkle). Pol γ is the only known DNA polymerase to be found in mammalian mitochondria and thus bears the burden of DNA replication and DNA repair functions (Graziewicz et al., 2006).
Mitochondrial diseases can be caused by genetic defects in mitochondrial DNA or in nuclear genes that encode proteins that function in the mitochondria (Wallace, 1999). The last ten years has defined a list of genes linked to instability of mtDNA, either due to mitochondrial DNA depletion syndrome (MDS) or disorders characterized by multiple deletions, with or without point mutations. Table 1 list the current genes associated with MDS and deletion syndromes. Disorders associated with deletions and point mutations comprise commonly known disorders including progressive external ophthalmoplegia (PEO) and ataxia-neuropathy syndromes but also some very rare disorders of TCA cycle abnormalities (Copeland, 2008). MDS includes early childhood disorders such as Alpers syndrome, hepatocerebral syndromes, myocerebrohepatopathy spectrum disorders, and fatal myopathies (Wong et al., 2008, Saneto and Naviaux, 2010).
Table 1.
Nuclear genes associated with instability of mitochondrial DNA in patients.
| Gene | Clinical Disorder* | Consequence of mtDNA | Chromosome locus | Function |
|---|---|---|---|---|
| POLG (POLG1) | Ar Alpers or ataxia ArPEO/adPEO |
Depletion Deletions |
15q25 | Mitochondrial DNA polymerase |
| POLG2 | AdPEO | Deletions | 17q23-24 | Pol γ accessory subunit |
| DNA helicase (C10orf2) | AdPEO Ar ataxia/encephalopathy |
Deletions Depletion |
10q24 | Mitochondrial DNA helicase, Twinkle |
| TP (TYMP) | Ar MNGIE | Depletion Deletions |
22q13.32 | Thymidine phosphorylase |
| DGUOK | Ar encephalohepatopathy | Depletion | 2p13 | Mitochondrial deoxyguanosine kinase |
| TK2 | Ar congenital myopathy | Depletion | 16q22 | Mitochondrial thymidine kinase |
| RRM2B | Ar encephalomyopathy Ad PEO |
Depletion | 8q23.1 | p53-inducible small subunit of ribonucleotide reductase |
| ANT1 (SLC25A4) | AdPEO | Deletions | 4q34-35 | Adenine nucleotide translocator |
| MPV17 | Ar neurohepatopathy | Depletion | 2p21-23 | Mitochondrial inner membrane protein |
| SUCLA2 | Ar encephalomyopathy | Depletion | 13q12.2-q13.3 | β-subunit, Succinate-CoA ligase |
| SUCLG1 | Ar encephalomyopathy | Depletion | 2p11.1 | α-subunit, Succinate-CoA ligase |
| OPA1 | Ad optic atrophy | Deletions | 3q28-q29 | Dynamin related GTPase |
Ad, autosomal dominant; Ar, autosomal recessive
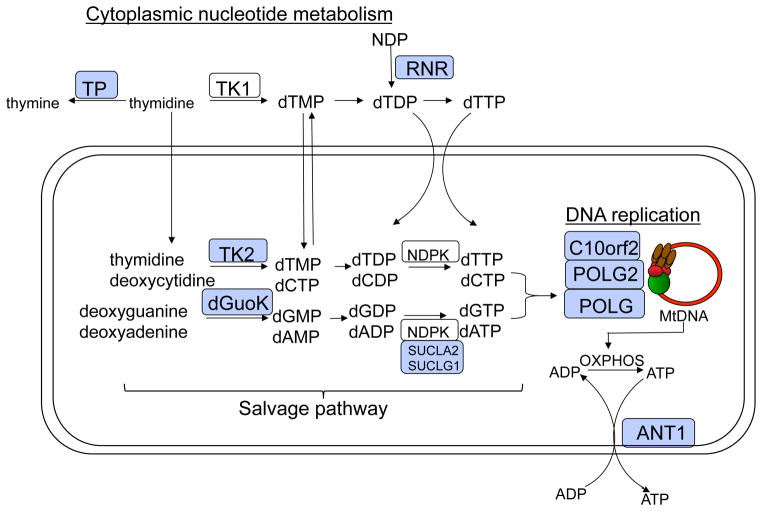
Defects in mtDNA and their associated disease can also arise in the production of mitochondrial nucleotide pools needed for DNA replication. Mitochondria depend heavily on either mitochondrial transport proteins or salvage pathway enzymes to supply deoxynucleotide triphosphates required for mtDNA replication (Figure 1). This review focuses on the genes and gene products identified as disease alleles and that function either directly at the replication fork or in metabolism of deoxynucleotide triphosphate pools used as the precursors for DNA replication. Other genes, such as OPA1, MPV17, ANT1, or SUCLA2, SUCLG1 that cause mitochondrial depletion or deletion syndromes but not directly involved in these two processes have been reviewed elsewhere (Copeland, 2008).
Figure 1.
Schematic diagram of the mitochondria representing enzyme pathways that cause mtDNA mutations or depletion when disrupted. Gene products associated with MDS or mtDNA mutations are labeled as light blue boxes.
Defects in proteins that act at the fork
Mitochondrial DNA is replicated and repaired by DNA polymerase γ (Pol γ the only known DNA polymerase to be found in animal cell mitochondria. The holoenzyme of pol γ in humans consists of a catalytic subunit (encoded by POLG at chromosomal locus 15q25) and a dimeric form of its accessory subunit (encoded by POLG2 at chromosomal locus 17q24.1). The catalytic subunit is a 140 kDa enzyme (p140) that has DNA polymerase, 3′-5′ exonuclease and 5′ dRP lyase activities (Kaguni, 2004, Graziewicz et al., 2006). The accessory subunit is a 55 kDa protein (p55) required for tight DNA binding and processive DNA synthesis (Lim et al., 1999). The pol γ holoenzyme functions in conjunction with the mitochondrial DNA helicase, Twinkle or C10orf2, and the mtSSB to form the minimal replication apparatus (Korhonen et al., 2004, Falkenberg et al., 2007). Gene mutations in POLG, POLG2 and C10orf2 have been implicated in mitochondrial disease and are discussed below.
DNA polymerase γcatalytic subunit, POLG
To date, nearly 200 mutations in POLG have been reported in association with mitochondrial diseases (http://tools.niehs.nih.gov/polg/(Longley et al., 2010, Walter et al., 2010, Copeland, 2008, Longley et al., 2005)Tang, 2011 #4939]). POLG-related disorders are currently defined by at least six major phenotypes of neurodegenerative disease that include: Alpers-Huttenlocher syndrome (AHS), childhood myocerebrohepatopathy spectrum (MCHS), myoclonic epilepsy myopathy sensory ataxia (MEMSA), the ataxia neuropathy spectrum (ANS), autosomal recessive progressive external ophthalmoplegia (arPEO), and autosomal dominant progressive external ophthalmoplegia (adPEO) (Wong et al., 2008, Cohen et al., 2010, Saneto et al., 2010, Saneto and Naviaux, 2010). Other signs and symptoms have been associated with POLG mutations such as male infertility, testicular cancer, and Parkinsonism and are reviewed in (Saneto and Naviaux, 2010).
POLG and PEO phenotypes
Mutations in POLG were first identified in 2001 by Van Goethem and colleagues as a locus for PEO (Van Goethem et al., 2001). PEO is a mitochondrial disorder associated with mtDNA depletion and/or accumulation of mtDNA point mutations and deletions (Zeviani et al., 1989, Hirano et al., 2001, Van Goethem et al., 2001). PEO is characterized by late onset (between 18 and 40 years of age) bilateral ptosis and progressive weakening of the external eye muscle, resulting in blepharoptosis and ophthalmoparesis, proximal muscle weakness, and wasting as well as exercise intolerance. Skeletal muscles of PEO patients present red ragged fibers and lowered activity of respiratory chain enzymes. Multiple large-scale deletions of mtDNA isolated from muscle biopsies were first demonstrated in Italian families with heritable autosomal dominant PEO (adPEO) by Zeviani and colleagues (Zeviani et al., 1989).
To date, with the exception of one mutation, all autosomal dominant POLG mutations responsible for developing PEO are mapped to the polymerase domain of pol γ. Two of the substitutions, R943H and Y955C, change side chains that interact directly with the incoming dNTP (Graziewicz et al., 2004). These enzymes retain less than 1% of the wild-type polymerase activity and display a severe decrease in processivity. The significant stalling of DNA synthesis and extremely low catalytic activities are the two most likely causes of the severe clinical presentation in R943H and Y955C heterozygotes (Graziewicz et al., 2004). The Y955C substitution also increases nucleotide misinsertion errors 10–100 fold in the absence of exonucleolytic proofreading (Ponamarev et al., 2002). In a yeast model developed to evaluate the homologous mutation in the yeast Mip1 gene we found that Y757C mutant (Y955C in humans) demonstrated enhanced mtDNA damage indicative of oxidative damage and very high petite frequency (Stuart et al., 2006). In a related study, Baruffini et al. showed that this high petite frequency could be rescued by treatment with antioxidants or up regulation of ribonucleotide reductase (Baruffini et al., 2006). In a mouse transgenic model in which the Y955C POLG was targeted to the heart, the mouse developed cardiomyopathty, loss of mtDNA, enlarged heart, and increased levels of 8-oxo-dG in their mtDNA (Lewis et al., 2007). Collectively, these phenotypes suggest that patients harboring the Y955C mutation may have elevated oxidative damage and may benefit from antioxidant therapy.
Significant cosegregation of parkinsonism with mutations in POLG gene was described in two individuals with adPEO (Luoma et al., 2004). Parkinsonism associated with POLG mutations manifested several years after initial disease symptoms. In addition many women with PEO from POLG mutations go through early menopause and suffer from high gonadotropin and low estrogen concentrations, indicative of premature ovarian failure (Luoma et al., 2004, Pagnamenta et al., 2006).
Most of the mutations found in POLG are associated with autosomal recessive PEO (arPEO) and patients with PEO are often compound heterozygotes with two arPEO alleles. For example, the A467T mutation has been found in trans with other POLG missense mutations in PEO, ataxia-neuropathy and Alpers syndrome (Van Goethem et al., 2003, Naviaux and Nguyen, 2004). The A467T mutation was found in two pedigrees as a homozygous mutation and associated with severe ataxia in mid-life (Van Goethem et al., 2004).
POLG and Alpers syndrome and myocerebrohepatopathy spectrum disorders
Alpers syndrome is a rare but severe, heritable, autosomal recessive MDS disease that afflicts young children. Within the first few years of life, patients exhibit progressive spastic quadriparesis, progressive cerebral degeneration leading to mental deterioration and seizures, cortical blindness, deafness, liver failure, and eventual death. Naviaux et al. reported an Alpers patient with reduced electron transport chain function, dicarboxylic aciduria, fulminant hepatic failure, refractory epilepsy, and lactic acidosis which resulted in death at 42 months (Naviaux et al., 1999). Analysis of mtDNA and pol γ activity from skeletal muscle biopsy indicated a reduction of mitochondrial DNA content to 30% of wildtype levels and no detectable pol γ activity (Naviaux et al., 1999). Sequencing of the POLG gene in two unrelated pedigrees revealed a heterozygous G to T nonsense mutation in POLG that converts Glu873 (GAG) to a stop codon and the heterozygous A467T substitution just after the exonuclease domain in the linker region (Naviaux and Nguyen, 2004). Pol γ mRNAs with the E873stop mutation are degraded from the pool of mRNAs by nonsense mediated decay resulting in mono-allelic expression of POLG containing only the A467T mutation (Chan et al., 2005b). To date the number of reported Alpers and other early childhood hepatocerebral syndromes associated POLG mutations has risen to nearly 100 different types of mutations (Ferrari et al., 2005, Nguyen et al., 2006, Horvath et al., 2006, Wong et al., 2008, Tang et al., 2011) (see Human DNA Polymerase Gamma Mutation Database: http://tools.niehs.nih.gov/polg/). In all cases, the POLG mutations in Alpers patients are recessive. Many of these mutations also occur in PEO patients as autosomal recessive mutations. The A467T mutation is the most frequently found pathogenic mutation in POLG. The frequency of the A467T mutation has been found to exist in approximately 0.6% in the Belgian normal population (Van Goethem et al., 2001), 0.69% in the British population (Horvath et al., 2006), 1% in the Norwegian populations (Winterthun et al., 2005), and less than 0.2% in 380 Finnish controls (Luoma et al., 2005). However, the A467T allele has been estimated to occur in 36% of all alleles associated with POLG disease, as shown by five large mitochondrial disease population studies identifying POLG mutations (Ferrari et al., 2005, de Vries et al., 2007, Horvath et al., 2006, Nguyen et al., 2006, Wong et al., 2008). Biochemical analysis has demonstrated that the recombinant A467T pol γ is unstable and retains only 4% activity compared to WT enzyme as a result of >5-fold decrease in kcat and a 4.7-fold increase in Km(dNTP) (Chan et al., 2005a). Additionally, the A467T pol γ is compromised for its interaction with the accessory subunit.
Analysis of a cluster of Alper’s mutations in the thumb-palm domain has shown a striking correlation with the severity of the defect and the degree of conservation (Kasiviswanathan et al., 2009). Mutations in the most conserved sites represented by G848S, T851A, R852C and R853Q exhibited less than 1% WT enzyme activity (Kasiviswanathan et al., 2009). Mutations in codons for less conserved amino acids (Gln879 and Thr885) resulted in only moderate reduction in activity (Kasiviswanathan et al., 2009).
Yeast has been a useful model system to study mutations in residues that are conserved between yeast (in the Mip1 gene) and man (Stuart et al., 2006, Baruffini et al., 2007, Baruffini et al., 2011, Baruffini et al., 2006, Stumpf et al., 2010). In a study of 31 mutations in the conserved regions in Mip1, 20 mutations were shown to disrupt mtDNA replication (Stumpf et al., 2010). The sporadic mutations, Q308H, R807C, G1076V, R1096H and S1104C, caused decreased polymerase activity leading to mtDNA depletion and mitochondrial dysfunction. Interestingly, point mutagenesis in mtDNA as a consequence of these POLG mutations, especially in the exonuclease region, were insufficient to explain pathogenesis (Stumpf et al., 2010). Instead, mutations in the exonuclease domain cause pathogenesis by disrupting polymerase activity and not by decreasing fidelity (Stumpf et al., 2010, Szczepanowska and Foury, 2010). For the majority of the disease substitutions that have been studied in vitro, the biochemical defects correlate with the severity and age of onset found in patients (Chan and Copeland, 2009).
POLG and ataxia neuropathy spectrum and myoclonic epilepsy myopathy sensory ataxia phenotypes
Mutations in POLG can also cause ataxia-neuropathy syndrome with onset in the early teens to late thirties. This ataxia, also termed mitochondrial associated ataxia syndrome (MIRAS) (Hakonen et al., 2005), spino-cerebellar ataxia-epilepsy syndrome (SCAE), or SANDO (Van Goethem et al., 2004) is caused by autosomal recessive mutations in POLG producing multiple mtDNA deletions in the affected individuals. Symptoms of ataxia involving mutations in POLG include peripheral neuropathy, dysarthria, mild cognitive impairment, involuntary movements, psychiatric symptoms, myoclonus, and epileptic seizures. Ataxic patients who are homozygous for the A467T mutation present with symptoms in their early to late teens (Van Goethem et al., 2004, Winterthun et al., 2005, Tzoulis et al., 2006). SANDO patients have also been found to have compound heterozygous mutations with the A467T mutation in one POLG allele and either R3P, L304R, or R627W in the other (Van Goethem et al., 2003). One patient with ataxia-myopathy syndrome was shown to have A467T in one POLG allele and R627Q mutation in cis with the Q1236H single nucleotide polymorphism in the other POLG allele (Luoma et al., 2005). Other patients with ataxia were found to be heterozygous with the A467T mutation on one allele and W748S in cis with the E1143G mutation in the other allele. The latter mutation, E1143G, was originally identified as a SNP in 4% of the general population (Longley et al., 2005). However, the accumulated reports of this mutation with other POLG mutations in mitochondrial disease suggest that E1143G may augment the disease process (Hudson and Chinnery, 2006, Hudson et al., 2006). The W748S mutation in combination with E1143G has been found as a frequent cause of ataxia (Winterthun et al., 2005, Hakonen et al., 2005). Haplotype analysis of the Finnish population demonstrates a carrier frequency of 1:125 for the W748S mutation with a common ancestor origin of this diseased allele in the ancient European population (Hakonen et al., 2005). We have found that the W748S mutation alone causes the polymerase to have a low catalytic activity and a severe DNA-binding defect (Chan et al., 2006). The E1143G substitution partially rescues the deleterious effects of the W748S mutation and appears to modulate the effects of disease mutations in POLG.
POLG2, the accessory subunit of pol γ
Mutation of the POLG2 gene is rarely described as compared to mutations in the gene for the catalytic subunit. The first mutation described (c.1352G>A; G451E) was in a late onset adPEO patient with multiple mtDNA deletions in muscle and ptosis (Longley et al., 2006). The G451E substitution was in a region of the p55 protein that interacts with the p140 subunit, and caused a decreased in processivity of the enzyme complex due to a compromised p55-p140 subunit interaction (Longley et al., 2006). The second case also involved a late onset adPEO patient with mtDNA deletions and harbored a c.1207–1208ins24 mutation, causing mis-splicing and skipping of exon 7, thus impairing the C-terminal domain required for enzyme processivity (Walter et al., 2010). More recently, in a cohort of 112 patients with mitochondrial disease and suspected of POLG involvement but absent of POLG mutations, 8 heterozygous mutations were identified where 7 mutations were novel (Young et al., 2011). Biochemical analysis of these 7 variant p55 proteins (G103S, L153V, P205R, R369G, D386E, S423Y, and L475DfsX2) revealed that G103S, L153V, D386E and S423Y where similar to wild-type, and the P205R and R369G p55 variants had reduced stimulation of processivity and decreased affinity for the catalytic subunit (Young et al., 2011). The L475DfsX2 protein was unable to bind the p140 catalytic subunit, unable to bind dsDNA and was generally unstable. The failure to enhance processivity in the catalytic subunit by these mutant variants would cause the complex to stall during DNA replication and is consistent with the accumulation of mtDNA deletions detected in PEO.
The mitochondrial DNA helicase, the TWINKLE or C10orf21 gene
The mitochondrial DNA helicase encoded by the PEO1 or TWINKLE gene is related to the phage T7 gp4 helicase-primase. This gene was first identified as a locus for PEO on chromosome 10, C10orf2 (Spelbrink et al., 2001). The derived amino acid sequence showed significant sequence homology to the C-terminal end of T7 gp4 helicase but lacked critical primase-associated sequences found in T7 gp4 (Spelbrink et al., 2001, Garrido et al., 2003). Screening of the C10orf2 gene in individuals with adPEO, associated with multiple mtDNA deletions, identified 11 different mutations that co-segregated with the disorder in 12 affected families (Spelbrink et al., 2001). At least 23 additional missense mutations in C10orf2 associated have been reported in adPEO (Van Hove et al., 2009, Fratter et al., 2010). Although mutations in C10orf2 are mainly associated with adPEO, several reports have described recessive mutations as a cause of either epileptic encephalopathy with mtDNA depletion or infantile-onset spinocerebellar ataxia (Hudson et al., 2005, Hakonen et al., 2007, Lonnqvist et al., 2009).
Mouse transgenic models that overexpressed several of the PEO mutations in C10orf2 recapitulated many of the symptoms observed in human PEO including multiple mtDNA deletions, progressive respiratory dysfunction, and cytochrome c oxidase deficiency (Tyynismaa et al., 2005, Tyynismaa and Suomalainen, 2009). Overexpression of dominant disease variants of the mtDNA helicase in cultured human or Schneider cells results in stalled mtDNA replication or depletion of mtDNA (Wanrooij et al., 2007, Matsushima and Kaguni, 2007, Goffart et al., 2009), which emulates the disease state. Two of five adPEO mutants exhibited a dominant negative phenotype with mtDNA depletion in Schneider cells (Matsushima and Kaguni, 2007). Disease mutations in the C10orf2 gene have been evaluated by study of the recombinant helicase harboring these mutations as a homogeneous population. Analysis of the recombinant helicase purified from baculoviral infected insect cells has demonstrated defects in the helicase due to disease mutations. Disease mutations in the linker region were shown to disrupt protein hexamerization and abolish DNA helicase activity (Korhonen et al., 2008). Four mutations in the N-terminal domain demonstrated a dramatic decrease in ATPase activity (Holmlund et al., 2009). A comprehensive study of recombinant disease variants overproduced and purified from E. coli has reveled that all of the disease variants display some level of activity where mild to moderate defects were seen variably over 20 different variants with defects in helicase activity, ATP hydrolysis and stability (Longley et al., 2010). All of the variants displayed efficient DNA binding. This study emphasises the need to optimize in vitro conditions for biochemical analysis of disease variants (Longley et al., 2010). The moderate defects demonstrated in vitro is consistent with the delayed onset the adPEO associated with mutation of C10orf2.
Defects in mitochondrial nucleotide metabolism
Mitochondrial dNTP pools arise either through active transport of cytsolic dNTPs or through the purine and pyrimidine salvage pathways by action of two mitochondrial deoxyribonucleoside kinases, thymidine kinase 2 (TK2) and deoxyguanosine kinase (DGUOK) (Figure 1). In non-dividing cells cytosolic TK1 and dNTP synthesis is down-regulated forcing the burden of mitochondrial dNTP pool synthesis on the two mitochondrial deoxyribonucleoside kinases. Mutations in the genes for enzymes involved in this salvage pathway cause several forms of mtDNA depletion syndromes. Computational models suggest that this salvage pathway is insufficient to support rapid mtDNA replication and the mitochondrial salvage pathway serves more as a backup role for the supply of nucleotide precursors for mtDNA replication (Gandhi and Samuels, 2011). This model helps to explain why defects in TK2 are restrictive to muscle and present as myopathic mtDNA depletion syndromes (Mancuso et al., 2002). An alternative pathway for mitochondrial pools of nucleotides involves transport from cytoplasmic pools, arising from reduction of ribonucleotides by ribonucleotide reductase (RNR). Nucleotide pools arising from RNR would then be imported into the mitochondria as dNTPs to mix with dNTPs derived from TK2/DGUOK pathways (Figure 1).
Thymidine Phosphorylase, TP or TYMP
Thymidine Phosphorylase (TP) is part of the pyrimidine salvage pathway required for the reversible reaction catalyzing thymidine and phosphate to thymine and deoxyribose-1-phosphate (Figure 1). A defect in this gene (TYMP) causes mitochondrial neurogastrointestinal encephalopathy (MNGIE) due to the accumulation of thymidine and uracil in the blood. Because mitochondria rely heavily on salvage pathways for generating intramitochondrial dNTP pools, the mitochondria will uptake the excess thymidine which stimulates the synthesis of unbalanced and elevated dTTP levels by thymidine kinase 2 in the mitochondria. The unbalanced mitochondrial deoxynucleotide pools result in mitochondrial DNA depletion, multiple deletions, and point mutations. MNGIE is an autosomal recessive disorder caused by mutations in the gene for thymidine phosphorylase (Nishino et al., 1999, Hirano et al., 2005). There are over 30 mutations in TP associated with MNGIE (Hirano et al., 2005). Similar to PEO, the disease is associated with multiple deletions and depletion of mtDNA in affected tissues (Hirano et al., 2001). The onset usually takes place between the second and fifth decade of life and typical clinical features include ptosis, progressive external ophthalmoparesis, gastrointestinal dysmotility, cachexia, peripheral neuropathy, myopathy, and leukoencephalopathy (Hirano et al., 2004, Nishino et al., 2000). TP deficiency leads to increased concentrations of circulating deoxythymidine (Spinazzola et al., 2001) and deoxyuridine (Marti et al., 2003), which generates imbalanced mitochondrial deoxyribonucleoside triphosphate pools, the effect of which can be responsible for increased mtDNA mutagenesis (Song et al., 2003). Analysis of thymidine and deoxyuridine levels in post-mortem samples of MNGIE patients have revealed that the absence of TP activity causes an accumulation of thymidine and nucleosides, higher than observed in the plasma of these patients (Valentino et al., 2007).
The majority (>80%) of mtDNA mutations found in tissues from MNGIE patients are T to C transitions preceded by a short run of As (Nishigaki et al., 2003). This signature mutation suggests a “next-nucleotide effect” caused by the more common T:dGMP misinsertion (Longley et al., 2001) which is quickly extended by the elevated dTTP concentration resulting from TP deficiency in the mitochondria of MNGIE cells (Nishigaki et al., 2003). Additionally, elevated concentrations of dTMP derived from the increased thymidine can inhibit the exonuclease activity of pol γ (Lim and Copeland, 2001). HeLa cells grown in the media supplemented with 50 μM thymidine causes mtDNA deletions and elevated mitochondrial pools of dTTP and dGTP, a result that recapitulated many of the genetic effects seen in MNGIE (Song et al., 2003). These results support a mutagenic mechanism involving competition between dGTP and dATP for incorporation opposite to template T reported by Nishigaki and colleagues (Nishigaki et al., 2003).
Several lines of therapy or treatment of MNGIE are underway. Hemodialysis has been shown to transiently reduce thymidine levels in blood (Spinazzola et al., 2002). Thymidine levels in blood can be reduced in MNGIE patients by repeated platelet infusions (Lara et al., 2006). Allogeneic stem cell transplantation has been shown to have some success in restoring TP activity and lowering plasma thymidine levels (Hirano et al., 2006) however this approach shows toxicity of myeloablative conditional and graft rejections. More recently, hematopoietic gene therapy by the introduction of TP with lentiviral vectors into TP-deficient B-lymphoblastoid cells and TP/Upp1 knockout mice was shown to restore TP activity with the concomitant reduction of nucleosides in peripheral blood (Torres-Torronteras et al.).
Mitochondrial thymidine kinase, TK2
In a study of four unrelated patients with fatal myopathy during infancy and mitochondrial depletion syndrome, Saada et al. found reduced mitochondrial respiratory chain function and reduced amounts of mtDNA in muscle (Saada et al., 2001). Sequence analysis of the thymidine kinase 2 (TK2) gene identified either the homozygous H90R mutation or the homozygous I181D mutation in these patients. Activity analysis in mitochondrial lysates from these patients revealed reduced TK2 activity (Saada et al., 2001). DNA sequence by other groups have identified over 20 missense mutations found either as recessive homozygous or as compound heterozygous mutation and one stop codon from a total (Mancuso et al., 2002, Mancuso et al., 2003, Carrozzo et al., 2003, Wang et al., 2005, Galbiati et al., 2006, Alberio et al., 2007, Gotz et al., 2008, Blakely et al., 2008, Collins et al., 2009, Lesko et al., 2010). TK2 defects due to two compound heterozygous mutations, R172W and R225W, were also found to be responsible for multi-tissue mtDNA depletion syndrome in five unrelated patients and two brothers (Gotz et al., 2008). A review of these mutations in the structure of the human TK2 shows a random distribution of these mutations over the TK2 gene (Eriksson and Wang, 2008). TK2 gene mutations primarily affect muscle tissue with no effect on liver, brain, heart, or skin. Quantitation of TK2 activity in various tissues relative to mitochondrial DNA or cytochrome c oxidase activity helps to explain the tissue specificity of this disorder (Saada et al., 2003).
Kinetic analysis of two TK2 mutants, I212N and H121N demonstrated that the I212N mutant enzyme has less than 1% activity while the H121N mutant enzyme has a 2–3 fold lower Vmax as compared to the wild type TK2 enzyme (Wang et al., 2003). Biochemical analysis of two other TK2 mutations, T77M and R161K, found as compound heterozygous mutation in one patient revealed that the recombinant enzyme only retained 2–4% activity (Wang et al., 2005). Introduction of the H126N mutation in mouse knockin mice caused rapid progressive weakness 10 days after birth followed by death between 2–3 weeks (Akman et al., 2008). These mice recapitulated many of the symptoms of MDS and showed TK2 deficiency, unbalanced dNTP pools and mtDNA depletion, along with electron chain respiratory defects (Akman et al., 2008).
Mitochondrial deoxyguanosine kinase, DGUOK
Deoxyguanosine kinase is the other mitochondrial deoxyribonucleoside kinase and phosphorylates the purine nucleosides into nucleotide monophosphates (Figure 1). Homozygosity mapping in three consanguineous kindreds with hepatocerebral MDS identified a region on chromosome 2p13 that included the deoxyguanosine kinase gene (Mandel et al., 2001). Sequence analysis of this gene identified a nucleotide deletion (204delA) that segregated with the disease (Mandel et al., 2001). In a screen of 21 patients with MDS, Salviati et al. identified 3 patients (14%) with DGUOK gene mutations (Salviati et al., 2002). Phenotypes of these three patients were highly variable, including one patient who developed liver failure but responded well to liver transplant (Salviati et al., 2002). To confirm that the DGUOK mutations did indeed disrupt enzymatic function, Wang et al. characterized recombinant DGUOK proteins with these substitutions (Wang and Eriksson, 2003). They found that the R142K variant had very low activity with dG and no activity with dA. The E227K protein had normal affinity for dG and dA but low catalytic efficiency indicating that this mutation disrupts catalysis without affecting substrate binding. The C-terminal truncated variants were inactive. Further analysis later by Wang et al. identified one patient with MDS, muscular weakness, and exercise intolerance due to a severe mitochondrial myopathy that harbored the L250S mutation in DGUOK (Wang et al., 2005). Examination of the recombinant L250S-DGUOK protein revealed <1% activity as compared to the wild type enzyme with differential competition between dC and dT as substrates (Wang et al., 2005). De Camaret et al., identified two missense mutations, N46S and L266R in a 10 yr old child with hepatocerebral form of mtDNA depletion syndrome (Mousson de Camaret et al., 2007). Kinetic analysis mitochondrial fractions from skin fibroblasts derived from this patient as compared to normal cells showed 10 and 14% kinase activity for dG and dA substrates, respectively (Mousson de Camaret et al., 2007). Freisinger also identified 5 new mutations and 2 previously described mutations in 6 children with infantile hepatoencephalopathies and MDS (Freisinger et al., 2006). There has been nearly 40 different mutations described in the DGUOK gene, mostly presented as homozygous mutations from over 25 different probands (Freisinger et al., 2006, Alberio et al., 2007, Dimmock et al., 2008). In a screen of 9 kindreds, Dimmock et al noted that mutations that result in truncated proteins (nonsense, splice site or frameshift mutations) predominante over missense mutations (Dimmock et al., 2008). Patients with null mutations usually present with early onset liver failure with significant neurological symptoms with death before 2 years of age (Dimmock et al., 2008). Interestingly, five patients were identified in new born screening with elevated tyrosine or phenylalaine levels (Dimmock et al., 2008).
Currently, two forms of deoxyguanosine kinase deficiency have been described, the hepatocerebral mitochondrial DNA depletion syndrome which presents as a multisystem disorder and an isolated hepatic disease later in infancy or childhood (Scaglia et al., 2009). The majority of affected individuals have the multisystem illness with hepatic disease (cholestasis) and neurologic dysfunction evident within weeks of birth (reviewed in (Scaglia et al., 2009)).
RRM2B, the p53 inducible small subunit of ribonucleotide reductase
In contrast to the mitochondrial salvage pathway, nucleotide precursors required for DNA replication can be directly obtained by reduction of ribonucleoside diphosphates to deoxyribonucloside diphosphates by ribonucleotide reductase. Ribonucleotide reductase is made up of two subunits, a large catalytic subunit, R1, and the smaller R2 subunit. Cells have two forms of the R2 subunit, a cell cycle regulated form that is maximally expressed in S-phase, and a p53-inducible form known as p53R2. The p53R2 from is required for a basal level of DNA repair and mtDNA synthesis in non-proliferating cells. In 2007 Bourdon et al. (Bourdon et al., 2007) identified the RRM2B gene encoding p532B as a candidate disease gene from a genome-wide linkage analysis in a family with severe muscle mtDNA depletion. Sequence analysis of RRM2B in this family and three other affected families identified nonsense, missense and splice-site mutations and in-frame deletions within the RRM2B gene that were not found in control chromosomes (Bourdon et al., 2007). Severe mtDNA depletion from RRM2B disruption was also confirmed in Rrm2b−/− mice demonstrating the essential role of this gene in mtDNA nucleotide metabolism and mitochondrial disease (Bourdon et al., 2007).
Similar to POLG related disorders the symptoms and signs associated with RRM2B mutations cover a wide spectrum by age and disease. A 42 year old women with mitochondrial neurogastrointestinal encephalopathy and mtDNA depletion in muscle was found to have two pathogenic mutations, R110H and R121H in the RRM2B gene (Shaibani et al., 2009). These two mutations are believed to affect the dimerization of the RNR small subunit. RRM2B mutations have also been described in PEO patients. A heterozygous stop codon mutation was described in two families with autosomal dominant PEO (Tyynismaa et al., 2009). The stop codon mutation resided in exon 9 and escaped nonsense-mediated decay resulting in a dominant negative protein truncated by 26 amino acids and. This C-terminal region is believed to be essential for interaction with the R1 large subunit of RNR. To determine the frequency of RRM2B mutations in patients with PEO, Fratter et al., studied 75 probands with mtDNA deletions and PEO symptoms (Fratter et al., 2011). They found that 16% of the subjects contained mutations in the RRM2B gene for a total of 10 different RRM2B variants in 12 subjects. Seven patients had a single heterozygous missense mutations resulting in truncated proteins and two patients harbored compound heterozygous mutation.
Conclusion
The role of nuclear genes and the disease mutations in these genes that function in the maintenance of our mitochondrial DNA has gained increased appreciation over the last ten years. Disease of mtDNA stability are found in core proteins of mtDNA replication or in genes involved in supplying the mitochondrial nucleotide precursors needed for DNA replication (Figure 1). With our awareness of disease mutations in these genes the frequency of mitochondrial patients due to mutation in these genes will continue to increase. As an example, the number of individual harboring a recessive pathogenic mutation in POLG has been estimated to approach 2% of the population (Cohen and Naviaux, 2010). However, the varied polymorphic nature of these diseases as well as the age of presentation due to these gene mutations stumps our understanding and challenges clinician and researchers. While in vitro biochemistry and yeast models have proven very effective in understanding the biochemical and pathological defects, animal models are becoming an essential tool for predicting the in vivo consequences of heritable mutations with these genes. The next ten years of research should be illuminating in this field.
Footnotes
Declaration of interest
The author reports no declarations of interests. This review was supported by intramural funds from the National Institute of Environmental Health Sciences, NIH (ES 065078 and ES 065080).
References
- Akman HO, Dorado B, Lopez LC, Garcia-Cazorla A, Vila MR, Tanabe LM, Dauer WT, Bonilla E, Tanji K, Hirano M. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum Mol Genet. 2008;17:2433–40. doi: 10.1093/hmg/ddn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberio S, Mineri R, Tiranti V, Zeviani M. Depletion of mtDNA: Syndromes and genes. Mitochondrion. 2007;7(1–2):6–12. 6–12. doi: 10.1016/j.mito.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim Biophys Acta. 2007;1772:1225–35. doi: 10.1016/j.bbadis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Horvath R, Dallabona C, Czermin B, Lamantea E, Bindoff L, Invernizzi F, Ferrero I, Zeviani M, Lodi T. Predicting the contribution of novel POLG mutations to human disease through analysis in yeast model. Mitochondrion. 2011;11:182–90. doi: 10.1016/j.mito.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Lodi T, Dallabona C, Puglisi A, Zeviani M, Ferrero I. Genetic and chemical rescue of the Saccharomyces cerevisiae phenotype induced by mitochondrial DNA polymerase mutations associated with progressive external ophthalmoplegia in humans. Hum Mol Genet. 2006;15:2846–55. doi: 10.1093/hmg/ddl219. [DOI] [PubMed] [Google Scholar]
- Blakely E, He L, Gardner JL, Hudson G, Walter J, Hughes I, Turnbull DM, Taylor RW. Novel mutations in the TK2 gene associated with fatal mitochondrial DNA depletion myopathy. Neuromuscul Disord. 2008;18:557–60. doi: 10.1016/j.nmd.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, De Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–80. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- Carrozzo R, Bornstein B, Lucioli S, Campos Y, De La Pena P, Petit N, Dionisi-Vici C, Vilarinho L, Rizza T, Bertini E, Garesse R, Santorelli FM, Arenas J. Mutation analysis in 16 patients with mtDNA depletion. Hum Mutat. 2003;21:453–4. doi: 10.1002/humu.9135. [DOI] [PubMed] [Google Scholar]
- Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: Understanding the consequence of POLG mutations. Biochim Biophys Acta. 2009;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SSL, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem. 2005a;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- Chan SSL, Longley MJ, Copeland WC. Modulation of the W748S mutation in DNA polymerase {gamma} by the E1143G polymorphism in mitochondrial disorders. Hum Mol Genet. 2006;15:3473–3483. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SSL, Longley MJ, Naviaux RK, Copeland WC. Mono-allelic POLG expression resulting from nonsense-mediated decay and alternative splicing in a patient with Alpers syndrome. DNA Repair. 2005b;4:1381–1389. doi: 10.1016/j.dnarep.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Cohen BH, Chinnery PF, Copeland WC. POLG-Related Disorders. GeneReviews at GeneTests: Medical Genetics Information Resource [database online] Copyright, University of Washington; Seattle: 2010. pp. 1997–2010. Available at http://www.genetests.org. [Google Scholar]
- Cohen BH, Naviaux RK. The clinical diagnosis of POLG disease and other mitochondrial DNA depletion disorders. Methods. 2010;51:364–373. doi: 10.1016/j.ymeth.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Collins J, Bove KE, Dimmock D, Morehart P, Wong LJ, Wong B. Progressive myofiber loss with extensive fibro-fatty replacement in a child with mitochondrial DNA depletion syndrome and novel thymidine kinase 2 gene mutations. Neuromuscul Disord. 2009;19:784–7. doi: 10.1016/j.nmd.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–46. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries MC, Rodenburg RJ, Morava E, Van Kaauwen EP, Ter Laak H, Mullaart RA, Snoeck IN, Van Hasselt PM, Harding P, Van Den Heuvel LP, Smeitink JA. Multiple oxidative phosphorylation deficiencies in severe childhood multi-system disorders due to polymerase gamma (POLG1) mutations. Eur J Pediatr. 2007;166:229–234. doi: 10.1007/s00431-006-0234-9. [DOI] [PubMed] [Google Scholar]
- Dimmock DP, Zhang Q, Dionisi-Vici C, Carrozzo R, Shieh J, Tang LY, Truong C, Schmitt E, Sifry-Platt M, Lucioli S, Santorelli FM, Ficicioglu CH, Rodriguez M, Wierenga K, Enns GM, Longo N, Lipson MH, Vallance H, Craigen WJ, Scaglia F, Wong LJ. Clinical and molecular features of mitochondrial DNA depletion due to mutations in deoxyguanosine kinase. Hum Mutat. 2008;29:330–1. doi: 10.1002/humu.9519. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Wang L. Molecular mechanisms of mitochondrial DNA depletion diseases caused by deficiencies in enzymes in purine and pyrimidine metabolism. Nucleosides Nucleotides Nucleic Acids. 2008;27:800–8. doi: 10.1080/15257770802146197. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–99. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Ferrari G, Lamantea E, Donati A, Filosto M, Briem E, Carrara F, Parini R, Simonati A, Santer R, Zeviani M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-{gamma}A. Brain. 2005;128:723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- Fratter C, Gorman GS, Stewart JD, Buddles M, Smith C, Evans J, Seller A, Poulton J, Roberts M, Hanna MG, Rahman S, Omer SE, Klopstock T, Schoser B, Kornblum C, Czermin B, Lecky B, Blakely EL, Craig K, Chinnery PF, Turnbull DM, Horvath R, Taylor RW. The clinical, histochemical, and molecular spectrum of PEO1 (Twinkle)-linked adPEO. Neurology. 2010;74:1619–26. doi: 10.1212/WNL.0b013e3181df099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratter C, Raman P, Alston CL, Blakely EL, Craig K, Smith C, Evans J, Seller A, Czermin B, Hanna MG, Poulton J, Brierley C, Staunton TG, Turnpenny PD, Schaefer AM, Chinnery PF, Horvath R, Turnbull DM, Gorman GS, Taylor RW. RRM2B mutations are frequent in familial PEO with multiple mtDNA deletions. Neurology. 2011;76:2032–4. doi: 10.1212/WNL.0b013e31821e558b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisinger P, Futterer N, Lankes E, Gempel K, Berger TM, Spalinger J, Hoerbe A, Schwantes C, Lindner M, Santer R, Burdelski M, Schaefer H, Setzer B, Walker UA, Horvath R. Hepatocerebral mitochondrial DNA depletion syndrome caused by deoxyguanosine kinase (DGUOK) mutations. Arch Neurol. 2006;63:1129–34. doi: 10.1001/archneur.63.8.1129. [DOI] [PubMed] [Google Scholar]
- Galbiati S, Bordoni A, Papadimitriou D, Toscano A, Rodolico C, Katsarou E, Sciacco M, Garufi A, Prelle A, Aguennouz M, Bonsignore M, Crimi M, Martinuzzi A, Bresolin N, Papadimitriou A, Comi GP. New mutations in TK2 gene associated with mitochondrial DNA depletion. Pediatr Neurol. 2006;34:177–85. doi: 10.1016/j.pediatrneurol.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Gandhi VV, Samuels DC. Enzyme Kinetics of the Mitochondrial Deoxyribonucleoside Salvage Pathway Are Not Sufficient to Support Rapid mtDNA Replication. PLoS Comput Biol. 2011;7:e1002078. doi: 10.1371/journal.pcbi.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido N, Griparic L, Jokitalo E, Wartiovaara J, Van Der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–96. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18:328–40. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz A, Isohanni P, Pihko H, Paetau A, Herva R, Saarenpaa-Heikkila O, Valanne L, Marjavaara S, Suomalainen A. Thymidine kinase 2 defects can cause multi-tissue mtDNA depletion syndrome. Brain. 2008;131:2841–50. doi: 10.1093/brain/awn236. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat Struct Mol Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in Mitochondrial DNA Replication and Repair. Chemical Reviews. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, Goethem GV, Lofgren A, Hackman P, Paetau A, Kaakkola S, Majamaa K, Varilo T, Udd B, Kaariainen H, Bindoff LA, Suomalainen A. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am J Hum Genet. 2005;77:430–41. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonen AH, Isohanni P, Paetau A, Herva R, Suomalainen A, Lonnqvist T. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain. 2007;130:3032–40. doi: 10.1093/brain/awm242. [DOI] [PubMed] [Google Scholar]
- Hirano M, Lagier-Tourenne C, Valentino ML, Marti R, Nishigaki Y. Thymidine phosphorylase mutations cause instability of mitochondrial DNA. Gene. 2005;354:152–6. doi: 10.1016/j.gene.2005.04.041. [DOI] [PubMed] [Google Scholar]
- Hirano M, Marti R, Casali C, Tadesse S, Uldrick T, Fine B, Escolar DM, Valentino ML, Nishino I, Hesdorffer C, Schwartz J, Hawks RG, Martone DL, Cairo MS, Dimauro S, Stanzani M, Garvin JH, Jr, Savage DG. Allogeneic stem cell transplantation corrects biochemical derangements in MNGIE. Neurology. 2006;67:1458–60. doi: 10.1212/01.wnl.0000240853.97716.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Marti R, Ferreiro-Barros C, Vila MR, Tadesse S, Nishigaki Y, Nishino I, Vu TH. Defects of intergenomic communication: autosomal disorders that cause multiple deletions and depletion of mitochondrial DNA. Semin Cell Dev Biol. 2001;12:417–27. doi: 10.1006/scdb.2001.0279. [DOI] [PubMed] [Google Scholar]
- Hirano M, Nishigaki Y, Marti R. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): a disease of two genomes. Neurologist. 2004;10:8–17. doi: 10.1097/01.nrl.0000106919.06469.04. [DOI] [PubMed] [Google Scholar]
- Holmlund T, Farge G, Pande V, Korhonen J, Nilsson L, Falkenberg M. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim Biophys Acta. 2009;1792:132–9. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Horvath R, Hudson G, Ferrari G, Futterer N, Ahola S, Lamantea E, Prokisch H, Lochmuller H, Mcfarland R, Ramesh V, Klopstock T, Freisinger P, Salvi F, Mayr JA, Santer R, Tesarova M, Zeman J, Udd B, Taylor RW, Turnbull D, Suomalainen A, Zeviani M, Chinnery PF. Phenotypic spectrum associated with mutations of the mitochondrial polymerase {gamma} gene. Brain. 2006;129:1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum Mol Genet. 2006;15(Spec No 2):R244–52. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- Hudson G, Deschauer M, Busse K, Zierz S, Chinnery PF. Sensory ataxic neuropathy due to a novel C10Orf2 mutation with probable germline mosaicism. Neurology. 2005;64:371–3. doi: 10.1212/01.WNL.0000149767.51152.83. [DOI] [PubMed] [Google Scholar]
- Hudson G, Deschauer M, Taylor RW, Hanna MG, Fialho D, Schaefer AM, He LP, Blakely E, Turnbull DM, Chinnery PF. POLG1, C10ORF2 & ANT1 mutations are uncommon in sporadic PEO with multiple mtDNA deletions. Neurology. 2006;66:1439–1441. doi: 10.1212/01.wnl.0000210486.32196.24. [DOI] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kasiviswanathan R, Longley MJ, Chan SS, Copeland WC. Disease mutations in the human mitochondrial DNA polymerase thumb subdomain impart severe defects in MtDNA replication. J Biol Chem. 2009;284:19501–19510. doi: 10.1074/jbc.M109.011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen JA, Pande V, Holmlund T, Farge G, Pham XH, Nilsson L, Falkenberg M. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J. 2004;23:2423–9. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A. 2011;108:13534–9. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara MC, Weiss B, Illa I, Madoz P, Massuet L, Andreu AL, Valentino ML, Anikster Y, Hirano M, Marti R. Infusion of platelets transiently reduces nucleoside overload in MNGIE. Neurology. 2006;67:1461–3. doi: 10.1212/01.wnl.0000239824.95411.52. [DOI] [PubMed] [Google Scholar]
- Lesko N, Naess K, Wibom R, Solaroli N, Nennesmo I, Von Dobeln U, Karlsson A, Larsson NG. Two novel mutations in thymidine kinase-2 cause early onset fatal encephalomyopathy and severe mtDNA depletion. Neuromuscul Disord. 2010;20:198–203. doi: 10.1016/j.nmd.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Lewis W, Day BJ, Kohler JJ, Hosseini SH, Chan SSL, Green E, Haase CP, Keebaugh E, Long R, Ludaway T, Russ R, Steltzer J, Tioleco N, Santoianni R, Copeland WC. MtDNA depletion, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase gamma. J Biol Chem. 2001;276:23616–23. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, Chinnery PF. Mutant POLG2 Disrupts DNA Polymerase gamma Subunits and Causes Progressive External Ophthalmoplegia. Am J Hum Genet. 2006;78:1026–34. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase γ. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Humble MM, Sharief FS, Copeland WC. Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis and helicase activity. J Biol Chem. 2010;285:29690–702. doi: 10.1074/jbc.M110.151795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The Fidelity of Human DNA Polymerase gamma with and without Exonucleolytic Proofreading and the p55 Accessory Subunit. J Biol Chem. 2001;276:38555–62. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Lonnqvist T, Paetau A, Valanne L, Pihko H. Recessive twinkle mutations cause severe epileptic encephalopathy. Brain. 2009;132:1553–62. doi: 10.1093/brain/awp045. [DOI] [PubMed] [Google Scholar]
- Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors PA, Rautakorpi I, Peltonen PL, Majamaa PK, Somer H, Suomalainen A. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–82. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, Kiechl S, Kaguni LS, Suomalainen A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum Mol Genet. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Filosto M, Bonilla E, Hirano M, Shanske S, Vu TH, Dimauro S. Mitochondrial myopathy of childhood associated with mitochondrial DNA depletion and a homozygous mutation (T77M) in the TK2 gene. Arch Neurol. 2003;60:1007–9. doi: 10.1001/archneur.60.7.1007. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Salviati L, Sacconi S, Otaegui D, Camano P, Marina A, Bacman S, Moraes CT, Carlo JR, Garcia M, Garcia-Alvarez M, Monzon L, Naini AB, Hirano M, Bonilla E, Taratuto AL, Dimauro S, Vu TH. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59:1197–202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, Shalata A, Anbinder Y, Berkowitz D, Hartman C, Barak M, Eriksson S, Cohen N. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet. 2001;29:337–41. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- Marti R, Nishigaki Y, Hirano M. Elevated plasma deoxyuridine in patients with thymidine phosphorylase deficiency. Biochem Biophys Res Commun. 2003;303:14–8. doi: 10.1016/s0006-291x(03)00294-8. [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Kaguni LS. Differential phenotypes of active site and human adPEO mutations in drosophila mitochondrial DNA helicase expressed in schneider cells. J Biol Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31:e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousson De Camaret B, Taanman JW, Padet S, Chassagne M, Mayencon M, Clerc-Renaud P, Mandon G, Zabot MT, Lachaux A, Bozon D. Kinetic properties of mutant deoxyguanosine kinase in a case of reversible hepatic mtDNA depletion. Biochem J. 2007;402:377–85. doi: 10.1042/BJ20060705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Nguyen KV. POLG Mutations associated with Alpers’ Syndrome and Mitochondrial DNA Depletion. Ann Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Nyhan WL, Barshop BA, Poulton J, Markusic D, Karpinski NC, Haas RH. Mitochondrial DNA polymerase gamma deficiency and mtDNA depletion in a child with Alpers’ syndrome. Ann Neurol. 1999;45:54–8. doi: 10.1002/1531-8249(199901)45:1<54::aid-art10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Nguyen KV, Sharief F, Chan SSL, Copeland WC, Naviaux RK. Molecular Diagnosis of Alpers Syndrome. J Hepatology. 2006;45:108–116. doi: 10.1016/j.jhep.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Nishigaki Y, Marti R, Copeland WC, Hirano M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J Clin Invest. 2003;111:1913–21. doi: 10.1172/JCI17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–92. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Papadimitriou A, Hammans S, Steiner I, Hahn CD, Connolly AM, Verloes A, Guimaraes J, Maillard I, Hamano H, Donati MA, Semrad CE, Russell JA, Andreu AL, Hadjigeorgiou GM, Vu TH, Tadesse S, Nygaard TG, Nonaka I, Hirano I, Bonilla E, Rowland LP, Dimauro S, Hirano M. Mitochondrial neurogastrointestinal encephalomyopathy: an autosomal recessive disorder due to thymidine phosphorylase mutations. Ann Neurol. 2000;47:792–800. [PubMed] [Google Scholar]
- Pagnamenta AT, Taanman JW, Wilson CJ, Anderson NE, Marotta R, Duncan AJ, Bitner-Glindzicz M, Taylor RW, Laskowski A, Thorburn DR, Rahman S. Dominant inheritance of premature ovarian failure associated with mutant mitochondrial DNA polymerase gamma. Hum Reprod. 2006;21:2467–2473. doi: 10.1093/humrep/del076. [DOI] [PubMed] [Google Scholar]
- Ponamarev MV, Longley MJ, Nguyen D, Kunkel TA, Copeland WC. Active Site Mutation in DNA Polymerase gamma Associated with Progressive External Ophthalmoplegia Causes Error-prone DNA Synthesis. J Biol Chem. 2002;277:15225–8. doi: 10.1074/jbc.C200100200. [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Elpeleg O. mtDNA depletion myopathy: elucidation of the tissue specificity in the mitochondrial thymidine kinase (TK2) deficiency. Mol Genet Metab. 2003;79:1–5. doi: 10.1016/s1096-7192(03)00063-5. [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29:342–4. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- Salviati L, Sacconi S, Mancuso M, Otaegui D, Camano P, Marina A, Rabinowitz S, Shiffman R, Thompson K, Wilson CM, Feigenbaum A, Naini AB, Hirano M, Bonilla E, Dimauro S, Vu TH. Mitochondrial DNA depletion and dGK gene mutations. Ann Neurol. 2002;52:311–7. doi: 10.1002/ana.10284. [DOI] [PubMed] [Google Scholar]
- Saneto RP, Lee IC, Koenig MK, Bao X, Weng SW, Naviaux RK, Wong LJ. POLG DNA testing as an emerging standard of care before instituting valproic acid therapy for pediatric seizure disorders. Seizure. 2010;19:140–6. doi: 10.1016/j.seizure.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneto RP, Naviaux RK. Polymerase gamma disease through the ages. Dev Disabil Res Rev. 2010;16:163–74. doi: 10.1002/ddrr.105. [DOI] [PubMed] [Google Scholar]
- Scaglia F, Dimmock D, Wong LJ. DGUOK-Related Mitochondrial DNA Depletion Syndrome, Hepatocerebral Form. GeneReviews at GeneTests: Medical Genetics Information Resource [database online] Copyright, University of Washington; Seattle: 2009. pp. 1997–2010. Available at http://www.genetests.org. [Google Scholar]
- Shaibani A, Shchelochkov OA, Zhang S, Katsonis P, Lichtarge O, Wong LJ, Shinawi M. Mitochondrial neurogastrointestinal encephalopathy due to mutations in RRM2B. Arch Neurol. 2009;66:1028–32. doi: 10.1001/archneurol.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Wheeler LJ, Mathews CK. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J Biol Chem. 2003;278:43893–6. doi: 10.1074/jbc.C300401200. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Marti R, Nishino I, Andreu AL, Naini A, Tadesse S, Pela I, Zammarchi E, Donati MA, Oliver JA, Hirano M. Altered thymidine metabolism due to defects of thymidine phosphorylase. J Biol Chem. 2001;3:3. doi: 10.1074/jbc.M111028200. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Marti R, Nishino I, Andreu AL, Naini A, Tadesse S, Pela I, Zammarchi E, Donati MA, Oliver JA, Hirano M. Altered thymidine metabolism due to defects of thymidine phosphorylase. J Biol Chem. 2002;277:4128–33. doi: 10.1074/jbc.M111028200. [DOI] [PubMed] [Google Scholar]
- Stuart GR, Santos JH, Strand MK, Van Houten B, Copeland WC. Mitochondrial DNA defects in S. cerevisiae with mutations in DNA polymerase gamma associated with Progressive External Ophthalmolplegia. Hum Mol Genet. 2006;15:363–374. doi: 10.1093/hmg/ddi454. [DOI] [PubMed] [Google Scholar]
- Stumpf JD, Bailey CM, Spell D, Stillwagon M, Anderson KS, Copeland WC. mip1 Containing mutations associated with mitochondrial disease causes mutagenesis and depletion of mtDNA in Saccharomyces cerevisiae. Hum Mol Genet. 2010;19:2123–33. doi: 10.1093/hmg/ddq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowska K, Foury F. A cluster of pathogenic mutations in the 3′-5′ exonuclease domain of DNA polymerase gamma defines a novel module coupling DNA synthesis and degradation. Hum Mol Genet. 2010;19:3516–3529. doi: 10.1093/hmg/ddq267. [DOI] [PubMed] [Google Scholar]
- Tang S, Wang J, Lee NC, Milone M, Halberg MC, Schmitt ES, Craigen WJ, Zhang W, Wong LJ. Mitochondrial DNA polymerase {gamma} mutations: an ever expanding molecular and clinical spectrum. J Med Genet. 2011;48:669–81. doi: 10.1136/jmedgenet-2011-100222. [DOI] [PubMed] [Google Scholar]
- Torres-Torronteras J, Gomez A, Eixarch H, Palenzuela L, Pizzorno G, Hirano M, Andreu AL, Barquinero J, Marti R. Hematopoietic gene therapy restores thymidine phosphorylase activity in a cell culture and a murine model of MNGIE. Gene Ther. 2011;18:795–806. doi: 10.1038/gt.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102:17687–92. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 2009;10:137–43. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Ylikallio E, Patel M, Molnar MJ, Haller RG, Suomalainen A. A heterozygous truncating mutation in RRM2B causes autosomal-dominant progressive external ophthalmoplegia with multiple mtDNA deletions. Am J Hum Genet. 2009;85:290–5. doi: 10.1016/j.ajhg.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulis C, Engelsen BA, Telstad W, Aasly J, Zeviani M, Winterthun S, Ferrari G, Aarseth JH, Bindoff LA. The spectrum of clinical disease caused by the A467T and W748S POLG mutations: a study of 26 cases. Brain. 2006;129:1685–1692. doi: 10.1093/brain/awl097. [DOI] [PubMed] [Google Scholar]
- Valentino ML, Marti R, Tadesse S, Lopez LC, Manes JL, Lyzak J, Hahn A, Carelli V, Hirano M. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) FEBS Lett. 2007;581:3410–4. doi: 10.1016/j.febslet.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Luoma P, Rantamaki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Lofgren A, Martin JJ, De Jonghe P, Suomalainen A, Udd B, Van Broeckhoven C. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Martin JJ, Dermaut B, Lofgren A, Wibail A, Ververken D, Tack P, Dehaene I, Van Zandijcke M, Moonen M, Ceuterick C, De Jonghe P, Van Broeckhoven C. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul Disord. 2003;13:133–142. doi: 10.1016/s0960-8966(02)00216-x. [DOI] [PubMed] [Google Scholar]
- Van Hove JL, Cunningham V, Rice C, Ringel SP, Zhang Q, Chou PC, Truong CK, Wong LJ. Finding twinkle in the eyes of a 71-year-old lady: a case report and review of the genotypic and phenotypic spectrum of TWINKLE-related dominant disease. Am J Med Genet A. 2009;149A:861–7. doi: 10.1002/ajmg.a.32731. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–8. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Walter MC, Czermin B, Muller-Ziermann S, Bulst S, Stewart JD, Hudson G, Schneiderat P, Abicht A, Holinski-Feder E, Lochmuller H, Chinnery PF, Klopstock T, Horvath R. Late-onset ptosis and myopathy in a patient with a heterozygous insertion in POLG2. J Neurol. 2010;257:1517–1523. doi: 10.1007/s00415-010-5565-9. [DOI] [PubMed] [Google Scholar]
- Wang L, Eriksson S. Mitochondrial deoxyguanosine kinase mutations and mitochondrial DNA depletion syndrome. FEBS Lett. 2003;554:319–22. doi: 10.1016/s0014-5793(03)01181-5. [DOI] [PubMed] [Google Scholar]
- Wang L, Limongelli A, Vila MR, Carrara F, Zeviani M, Eriksson S. Molecular insight into mitochondrial DNA depletion syndrome in two patients with novel mutations in the deoxyguanosine kinase and thymidine kinase 2 genes. Mol Genet Metab. 2005;84:75–82. doi: 10.1016/j.ymgme.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Wang L, Saada A, Eriksson S. Kinetic properties of mutant human thymidine kinase 2 suggest a mechanism for mitochondrial DNA depletion myopathy. J Biol Chem. 2003;278:6963–8. doi: 10.1074/jbc.M206143200. [DOI] [PubMed] [Google Scholar]
- Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–51. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterthun S, Ferrari G, He L, Taylor RW, Zeviani M, Turnbull DM, Engelsen BA, Moen G, Bindoff LA. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology. 2005;64:1204–8. doi: 10.1212/01.WNL.0000156516.77696.5A. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, Basinger AA, Burton BK, Swoboda K, Gilbert DL, Vanderver A, Saneto RP, Maranda B, Arnold G, Abdenur JE, Waters PJ, Copeland WC. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum Mutat. 2008;29:E150–E172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MJ, Longley MJ, Li FY, Kasiviswanathan R, Wong LJ, Copeland WC. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20:3052–66. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, Dimauro S, Didonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–11. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]