Abstract
Purpose
The specific role of Chromosomal Instability (CIN) in tumorigenesis has been a matter of conjecture. In part, this is due to the challenge of directly observing chromosome mis-segregation events as well as the inability to distinguish the role of CIN, which consists of increased rates of chromosome mis-segregation, from that of aneuploidy, which is a state of non-diploid chromosome number.
Experimental design
Here, we examine the contribution of CIN to the prognosis of patients diagnosed with Diffuse Large B-Cell Lymphoma (DLBCL) by directly surveying tumor cells, fixed while undergoing anaphase, for evidence of chromosome mis-segregation. H&E-stained samples from a cohort of 54 patients were used to examine the relationship between frequencies of chromosome mis-segregation and patient prognosis, overall survival, and response to treatment.
Results
We show that a two-fold increase in the frequency of chromosome mis-segregation led to a 24% decrease in overall survival and 48% decrease in relapse-free survival after treatment. The hazard ratio of death in patients with increased chromosome mis-segregation was 2.31 and these patients were more likely to present with higher tumor stage, exhibit tumor bone marrow involvement, and receive a higher International Prognostic Index (IPI) score.
Conclusions
Increased rates of chromosome mis-segregation in DLBCL substantiate inferior outcome and poor prognosis. This is likely due to increased heterogeneity of tumor cells leading to a larger predilection for adaptation in response to external pressures such as metastasis and drug treatments. We propose that targeting CIN would yield improved prognosis and improved response to chemotherapeutic drugs.
Introduction
Chromosomal instability (CIN) is a hallmark of human neoplasms (1-3). Most solid and many hematopoeitic tumors have evidence of elevated frequencies of chromosome mis-segregation (1, 4-11). By definition, CIN leads to aneuploidy however not all aneuploid tumors are chromosomally unstable as is the case with many hematopoeitic malignancies (12, 13). In these malignancies, aneuploidy – generated by single events of chromosome mis-segregation or chromosomal translocation – confers tumorigenic potential independently of CIN (14). Similarly, patients with global constitutional aneuploidy – such as Down syndrome, Turner syndrome, and Mosaic Variegated Aneuploidy – exhibit increased incidence of malignancies (15-17). This close relationship between CIN and aneuploidy has significantly complicated the endeavor of identifying the independent role of CIN in cancer. In theory, these elevated rates of chromosome mis-segregation has the potential to increase heterogeneity in the tumor cell population thereby leading to increased incidence of metastasis, drug-resistance, and inferior outcome (3). Yet, studies report both beneficial and adverse effects of CIN in cancer and the precise role of chromosome mis-segregation in tumor prognosis remains unclear. Furthermore, aneuploidy is frequently used as a surrogate marker for CIN without directly measuring chromosome mis-segregation events (18). Work in mouse models reveals that inducing CIN – and therefore aneuploidy – in normal cells can act to either promote or inhibit tumor formation (19-22). Similarly, studies using genetic signatures associated with aneuploidy as a marker for CIN infer both positive as well as negative contributions of CIN to tumor prognosis (18, 23-27). The lack of direct measurements of chromosome mis-segregation, however, increases the probability of confounders while considering the conclusions about the respective roles of CIN and aneuploidy in cancer.
Many mechanisms of CIN and chromosome mis-segregation have recently been proposed (28). They range from faulty sister chromatid cohesion (29), to defects in the spindle assembly checkpoint (30), centrosome duplication (31-33), telomere dysfunction (34) and the regulation of microtubule attachments to chromosomes at kinetochores (35, 36). Interestingly, most of these mechanisms yield an observable phenotype during anaphase, and experimental evidence shows that the most common indicators of chromosome mis-segregation are lagging chromosome and chromatin bridges (11), which are largely caused by deregulation in kinetochore-microtubule attachments (35, 36), supernumerary centrosomes (32, 33), and telomere fusion (34, 37). They are largely the result of persistent attachment errors between microtubules and chromosomes at kinetochores whereby individual chromosomes are attached to microtubule emanating from both spindle poles (38, 39). This error is called merotelic attachment and it leads to abnormal chromosome movement during anaphase as well as the physical separation of lagging chromosomes from the rest of the properly segregating chromosomes. Alternatively, merotelic attachments in addition to other causes of chromosome mis-segregation can also lead to chromatin breakage leading to a visible chromatin bridge spanning the spindle mid-zone during anaphase. The direct link between lagging chromosomes, chromatin bridges and chromosome mis-segregation is firmly established (11, 35, 38).
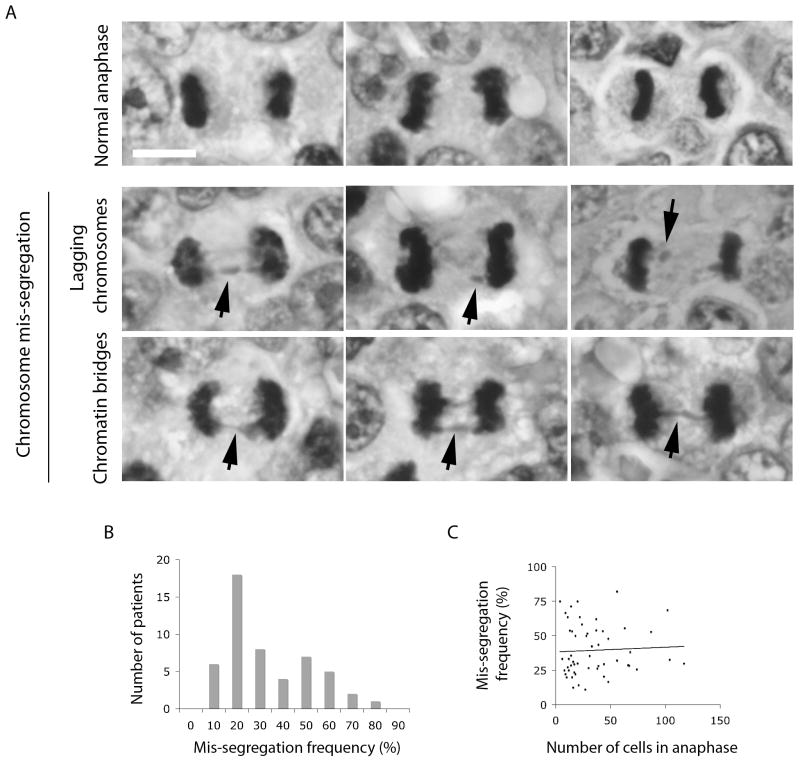
Since CIN most commonly manifests itself through lagging chromosomes and chromatin bridges, direct observation of cells undergoing anaphase in fixed tumor samples can provide insight into the role of CIN in the overall tumor prognosis. Here, we score chromosome segregation defects in anaphase as a direct marker of the dynamic process of chromosome mis-segregation associated with CIN in cancer. We elect to study samples from patients diagnosed with Diffuse Large B-Cell Lymphoma (DLBCL) because this cancer is known to be heterogeneously aneuploid, and CIN is postulated to play in important role in the natural evolution and aggressiveness of the tumor (9, 40-42). Furthermore, DLBCL samples provide adequate resolution to analyze individual cells during anaphase (Figure 1A) and exhibit a sufficiently elevated mitotic index necessary for our study.
Figure 1.
A, images of H&E stained samples showing cells during anaphase. Examples of normal anaphase, anaphase with lagging chromosomes and chromatin bridges (black arrows) are shown. Scale bar, 5-μm. B, histogram showing the distribution of patient samples based on frequency of anaphase cells which exhibit evidence of chromosome mis-segregation. C, Black circles depict the relationship between the number of anaphase cells and frequencies of chromosome mis-segregation observed in specimens taken from individual patients. Black line represents the linear trend, R2 = 0.0026.
Materials and methods
Samples and clinical data
Patients were included in this study based on the following criteria: 1) They were diagnosed as adults with DLBCL, 2) This diagnosis was the first of a kind and that patients had not received any prior treatment for lymphomas, 3) Their H&E stained samples from biopsied specimens were amenable to high-resolution microscopy to evaluate cells undergoing anaphase using a 100× objective. 73 cases of de novo DLBCL diagnosed between 1995 and 2008 were collected from the Department of Pathology at Dartmouth Medical School. Of the 73 cases only 54 were amenable to high-resolution analysis of anaphase cells. Samples were formalin-fixed, paraffin-embedded and stained with the standard H&E method. All anaphase cells present in available clinical samples were scored for evidence of chromosome mis-segregation (Figure 1B). An average number of 35 (up to 117) anaphases were scored in each sample (Figure 1C). Lagging chromosomes were defined as an area of hematoxylin staining completely isolated in between the remaining segregating chromosomes during anaphase. Chromatin bridges were defined as at least one continuous band of hematoxylin staining linking the remaining segregating chromosomes. The scoring of chromosome mis-segregation was performed in a blinded fashion prior to the statistical correlation with clinical variables. Clinical and follow-up data (summarized in Table 1) were obtained reviewing the charts. Retrieval of tissue and clinical data were done according to the regulations of the local institutional review board and data safety laws (IRB # 22910). Patients were staged by radiologic imaging (PET scan or CT chest/abdomen/pelvis) and bone marrow biopsy, IPI and R-IPI were assigned according to accepted criteria (43). The three parameters were analyzed: overall survival, progression-free survival and requirement for treatment. Overall survival was calculated by time period from the diagnosis to the endpoint (death, complete remission or loss to follow-up). Complete remission was defined as no evidence of disease clinically and by imaging for at least 6 months after the cessation of the last treatment regimen. Progression-free survival was calculated as time period from the completion of the first treatment to the need for additional treatment due to the progression/relapse of the disease. A list of patients with their clinical variables and details of treatment types are shown in Supplementary Table 1. Evidence of chromosome mis-segregation was scored using a 100× oil-immersion objective on a Nikon microscope (Lapophot 2). Images were collected on an Olympus BH-2 microscope with an Olympus DP25 camera camera using a 100× oil immersion objective.
Table 1.
Patient characteristics with respect to frequencies of chromosome mis-segregation.
| Patient characteristics | Number known | Mis-segregation frequency | P | |
|---|---|---|---|---|
| Low (<31.3%) | High (>31.3%) | |||
| Mean age at diagnosis | 52 | 59 | 66 | 0.77* |
| Sex | 54 | 0.57 | ||
| Male | 25 | 11 | 14 | |
| Female | 29 | 15 | 14 | |
| Primary organ of involvement | 53 | 0.38 | ||
| Nodal | 14 | 8 | 6 | |
| Extranodal | 39 | 17 | 22 | |
| Stage (I-IV) | 53 | 0.07 | ||
| I | 9 | 7 | 2 | |
| II | 17 | 8 | 9 | |
| III | 11 | 3 | 8 | |
| IV | 13 | 4 | 9 | |
| Stage (I/II or III/IV) | 53 | 0.042 | ||
| I/II | 26 | 15 | 11 | |
| III/IV | 24 | 7 | 17 | |
| IPI score | 47 | 0.0003 | ||
| Low (0-1) | 16 | 10 | 6 | |
| Low intermediate (2) | 12 | 5 | 7 | |
| High intermediate (3) | 3 | 1 | 2 | |
| High (4-5) | 16 | 5 | 11 | |
| Bone marrow involvement | 50 | 0.013 | ||
| Negative | 38 | 22 | 16 | |
| Positive | 12 | 2 | 10 | |
| Germinal phenotype | 42 | 0.57 | ||
| Germinal center | 17 | 6 | 11 | |
| Non-germinal center | 25 | 11 | 14 | |
| Response to treatment | 51 | 0.15 | ||
| Complete remission | 27 | 15 | 12 | |
| Failure to complete remission | 25 | 9 | 16 | |
T-test. For the remaining P-values, Pearson χ2-test was used.
Statistical analysis
Statistical analysis was done using XLSTAT version 2011.2.01 (Addinsoft, NY, USA). The median value of mis-segregation frequency (31.3%) was used as a cutoff to stratify tumor samples as having either low or high mis-segregation frequencies. This cutoff was used throughout unless otherwise noted. The Pearson χ2 statistic was used to analyze relationships between chromosome mis-segregation frequencies and clinical variables. The Kaplan-Meier method was used to estimate the probability of overall survival after diagnosis as well as treatment-free survival. The effect of chromosome mis-segregation and clinical variables on overall survival and treatment-free survival were determined using the log-rank test. Hazard ratios were calculated using the Cox multivariate proportional-hazard regression method. P < 0.05 were considered significant throughout. Two-sided tests were used throughout.
Results
All cells fixed undergoing anaphase in samples taken from tumor biopsies from patients diagnosed with DLBCL were surveyed for evidence of chromosome mis-segregation. Mis-segregation was defined by the existence of either lagging chromosomes or chromatin bridges (Figure 1A, black arrows), while normal chromosome segregation was defined by the absence of any chromatin staining between the segregating chromosome masses. Only 3 cells in all samples examined exhibited multi-polar anaphase and these were excluded from further analysis. The percentage of anaphase cells exhibiting mis-segregation was recorded (Figure 1B) and the mean mis-segregation frequency was 39.5 ± 18.1%. The distribution had a positive skew; therefore, a median mis-segregation frequency value of 31.1% was used to stratify patients into groups of high and low mis-segregation. These two groups had chromosome mis-segregation rates of 24.1 ± 5.7% and 48.6 ± 11.7%, respectively. There was no correlation between the number of anaphase cells observed and the frequencies of chromosome mis-segregation in a given specimen (Figure 1C, R2 = 0.0026).
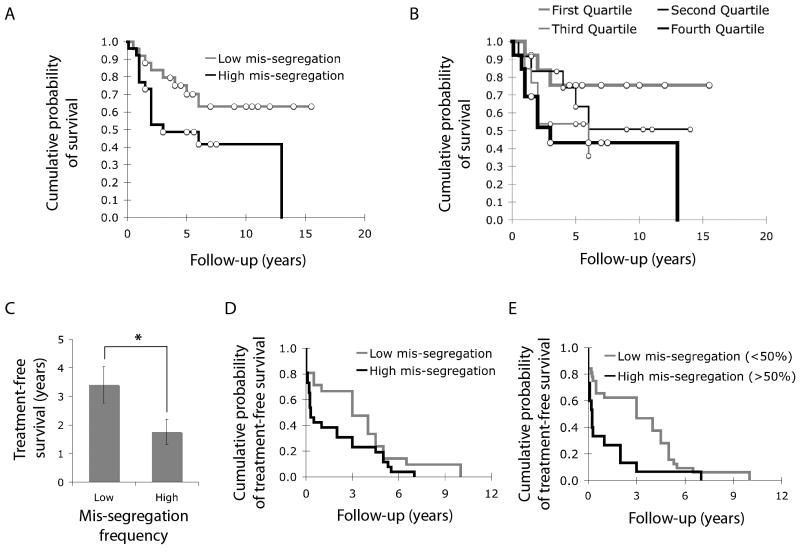
We then asked if rates of chromosome mis-segregation observed in tumor samples correlated with overall survival of patients. We used the Kaplan-Meier method to estimate overall survival probability after the initial diagnosis. Increased frequency of chromosome mis-segregation led to a significant decrease in overall survival by 2.14 years as the median survival time was 8.76 ± 1.01 years and 6.62 ± 1.18 years for the low and high mis-segregation groups, respectively (p < 0.05) (Figure 2A). Significant differences were also observed when comparing patients in the highest quartile of chromosome mis-segregation frequency with others in the lowest quartile (Figure 2B). In addition to overall survival, chromosome mis-segregation frequency also correlated with treatment-free survival after the first treatment regimen. The average treatment-free survival decreased by 48% in patients who had samples with higher frequency of chromosome mis-segregation (Figure 2C, p < 0.05, t-test). For this parameter, Kaplan-Meier analysis of treatment free survival probability was only significant (p < 0.05) when comparing patients based on a chromosome mis-segregation frequency of 50% (Figure 2D and E). This value corresponds to a single chromosome mis-segregation event every two cell divisions. And it was shown that this chromosome mis-segregation frequency is at the higher range of tolerable mis-segregation frequencies based on observations in cell lines and theoretical computations work (11, 44).
Figure 2.
A, Kaplan-Meier DSS analysis comparing the overall survival probability of patients with low (n = 25) and high (n = 26) chromosome mis-segregation, where median survival time was 8.76 ± 1.01 and 6.62 ± 1.18 years for low and high frequencies of mis-segregation groups, respectively. P < 0.05. White circles denote censored data. B, Kaplan-Meier DSS analysis comparing the overall survival probability of patients in the lowest (n = 13, thick grey line), second (n = 12, thin black line), third (n = 13, thin grey line) and highest (n = 13, thick black line) quartiles of chromosome mis-segregation frequencies. Statistical significance was achieved only when comparing lowest and highest quartiles where median survival time was 8.05 ± 1.21 and 6.45 ± 1.77 years for low and high mis-segregation groups, respectively. P < 0.05. White circles denote censored data. C, years of treatment-free survival for patients with low and high frequencies of chromosome mis-segregation. Bars represent mean ± s.e.m.. *, p < 0.05, t-test, n = 47 patients. D, Kaplan-Meier DSS analysis comparing treatment-free survival probability of patients with low (n = 21) and high (n = 26) frequencies of chromosome mis-segregation, where median treatment-free survival time was 3.41 ± 0.65 and 1.76 ± 0.44 years for low and high mis-segregation groups, respectively. P = 0.08. E, Kaplan-Meier DSS analysis comparing treatment-free survival probability of patients with low (n = 32) and high (n = 15) frequencies of chromosome mis-segregation categorized based on a chromosome mis-segregation frequency of 50%, where median treatment-free survival time was 3.16 ± 0.49 and 1.08 ± 0.49 years for low and high mis-segregation groups, respectively. P < 0.01.
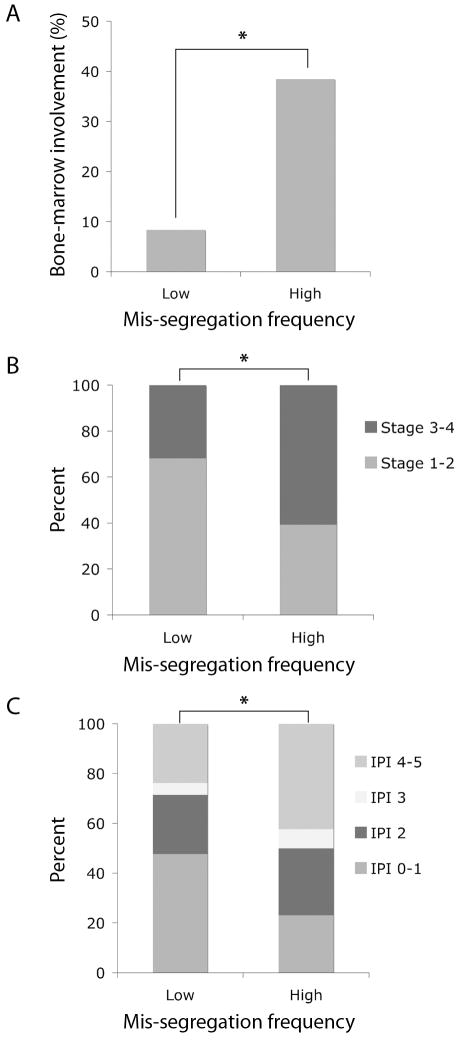
It is postulated that part of the effect of chromosome mis-segregation on tumor prognosis might be due to the contribution of CIN to tumor cell heterogeneity, making tumors more likely to spread and invade distant organs. Indeed, increased chromosome mis-segregation frequency significantly correlated with the likelihood of tumor bone marrow involvement upon presentation. Patients with tumors containing high mis-segregation rates were 4.6 times more likely to have tumor bone marrow involvement than patients with lower chromosome mis-segregation rates (Figure 3A, p < 0.05, Pearson's χ2-test) and as a consequence, these patients were also more likely to present with tumors that were either stage 3 or 4 (Figure 3B, p < 0.05, Pearson's χ2-test). We then asked if chromosome mis-segregation frequency was a predictor of the broader clinical picture upon presentation so we compared both groups of patients based on their International Prognostic Index (IPI) score. This score takes into account important prognostic indicators such as age upon diagnosis, stage of the disease, serum LDH levels, clinical performance status, and the presence of tumor in extranodal sites. Patients with tumors that exhibited increased chromosome mis-segregation frequency were more likely to present with elevated IPI scores than those with lower rates of chromosome mis-segregation. In fact 42% patients in the high mis-segregation group received a high IPI score of 4 or 5 whereas 24% of patients in the low mis-segregation group received the same IPI scores (Figure 3C, p < 0.001, Pearson's χ2-test).
Figure 3.
A, the percentage of patients with tumor bone marrow involvement in groups with low and high frequencies of chromosome mis-segregation. *, p < 0.05, Pearson's χ2-test, n = 24 and 26 for low and high mis-segregation groups, respectively. B, the percentage of patients stratified based on their tumor stage upon diagnosis and with respect to frequencies of chromosome mis-segregation. *, p < 0.05, Pearson's χ2-test, n = 22 and 28 for low and high mis-segregation groups, respectively. C, the percentage of patients stratified based on their IPI score upon diagnosis and with respect to frequencies of chromosome mis-segregation. *, p < 0.001, Pearson's χ2-test, n = 21 and 26 for low and high mis-segregation groups, respectively.
To determine if chromosome mis-segregation frequencies imparted an independent prognostic values, we segregated patients based on the aforementioned clinical variables. The overall survival of patients stratified based on IPI score, tumor stage, or bone marrow involvement was affected by chromosome mis-segregation frequencies, although these differences did not reach statistical significant as the study was not powered to detect such differences (not shown). However, chromosome mis-segregation frequencies had an independent prognostic value for bone marrow involvement and tumor stage when patients were stratified based on their IPI scores (Supplementary Table 2).
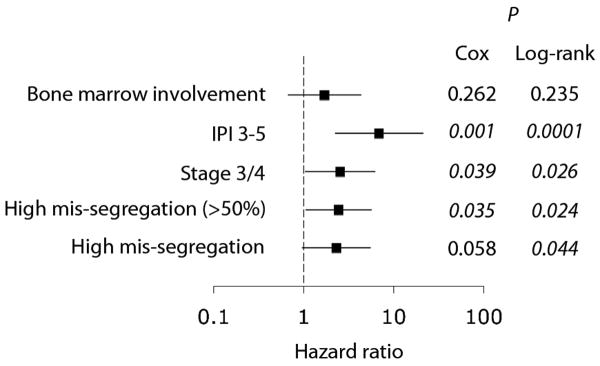
Overall, the frequency of chromosome mis-segregation correlated with an increased hazard ratio of death when patients were compared using either cutoff values for chromosome mis-segregation frequency of 31.3% and 50% (Figure 4). Furthermore, there was a significant correlation between the hazard ratio of death and rates of chromosome mis-segregation when patients were divided into four quartiles and a quantitative linear Cox regression model was applied (p < 0.05). The hazard ratio of death in patients with increased chromosome mis-segregation frequencies was comparable to those patients with tumors of stage 3 or 4. It was however smaller than that in patients with IPI scores of 3-5 (Figure 4).
Figure 4.
Forest plot showing hazard ratio for death with respects to patients with high frequencies of chromosome mis-segregation, tumor stage 3/4, IPI score 3-5, and positive tumor bone marrow involvement when compared with low frequencies of chromosome mis-segregation, tumor stage 1/2, IPI score 0-2, and negative tumor bone marrow involvement, respectively.
Discussion
Our work directly examines the correlation between chromosome mis-segregation and tumor outcome. It has been established that the most common causes of CIN manifest themselves as lagging chromosomes and chromatin bridges (11, 33, 35) and as such, we scored the frequencies of these two markers in tumor samples taken from patients with DLBCL. We show that the rate of chromosome mis-segregation significantly correlates with poor prognosis in these patients, at least over the range of the mis-segregation frequencies reported in this study. Interestingly, elevated chromosome mis-segregation was strongly tied to the ability of tumors to spread to distal lymph nodes and to the bone marrow. Furthermore, increased mis-segregation rates correlated with higher IPI scores and to tumor relapse after successful treatment.
This positive correlation is likely the consequence of the role of CIN in providing tumor cells with sufficient heterogeneity necessary to adapt to external pressures (45) such as chemotherapeutic agents or to new environments as is the case with metastasis and distal tumor spread. Chromosome mis-segregation rates cannot, however, be infinitely high otherwise that would strip tumor cells from the ability to retain beneficial phenotypes (46). Thus it is evident that there exists an optimal rate of chromosome mis-segregation below which, too little heterogeneity is generated to allow for tumor adaptation yet above which, tumors cannot maintain favorable phenotypes. Theoretical work indicates that the optimal rate of chromosome loss is between 10-3 and 10-2 (44). In human cells, this corresponds to a mis-segregation frequency of roughly 5% to 50%, which is similar to what we have observed in our study as well as to what has been reported in recent studies directly measuring chromosome mis-segregation in multiple cancer cell lines (11).
Recent work has identified major mechanisms by which chromosomes mis-segregate (1, 11, 32, 33, 35, 36, 38). Understanding these mechanisms has provided numerous potential drug targets, which can be used to modulate the frequency of chromosome mis-segregation (28, 47). Our potential ability to therapeutically increase or decrease chromosome mis-segregation frequency could prove clinically useful. As evidenced by our study, we predict that reducing mis-segregation could improve the response to treatment and delay or prevent tumor relapse. On the other hand, if chromosome mis-segregation frequency is already elevated, further increasing it might render tumor cells less viable. This approach can potentially be coupled with therapeutic options, such as radiation therapy, which rely, in part, on inducing genomic instability (48). The kinetochore-microtubule interface presents a prime pharmacological target for modulating chromosome mis-segregation for two important reasons: first, the kinetochore is a macromolecular organelle that contains several dozen proteins whose function is to regulate the stability of microtubule attachments. Second, it was recently shown that slight reductions in kinetochore-microtubule attachment stability, by overexpressing microtubule destabilizers Kif2b and MCAK, led to a significant decrease in chromosome mis-segregation frequencies and suppression of CIN (35). Interestingly, this was observed in multiple chromosomally unstable cells lines derived from different human cancers indicating that perturbing kinetochore-microtubule attachments might be an effective therapeutic approach to influence CIN regardless of its original cause. Indeed, small molecule inhibitors of the microtubule destabilizing kinase, aurora B, are currently being evaluated in clinical trials (49). Such an inhibitor would be expected to lead to increased chromosome mis-segregation in tumors beyond tolerable range. The strategy to suppress CIN pharmacologically is not yet clear. This can theoretically be achieved either by inhibiting a microtubule stabilizer, such as astrin, or activating a microtubule destabilizer. It is also possible to pharmacologically perturb interactions among kinetochore proteins to prevent the targeted recruitment of microtubule regulating proteins to kinetochores (50). Regardless of the approach, we propose that pharmacologically suppressing CIN would present an important adjuvant therapy to standard treatment that would limit tumor relapse and drug-resistance.
In summary, our work presents a strong case to develop therapeutic strategies that can modulate chromosome mis-segregation frequencies in tumors as an adjunct to standard therapy as well as a means to improve prognosis and limit tumor spread. Although our study focused on DLBCL, work on several cancer cell lines as well as mouse-models (11, 19, 21, 28, 35, 36) would predict that the conclusions presented herein would be valid in other chromosomally unstable malignancies.
Supplementary Material
Translational relevance.
Given the widespread prevalence of chromosomal instability (CIN) in human cancers, it is necessary to understand the contribution of chromosome mis-segregation to tumor prognosis and response to therapy. Here we show that a two-fold increase in chromosome mis-segregation rates in tumors taken from patients diagnosed with Diffuse Large B-Cell Lymphoma (DLBCL) leads to poor prognosis, inferior response to standard therapy, and increased tumor spread. These results present a strong case to target CIN in cancer. The feasibility of this approach is further corroborated by the emergence of candidate cellular targets that are involved in chromosome segregation fidelity and that are amenable to pharmacologic interventions. Suppressing CIN has the potential to significantly improve the response to treatment and limit tumor spread, presumably by restraining tumor cell heterogeneity.
Acknowledgments
We sincerely thank Rand Swenson, M.D., Ph.D., Dartmouth Medical School, for providing the Nikon microscope used in this study and for Alexey V. Danilov, M.D., Ph.D., Hematology and Oncology at Dartmouth Medical School, for his clinical expertise and assistance while writing this manuscript.
Grant support: This work was supported by the Hitchcock-Foundation Grant 250-4041 (S.F.B.) and the National Institutes of Health grant GM51542 (D.A.C)
Footnotes
Disclosure of potential conflict of interest: Authors claim no potential conflicts of interest
References
- 1.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Chung DC, Pino MS. The Chromosomal Instability Pathway in Colon Cancer. Gastroenterology. 2010;138:2059–72. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lingle WL, Lukasiewicz K, Salisbury JL. Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer. Genome Instability in Cancer Development. 2005;570:393–421. doi: 10.1007/1-4020-3764-3_14. [DOI] [PubMed] [Google Scholar]
- 6.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–50. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 7.M'kacher R, Andreoletti L, Flamant S, Milliat F, Girinsky T, Dossou J, et al. JC human polyomavirus is associated to chromosomal instability in peripheral blood lymphocytes of Hodgkin's lymphoma patients and poor clinical outcome. Ann Oncol. 2010;21:826–32. doi: 10.1093/annonc/mdp375. [DOI] [PubMed] [Google Scholar]
- 8.Knudson CM, van de Wetering CI. Chromosomal instability and supernumerary centrosomes represent precursor defects in a mouse model of T-cell lymphoma. Cancer Res. 2007;67:8081–8. doi: 10.1158/0008-5472.CAN-07-1666. [DOI] [PubMed] [Google Scholar]
- 9.Kramer A, Schweizer S, Neben K, Giesecke C, Kalla J, Katzenberger T, et al. Centrosome aberrations as a possible mechanism for chromosomal instability in non-Hodgkin's lymphoma. Leukemia. 2003;17:2207–13. doi: 10.1038/sj.leu.2403142. [DOI] [PubMed] [Google Scholar]
- 10.Swanton C, Tomlinson I, Downward J. Chromosomal instability, colorectal cancer and taxane resistance. Cell Cycle. 2006;5:818–23. doi: 10.4161/cc.5.8.2682. [DOI] [PubMed] [Google Scholar]
- 11.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsson K, Johansson B. Trisomy 8 as the sole chromosomal aberration in acute myeloid leukemia and myelodysplastic syndromes. Pathologie Biologie. 2007;55:37–48. doi: 10.1016/j.patbio.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Mugneret F, Callier P, Favre-Audry B. Chromosomal abnormalities in acute myeloid leukaemias. Pathologie Biologie. 2003;51:314–28. doi: 10.1016/s0369-8114(03)00114-7. [DOI] [PubMed] [Google Scholar]
- 14.Gulley ML, Shea TC, Fedoriw Y. Genetic Tests To Evaluate Prognosis and Predict Therapeutic Response in Acute Myeloid Leukemia. Journal of Molecular Diagnostics. 2010;12:3–16. doi: 10.2353/jmoldx.2010.090054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi K, Usami I, Kubota M, Nishio T, Kakazu N. Chromosome 7 abnormalities in acute megakaryoblastic leukemia associated with Down syndrome. Cancer Genetics and Cytogenetics. 2005;158:184–7. doi: 10.1016/j.cancergencyto.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 16.Jacquemont S, Boceno M, Rival JM, Mechinaud F, David A. High risk of malignancy in mosaic variegated aneuploidy syndrome. Am J Med Genet. 2002;109:17–21. doi: 10.1002/ajmg.10281. [DOI] [PubMed] [Google Scholar]
- 17.Rahman N, Hanks S, Coleman K, Reid S, Plaja A, Firth H, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 18.Szallasi Z, Carter SL, Eklund AC, Kohane IS, Harris LN. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 19.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Benezra R, Sotillo R, Schvartzman JM, Socci ND. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–U138. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benezra R, Schvartzman JM, Sotillo R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–15. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Deursen JM, Baker DJ, Jin F, Jeganathan KB. Whole Chromosome Instability Caused by Bub1 Insufficiency Drives Tumorigenesis through Tumor Suppressor Gene Loss of Heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanton C, Birkbak NJ, Eklund AC, Li QY, McClelland SE, Endesfelder D, et al. Paradoxical Relationship between Chromosomal Instability and Survival Outcome in Cancer. Cancer Res. 2011;71:3447–52. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanton C, Juul N, Li Q, Eklund A, Richardson A, Szallasi Z. Paradoxical Relationship of Chromosomal Instability with Breast Cancer Outcome: Identification of a Good Prognostic ER Negative/HER2 Negative Breast Cancer Cohort with Extreme Chromosomal Instability. Cancer Res. 2009;69:656S–S. [Google Scholar]
- 25.Swanton C, Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martens JWM, Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, et al. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Research and Treatment. 2011;128:23–30. doi: 10.1007/s10549-010-1026-5. [DOI] [PubMed] [Google Scholar]
- 27.Benezra R, Schvartzman JM, Duijf PHG, Sotillo R, Coker C. Mad2 Is a Critical Mediator of the Chromosome Instability Observed upon Rb and p53 Pathway Inhibition. Cancer Cell. 2011;19:701–14. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of Chromosomal Instability. Curr Biol. 2010;20:R285–R95. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pati D, Zhang NG, Ge GQ, Meyer R, Sethi S, Basu D, et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci USA. 2008;105:13033–8. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–3. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 31.Di Leonardo A, Lentini L, Amato A, Schillaci T. Simultaneous Aurora-A/STK15 overexpression and centrosome amplification induce chromosomal instability in tumour cells with a MIN phenotype. Bmc Cancer. 2007;7 doi: 10.1186/1471-2407-7-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. Plos One. 2009;8:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganem NJ, Godinho SA, Pellman D. A Mechanism Linking Extra Centrosomes to Chromosomal Instability. Nature. 2009 doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusell L, Pampalona J, Soler D, Frias C, Genesca A. Different outcomes of telomere-dependent anaphase bridges. Biochemical Society Transactions. 2010;38:1698–703. doi: 10.1042/BST0381698. [DOI] [PubMed] [Google Scholar]
- 35.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakhoum SF, Genovese G, Compton DA. Deviant Kinetochore Microtubule Dynamics Underlie Chromosomal Instability. Curr Biol. 2009;19:1937–42. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung ALM, Deng W. Telomere dysfunction, genome instability and cancer. Frontiers in Bioscience. 2008;13:2075–90. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 38.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimini D, Howell BJ, Degrassi F, Salmon ED. Merotelic attachment of single kinetochores to microtubules from opposite poles is not detected by the mitotic spindle checkpoint and induces chromosome loss during mitosis. Mol Biol Cell. 2000;11:430a–a. [Google Scholar]
- 40.Tornillo L, Bernasconi B, Karamitopolou-Diamantiis E, Lugli A, Vizio D, Dirnhofer S, et al. Chromosomal instability in gastric mucosa-associated lymphoid tissue lymphomas: a fluorescent in situ hybridization study using a tissue microarray approach. Human Pathology. 2008;39:536–42. doi: 10.1016/j.humpath.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Tibiletti MG, Milani K, Martin V, Zucca E, Motta T, Cortelazzo S, et al. Chromosome instability and translocation t(11;18) in primary gastric marginal zone B-cell lymphoma of MALT-type. Hematological Oncology. 2007;25:184–8. doi: 10.1002/hon.825. [DOI] [PubMed] [Google Scholar]
- 42.Tzankov A, Gschwendtner A, Augustin F, Fiegl M, Obermann EC, Dirnhofer S, et al. Diffuse large B-cell lymphoma with overexpression of cyclin E substantiates poor standard treatment response and inferior outcome. Clinical Cancer Research. 2006;12:2125–32. doi: 10.1158/1078-0432.CCR-05-2135. [DOI] [PubMed] [Google Scholar]
- 43.A predictive model for aggressive non-Hodgkin's lymphoma. The International Non-Hodgkin's Lymphoma Prognostic Factors Project. The New England Journal of Medicine. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 44.Komarova NL, Wodarz D. The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc Natl Acad Sci USA. 2004;101:7017–21. doi: 10.1073/pnas.0401943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woude GFV, Gao CF, Furge K, Koeman J, Dykema K, Su YL, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci USA. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kops GJPL, Janssen A, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–13. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- 48.Marcon F, Palli D, Zufferli A, Mazzoli E, Siniscalchi E, Sera F, et al. Evaluation of radiation-induced chromosome instability in subjects with a family history of gastric cancer. Biomarkers. 2009;14:226–34. doi: 10.1080/13547500902968538. [DOI] [PubMed] [Google Scholar]
- 49.Schellens JH, Boss DS, Witteveen PO, van der Sar J, Lolkema MP, Voest EE, et al. Clinical evaluation of AZD1152, an i.v. inhibitor of Aurora B kinase, in patients with solid malignant tumors. Ann Oncol. 2011;22:431–7. doi: 10.1093/annonc/mdq344. [DOI] [PubMed] [Google Scholar]
- 50.Compton DA, Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. Embo Journal. 2010;29:3531–43. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.