Abstract
The extraction and purification of nucleic acids from complex samples (e.g. blood, biopsied tissue, cultured cells, food) is an essential prerequisite for many applications in biology including genotyping, transcriptional analysis, systems biology, epigenetic analysis, and virus/bacterial detection. In this report, we describe a new process of nucleic acid extraction that utilizes “pinned” aqueous/organic liquid interfaces in microchannels to streamline the extraction mechanism, replacing all washing steps with a single traverse of an immiscible fluid barrier, termed Immiscible Filtration Assisted by Surface Tension (IFAST). Nucleic acids in biological samples are bound to paramagnetic particles and then drawn across the IFAST device (or array of IFAST devices) using a magnet. While the strength of the IFAST barrier is suitable for separation of nucleic acids from lysate in its current embodiment, its permeability can be selectively adapted by adjusting the surface tensions/energies associated with the cell lysate, the immiscible phase, and the device surface, enabling future expansion to other non-nucleic acid applications. Importantly, processing time is reduced from 15–45 minutes to less than 5 minutes while maintaining purity, yield, and scalability equal to or better than prevailing methods. Operation is extremely simple and no additional lab infrastructure is required. The IFAST technology thus significantly enhances researchers’ abilities to isolate and analyze nucleic acids, a process which is critical and ubiquitous in an extensive array of scientific fields.
Introduction
Worldwide, over 400 000 molecular biology scientists engage in nucleic acid (NA) purification,1 including researchers from a wide variety of fields including biomedical research, molecular diagnostics, pharmacology, and forensics. While great advances have been made in the detection and analysis of NAs over the past two decades, improvements in sample preparation have been relatively modest, generating a potential “bottleneck” in the nucleic acid analysis process flow.2 Boom et al. introduced silica particles almost 20 years ago, and changes in this basic purification method consist primarily of the use of paramagnetic particles (PMPs) and improved surface coatings,3–5 both of which being embodiments of the multi-step solid phase extraction (SPE) method. A typical SPE protocol can be summarized as follows: (1) binding NAs to an immobilized solid phase; (2–5) repeatedly washing bound NAs to remove contaminants; and (6) eluting NAs for downstream processing. High throughput versions of the SPE process based on microtiter plate architectures are commercially available (e.g. Qiagen TurboCapture, Invitogen FastTrack MAG 96), but these processes are labor intensive and require expensive robotics to facilitate the extensive washing that must be performed on individual samples, limiting widespread adoption of these technologies.
Other derivations of the technology exploit the intrinsic advantages of microfluidics in miniaturized SPE devices. These advantages include low reagent consumption, parallel fabrication, predictable laminar flow, high integration potential, and increased throughput. The devices employ either functionalized surfaces6–8 or immobilized particles9–12 to reversibly bind NAs, while wash buffers remove contaminants. A limitation of the miniature SPE protocols is that they require an intricate device configuration, often incorporating advanced components including pumps, valves, mixers, sensors, or other electronics, making such devices unappealing to non-microfluidic scientists and difficult to manufacture using traditional methods. Moreover, as direct embodiments of the traditional three-step SPE protocol, these devices do not overcome the inherent process flow limitations.
In this manuscript, we present a distinctly different process, termed IFAST, which employs an immiscible phase (e.g. oil, paraffin wax) to effectively filter contaminants in a single step, thereby eliminating multiple washing, centrifugation, and/or magnetic bead capture steps. Sur et al. introduced the concept of immiscible phase filtration for NA purification in a cartridge-based device for point of care diagnostics.13 Here we leverage the dominance of surface tension over gravity at the microscale, to establish “virtual walls”14,15 between each region of the IFAST device to greatly simplify operation and enable arrayed processing for NA purification. These advances reduce time, effort, and cost associated with NA purification in a general laboratory setting and thus will greatly facilitate the multiple facets of biological research that depend on nucleic acid and gene expression analysis.
Results
IFAST design
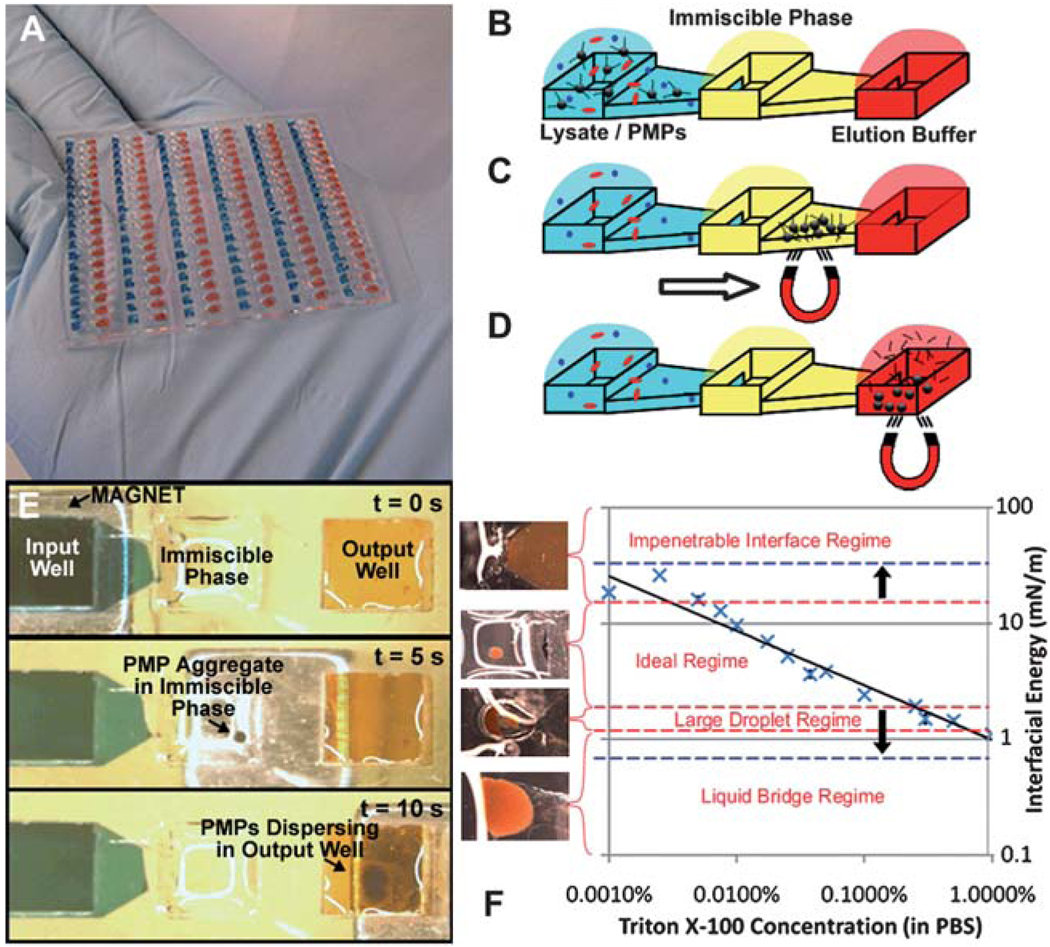
SPE devices are utilized by a broad spectrum of scientists with varying operational capabilities. With this in mind, the IFAST device was engineered to operate using only basic laboratory equipment (pipette and magnet), without the need for any external hardware or physical connections of any kind. Comparisons of the SPE and IFAST protocols are given in Fig. 1 and Fig. S1†. The platform footprint was arrayed to mimic the standardized configurations of 384- and 1536-well microtiter plates, ensuring compatibility with existing high-throughput laboratory infrastructure (e.g. liquid handling robots, multichannel pipettes) (Fig. 2A). Fabrication of large arrays (up to 512 devices on a standard well plate footprint) of IFAST devices is compatible with a broad range of polymeric materials and standard high volume manufacturing processes (e.g. injection molding). Each IFAST device consists of three wells (well volumes = 5 to 10 µl for 384 well configuration, 2 to 4 µl for 1536 well configuration) in a linear configuration connected by two trapezoidal microfluidic conduits. One end well, termed the input well, is loaded with cell suspension, appropriate lysis buffer, and NA-binding PMPs while the other end well, termed the output well, is loaded with nuclease-free elution buffer and/or downstream process reagents. The center well is then filled with an immiscible liquid to complete the IFAST setup through formation of stable aqueous/organic liquid interfaces (described in more detail in the next section). To operate the device, a permanent magnet draws the PMP-bound nucleic acids through the immiscible liquid and into the elution buffer (Video S1†). Without the aid of bound magnetic PMPs, hydrophilic contaminants are unlikely to traverse the aqueous/oil interface, and are sequestered in the input well. Hydrophobic contaminants may enter the immiscible phase, but passage through the second oil/aqueous interface into the elution buffer is energetically unfavorable. In this manner, NAs are effectively isolated from the remainder of the lysate using a simple, streamlined protocol: (1) cell suspension is lysed and NAs attach to surrounding PMPs, (2) PMPs are transported across the immiscible phase using magnetic field, and (3) NAs are eluted in pure buffer (Fig. 2B–E). In addition to minimizing the number of steps and time required for NA extraction, moving PMPs instead of fluids also reduces costs by decreasing the number of consumables required to process a sample. Following traverse of the immiscible liquid, the elution buffer and purified NAs are collected via pipette for downstream processing, such as cDNA synthesis and PCR. Alternatively, RT reagents can be directly added to the output well and the RT reaction can be performed on-chip. The simplicity of this device is particularly well suited for high-throughput operations. Arrayed devices as shown in Fig. 2A can be simultaneously processed by moving a magnetic bar underneath the IFAST array and simultaneously drawing the NA-bound PMPs across each IFAST device. This is in contrast with most commercial high throughput processes, where each sample well must be independently addressed multiple times to accommodate washing. Therefore, an IFAST array allows simultaneous purification of 100+ samples with a single traverse of a magnet (t = 10 s).
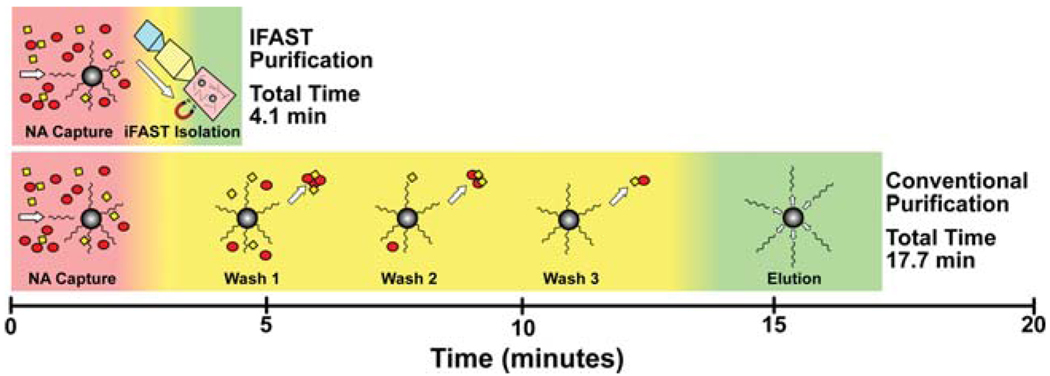
Fig. 1.
Comparison of the protocols for conventional PMP-based NA purification and IFAST device. The IFAST method reduces the number of processing steps from 18 to 6 (67%) and the total purification time from 17.7 minutes to 4.1 minutes (77%), largely through the elimination of repetitive washing steps. More details on each step are given in Fig. S1†.
Fig. 2.
IFAST device design and operation. Large arrays of devices, such as the 114 device array shown in (A), can be operated in parallel using arrays of magnetic strips. The IFAST platform consists of three wells connected in series by microchannels. The central well is loaded with an immiscible oil phase to separate cell lysate containing NA-binding magnetic particles from nuclease-free elution buffer (B). A magnet is utilized to draw NA-bound particles through the immiscible phase (C) and into elution buffer (D). The total time to operate actual device is only 10 seconds (E). Increasing the concentration of a detergent (Triton X-100) reduces the interfacial energy with the oil phase (Chill-Out Liquid Wax), promoting carryover of liquid across the immiscible phase barrier (F). The various regimes of PMP transfer are defined by the red dashed lines and photographs of representative traverses are given in the graph, with an interfacial energy of −3 to 15 mN m−1 resulting in ideal transfer of PMPs. By reducing the surface energy between the oil and the device surface, the ideal regime can be expanded, as defined by the blue dashed lines, to encompass the entire tested range of detergent concentrations.
IFAST operational principle
The microfluidic IFAST platform relies upon the dominance of surface tension over gravity at the microscale to establish “virtual walls” between each region of the IFAST device. This enables side-by-side loading of liquids that is not possible on the macroscale. This phenomenon is quantified by the dimensionless Bond number (Bo = ρgL2/γ), where ρ is the density of liquid, g is the acceleration of gravity, L is a characteristic length scale of the device, and γ is the surface energy of the liquid. A Bo ≪ 1 indicates a system in which surface tension forces are sufficiently large to marginalize the effects of gravity. For larger Bo devices, including the one presented by Sur et al.,13 gravity dominance mandates positioning of the denser lysate and elution buffer compartments below the oil phase, constraining device geometry into a three-dimensional architecture. Because Bo scales with L2, a reduction in device dimensions rapidly reduces Bo into the surface tension-dominant regime. Microfluidic constrictions with very small characteristic length scales selectively impede liquid motion, enabling serial loading of all three device liquids (lysate, oil phase, and elution buffer) into their respective compartments without intermixing or density-driven stratification (Video S2†). This facilitates planarization of the device layout, which simplifies both device fabrication and operation while also enabling high-throughput arrays in well plate-like configurations. We have experimentally observed that a microfluidic constriction measuring 250 µm high by 500 µm wide is sufficient to arrest flow as long as the contact angle between the aqueous phases and the device surface is greater than >34° (Table S1†). This observation is in agreement with past observations regarding the wetting of a liquid around an angled geometry.16 Device loading is very robust in that interfaces between each liquid are precisely positioned independent of variations in liquid volume, dispensing rate, or pipette orientation.
The purification effectiveness of the IFAST depends upon a stable immiscible phase barrier that allows passage of PMP-bound NAs in a magnetic field while minimizing carryover of lysate. The “strength” of this barrier relies upon an interfacial energy between the lysate and oil, which resists deflection or displacement from the microfluidic constriction in order to minimize contact area between the two phases. While the interfacial energy between two immiscible phases is generally high, detergents present in some buffers could reduce this energy and potentially allow carryover of undesirable quantities of lysate during PMP traverse. To address this possibility, input wells were loaded with a series of detergent-containing solutions with decreasing interfacial energies with oil as assessed by the drop-weight method19 and PMP traverse across the immiscible phase was evaluated. For a given oil (Chill-Out Liquid Wax) and concentration of PMPs (700 µg ml−1), an interfacial energy of 3 to 15 mN m−1 (corresponding to Triton X-100 concentrations of 0.001% to 0.25%) was required to transfer a tightly packed PMP aggregate. However, when interfacial energy was decreased below this range, larger droplets became associated with the PMP aggregates and transferred the unwanted lysate across the oil phase into the elution buffer. Further reduction in interfacial energy caused the formation of a continuous aqueous-phase connecting the input and output wells, and resulted in complete mixing of the lysate and elution buffer. In contrast, interfaces with excessively high energies (>15 mN m−1) were found to be too rigid to facilitate PMP transfer, permanently trapping the PMP-bound NAs in the lysate. The various regimes of PMP transfer are defined for the Chill-Out Liquid Wax by the red dashed lines in Fig. 2F.
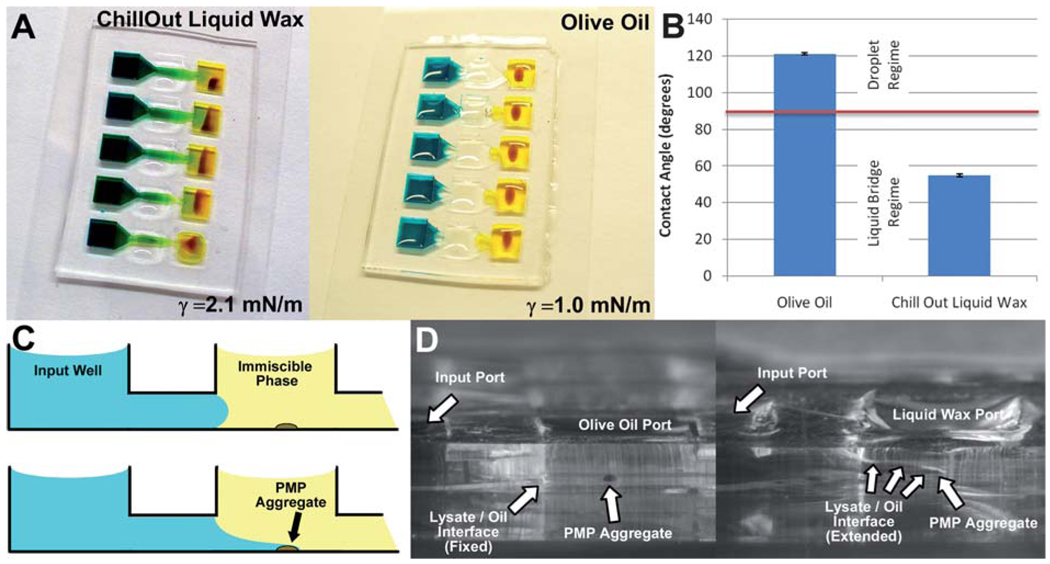
PMP transfer was further improved by reducing the surface energy between the device surface and the oil phase, thus stabilizing the oil barrier and enabling expansion of the functional range of aqueous/oil interfacial energies such that detergent concentrations used in common lysis and elution buffers (e.g. lysis buffers containing 1% Triton, 1% LiDS, or 2% SDS) become compatible with the system. In fact, successful transfer of the PMP aggregate was obtained with the entire range of tested Triton X-100 concentrations (0.001% to 1.0%), as illustrated by the blue dashed lines in Fig. 2F. The left panel of Fig. 3A shows the continuous aqueous carryover that occurs during the transfer of PMPs across a liquid wax immiscible phase (Chill-Out Liquid Wax) with a buffer containing high concentration of detergents (i.e., less than 3 mN m−1 interfacial energy). Replacement of the liquid wax with olive oil (Unilever) slightly reduced the lysate/oil interfacial energy for lysates with high detergent concentrations (e.g. the energy of an interface between oil and 0.25% Triton X-100 decreased from 2.1 mN m−1 to 1.0 mN m−1 when olive oil was substituted), but resulted in a successful PMP transfer. This seemingly paradoxical outcome is due to changes in the device surface/oil interfacial energy. The propensity of the lysate to form an aqueous bridge upon PMP transfer is a function of the change in total energy associated, ΔE, with the formation of the liquid bridge surfaces. This change in energy is given by the sum of the increases in energy associated with increased lysate/oil and lysate/device surface contact and the decrease in energy associated with the reduction in device surface/oil contact, or:
| (1) |
where A is the footprint area occupied by the bridge, assuming it is relatively flat. If the net change in energy is positive, aqueous bridge formation is unfavorable while a negative value of ΔE promotes aqueous bridge formation. The practical implications of eqn (1) can be elucidated by noting its similarity with Young’s equation for a droplet of lysate on a surface covered in oil:
| (2) |
where θ is the contact angle of the lysate in oil. Identifying that A and each value of γ are positive and −1 < cos θ < 1, it can be deduced that ΔE will be positive only when cos θ is negative, or when θ > 90°. This concept enables the prediction of liquid bridge formation using only a simple contact angle measurement (Fig. 3B) (i.e., the purification will contain minimal or no detectable carryover when θ > 90°). Side imaging of the IFAST device during PMP traverse (shown schematically as Fig. 3C and photographically as Fig. 3D) confirms the formation/repression of a liquid bridge for the two conditions given in Fig. 3A. Importantly, while a single embodiment operates robustly across a wide range of commonly used lysate compositions, simple modifications to device geometry (thus changing Bo) can be made to accommodate lysates with extreme surface tensions.
Fig. 3.
Optimization of the immiscible phase interface. Using olive oil as the immiscible phase results in a favorable outcome relative to liquid wax despite its lower interfacial energy with a detergent-rich lysis buffer (A). The difference in contact angle between lysis buffer and glass submerged in both oils elucidates differences in interfacial energies. Data encompass 6 measurements for each oil and error bars represent standard deviations (B). Side view of PMP aggregate traverse is shown both schematically (C) and photographically (D) to illustrate the selective extension of lysate between the oil and the floor of the device.
IFAST performance
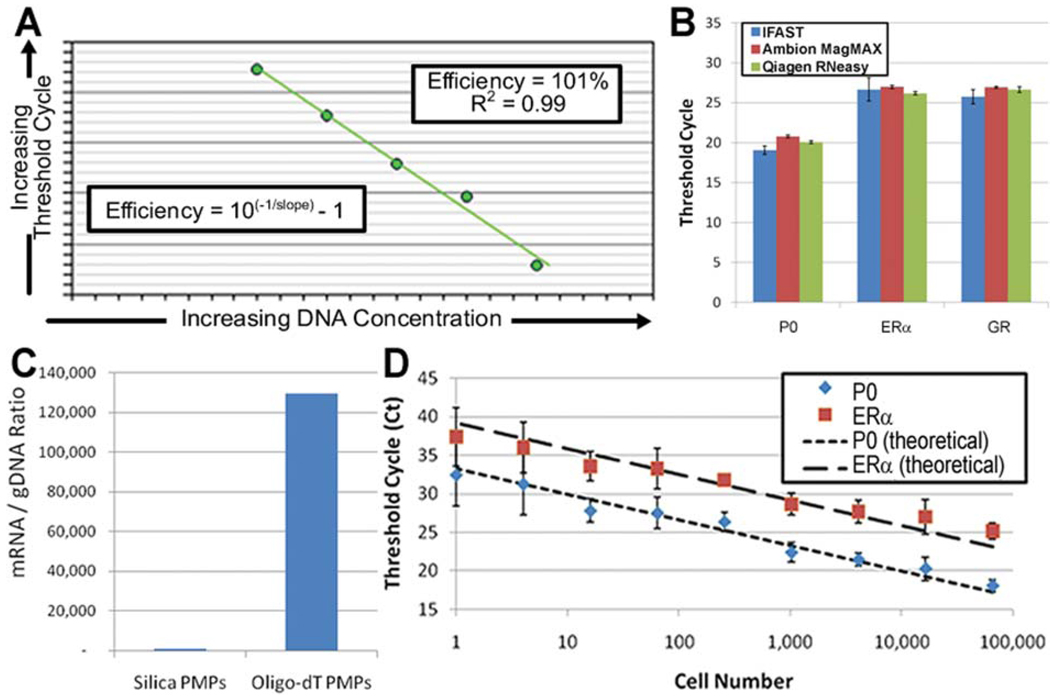
The performance of the IFAST platform was quantified using four metrics commonly applied to nucleic acid purification: (1) amplification efficiency of a downstream qRT-PCR reaction, (2) nucleic acid yield, (3) scalability to small sample sizes, such as those associated with biopsied tumors, cheek swabs, and forensic samples, and (4) overall time to complete the process. Amplification efficiencies of sample prepared using the IFAST platform were found to be between 90% and 110% for multiple genes, indicating that the nucleic acids have been sufficiently separated from the PCR interferents present in whole cell lysate (Fig. 4A) as the presence of significant carryover lysate would impede amplification and reduce efficiency. The nucleic acid yield was assessed through a direct comparison between the IFAST platform and popular commercially available kits (Qiagen RNeasy Mini Kit and Ambion MagMAX Kit), in which samples containing ~56 000 cells were processed by each technique and multiple genes were amplified and detected via qRT-PCR (Fig. 4B). Comparison of cycle threshold (CT) values of large ribosomal protein (P0), and two low abundance genes, estrogen receptor-α (ERα) and glucocorticoid receptor (GR), in lysates from cancer cells prepared via IFAST, silica membrane-based commercial kits, or PMP-based commercial kits indicated that the NA yield was comparable to or better than the commercial kits.
Fig. 4.
IFAST performance. Efficiency was calculated by amplifying multiple genes (P0 and ERα) from a serial dilution of IFAST-purified NA and fitting the given equation to the data (A). The yield was determined by amplifying three genes (P0, ERα, and GR) from mRNA from ~56 000 breast cancer epithelial cells (MCF-7) using both the IFAST and commercial kits to isolate the NA. Data represent three independent experiments performed in duplicate and error bars represent standard deviation (B). The implementation of PMPs coated with oligo-dT nucleotides selectively captured mRNA (as quantified by amplifying the P0 gene) from cell lysate (C). Sensitivity was measured by purifying a series of cell concentration via IFAST and amplifying multiple genes (P0 and ERα). Data represent three experiments for each cell concentration and error bars represent standard deviations of the data (D).
Additionally, to directly assess the possibility of loss of mRNA during IFAST purification, the recovery of pure mRNA was quantified as it was drawn across the IFAST device. Briefly, mRNA was purified from lysate via IFAST and this mRNA was either directly amplified or processed a second time with the IFAST device and then amplified. It was found that the CT values for P0 amplified from these two samples were within 0.2 cycles of each other (p = 0.82 by paired t-test, n = 3), suggesting that virtually no mRNA was lost during PMP binding, immiscible phase traverse, and elution. Furthermore, mRNA was effectively isolated from genomic DNA (gDNA) using oligo-dT-bound PMPs (Invitrogen oligo-dT Dynabeads) that selectively capture the poly-A tails of mRNA. Using these PMPs, the ratio of RNA to gDNA for the P0 gene was found to be ~130 000 : 1 as measured by no-RT controls. Additionally, the post-amplification melt curve from NA isolated with the oligo-dT PMPs contained no noticeable gDNA contamination. This is in contrast to silica PMPs (Ambion MagMAX) that capture total NA and generated an RNA to gDNA ratio of ~1000 : 1 for the same gene (Fig. 4C). The scalability of the IFAST platform was evaluated by preparing a variety of sample sizes ranging from 1 to 65 000 cells and amplifying multiple genes after IFAST purification. Minimal sample loss was observed as the device was scaled down as indicated by the similarity of the experimental data with the theoretical amplification time when complete scalability is assumed (i.e. reducing the sample size by a factor of 2 will require 1 more cycle to achieve detection) (Fig. 4D). Importantly, total time-to-preparation was reduced by >75% over both commercial kits tested.
Discussion
The IFAST device described here provides a new and efficient method of recovering maximum NA, with scalability for small sample sizes, and with a total operation time of less than 5 minutes from device filling to nucleic acid recovery. This is in sharp contrast to the tested kits, which require approximately 15 to 45 minutes due to the increased number of processing steps associated with washing. Replacement of these steps with a single immiscible liquid traverse (t ≈ 10 seconds) drastically simplifies and expedites nucleic acid extraction and purification without sacrificing any aspect of performance, thus significantly improving a process which is essential to an extensive array of scientific fields. The IFAST platform represents a considerable simplification compared to conventional processes and other microfluidic approaches, offering facile operation with only basic laboratory tools while also being directly compatible with existing high throughput systems. It reduces the number of processing steps by 67% and the total purification time by 77% relative to conventional PMP-based purification, which is among the fastest available commercial methods, while also decreasing reagent and consumables (e.g. pipette tips, sample tubes) usage.
The ability to expedite and simplify NA sample preparation will have immediate practical relevance since it will enable researchers, scientists and technicians to answer more biological questions with a limited set of resources (e.g. funding, labor, lab space). This enhancement will be of particular importance for applications where NA analysis is required for each individual, such as in molecular diagnostics, forensics, and personal genotyping. Furthermore, simplified and accelerated NA isolation will facilitate studies in cellular heterogeneity (such as the method described by Janes et al., 201017) by enabling parallel isolation of multiple fractions of a single sample.
The IFAST platform exploits the surface tension-dominant fluid mechanics of the microscale to efficiently separate PMP-bound NAs from bulk cell lysate. The low Bond number of the system facilitates precise positioning of each liquid while making the device resistant to unintended variations in loading conditions. The stability of the immiscible phase barrier was increased through the modulation of geometry, device surface energy, and the surface energies of the aqueous and oil phases, thus making this platform compatible with a variety of lysis and extraction buffers, including those containing a high concentration of detergent or low-surface tension organic solvents, such as TRIzol.
Recent estimates indicate that molecular biology workers devote an average of 15% of their time to nucleic acid purifications1 and the size of the US market alone exceeds $300 million.18 The level of time savings associated with the IFAST suggests that adoption of this technology could potentially result in a reduction of 200 work-hours per person per year. Worldwide with ~400 000 molecular biology scientists,18 this reduction translates into a potential yearly savings of 80 million work-hours. The benefits afforded by this new technology which streamlines NA purification process thus have the potential to broadly impact life sciences research.
Materials and methods
Device fabrication
IFAST devices were patterned from polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning) via soft lithography. Briefly, master molds comprising relief representations of the IFAST devices were generated through selective curing of a UV-sensitive epoxy (SU-8, Microchem) followed by development in propylene glycol. PDMS monomer was mixed with hardener at 10 : 1 ratio, degassed for 1 hour under vacuum, poured onto the master, and cured for 4 hours at 85 °C. To complete channel formation, PDMS was plasma treated in O2 for 1 minute, then bonded to glass cover slip substrate at 135 °C for 2 minutes. Test devices possessed wells measuring 3 mm by 3 mm, with 4.5 mm well-to-well spacing, thereby mimicking the footprint of a 384-well plate. Devices mimicking the footprint of 1536-well plates were also fabricated and were found to perform equally well (p = 0.43 for the number of threshold cycles (CT) required to detect exponential amplification of P0 mRNA as measured by Student’s t-test). The heights of the microchannel and well regions are 250 µm and 750 µm, respectively, for both configurations.
Contact angle and surface energy measurements
Contact angles were measured using a goniometer to capture images of a droplet on a leveled surface (Ramé-Hart Instrument Co.) followed by angle determination using DropImage software (Ramé-Hart). Interfacial surface energies between liquids were measured using the drop weight method as outlined by Campbell. 19 Briefly, a syringe pump (Cole-Parmer) was utilized to eject droplet of aqueous phase liquid from a length of 100 µm diameter (I.D.) tubing into a cuvette of oil. The goniometer was used to record the radius of the droplets at the instant they detached from the tubing due to gravity. This radius, r, is correlated to interfacial energy, γ, by the equation:
| (3) |
where Δρ is the difference in density between the oil and aqueous phases and R is the radius of the ejecting tube. Each contact angle and interfacial energy were measured at least three times.
Cell culture
Breast cancer epithelial cells (MCF-7) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) at 37 °C in polystyrene flasks until confluence. Cells were released using a 0.05% trypsin/EDTA solution and collected via centrifugation. Cell pellets were frozen at −80 °C until NA isolation procedures were performed.
IFAST loading and operation
Cell pellets were mixed with a solution containing 0.7 mg ml−1 oligo-dT PMPs (from Dynabeads mRNA Direct Kit, Invitrogen) in lysis buffer (1% LiDS, 100 mM Tris–HCl, 500 mM LiCl, 10 mM EDTA, 5 mM DTT) and incubated for five minutes at room temperature to allow lysis and binding. For the devices based on 384-well plate architecture, 8.5 µl of the lysate/PMP mixture and elution buffer (10 mM Tris–HCl) were added to the input and output wells, respectively. These solutions immediately filled their individual compartment, but resisted filling through the microfluidic constrictions (virtual walls). Next 8.5 µl of oil was added to the middle well, filling the remaining device area between the input and output wells. For devices based on the 1536-well plate architecture, volumes were reduced to 3 µl. The filled devices, which were typically operated in arrays of five, were placed on top of a magnet (K&J Magnetics B333, N52-grade neodymium cube (0.64 T) for single IFAST operation; BX041, N52-grade neodymium bar (1.48 T) for arrayed IFAST operation), aligning the input well with the magnet surface. Magnets that were wider than the IFAST device(s) were chosen such that the magnetic field was distributed relatively uniformly across the device (i.e., no edge effects), causing the PMPs to be pulled against the glass device bottom in a uniform line spanning the width of the device, thus preventing coagulation at maxima in the magnetic field. The magnet was moved at a velocity of −5 mms−1, drawing the PMPs across the oil barrier and into the elution buffer. The eluent was collected via pipette for RT-PCR. Varying the moving speed of the magnet did not affect operation except in the case of viscous samples (>10 cP), which required slowing the magnet velocity to −1 mm s−1.
Conventional nucleic acid purification
In order to benchmark IFAST performance, commercial NA isolation kits were run in parallel with the IFAST device. Cell pellets were re-suspended and split into aliquots containing equal cell numbers, which were then purified using either silica membrane spin column-based (Qiagen RNeasy) or PMP-based (Ambion MagMAX) NA isolation kits as per the manufacturers protocol. In the case of the silica membrane kit, a homogenizer (Qiagen QiaShredder) was used to pre-process the cultured cells as per the manufacturer’s recommendation.
RT-PCR
Isolated mRNA was reverse transcribed using a cDNA synthesis kit (iScript cDNA Synthesis Kit, Bio-Rad) at 42 °C for 30 minutes followed by 85 °C for 5 minutes. This cDNA was then mixed with qPCR master mix (SYBR® Green PCR Master Mix, Bio-Rad) and amplified for 40 cycles (95 °C for 15 seconds, then 57 to 60 °C for 1 minute) using a thermal cycler (MyiQ Thermal Cycler, Bio-Rad). Primers and annealing temperatures are detailed in Table S2†. Primers were chosen to span introns such that gDNA contamination could be identified from an analysis of a post-amplification melt curve. Gene expression levels were quantified by determining the CT at which exponential amplification was observed. Standard curves of PCR amplification efficiency were generated by preparing five serial dilutions of cDNA, plotting these values against CT, then using the slope of this plot to calculate efficiency.
Supplementary Material
Acknowledgements
This work was funded by the UW Stem Cell and Regenerative Medicine Center, the Walter H. Coulter Translational Research Partnership, NIH Grant #5R33CA137673, and DOD BCRP Concept Award #W81XWH-08-1-0525.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c1lc00004g
References
- 1.Qiagen Annual Report. 2008
- 2.Zhang C, Xu J, Ma W, Zheng W. PCR microfluidic devices for DNA amplification. Biotechnol. Adv. 2006;24(3):243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Cady NC, Stelick S, Kunnavakkam MV, Batt CA. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens. Actuators, B. 2005;107(1):332–341. [Google Scholar]
- 7.Anderson RC, Su X, Bogdan GJ, Fenton J. A miniature integrated device for automated multistep genetic assays. Nucleic Acids Res. 2000;28:e60. doi: 10.1093/nar/28.12.e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witek MA. 96-well polycarbonate-based microfluidic titer plate for high-throughput purification of DNA and RNA. Anal. Chem. 2008;80:3483–3491. doi: 10.1021/ac8002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J-G, Cheong KH, Huh N, Kim S, Choi J-W, Ko C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip. 2006;6:886–895. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
- 10.Lindstrom S, Hammond M, Brismar H, Andersson-Svahn H, Ahmadian A. PCR amplification and genetic analysis in a microwell cell culturing chip. Lab Chip. 2009;9:3465–3471. doi: 10.1039/b912596e. [DOI] [PubMed] [Google Scholar]
- 11.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal. Chem. 2004;76(7):1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 12.Marcus JS, Anderson WF, Quake SR. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 2006;78(9):3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 13.Sur K, McFall SM, Yeh ET, Jangam SR, Hayden MA, Stroupe SD, Kelso DM. Immiscible phase nucleic acid purification eliminates PCR inhibitors with a single pass of paramagnetic particles through a hydrophobic liquid. J. Mol. Diagn. 2010;12(5):620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atencia J, Beebe DJ. Controlled microfluidic interfaces. Nature. 2005;437(7059):648–655. doi: 10.1038/nature04163. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B, Moore JS, Beebe DJ. Surface-directed liquid flow inside microchannels. Science. 2001;291:1023–1026. doi: 10.1126/science.291.5506.1023. [DOI] [PubMed] [Google Scholar]
- 16.Darhuber A, Troian SM. Principles of microfluidic actuation by modulation of surface stresses. Annu. Rev. Fluid Mech. 2005;37:425–455. [Google Scholar]
- 17.Janes KA, Wang C-C, Holmberg KJ, Cabral K, Brugge JS. Identifying single-cell molecular programs by stochastic profiling. Nat. Methods. 2010;7(4):311–317. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.US Nucleic Acid Purification and Isolation Markets. Palo Alto, CA: Frost & Sullivan; 2008. [Google Scholar]
- 19.Campell J. Surface tension measurement by the drop weight method. J. Phys. D: Appl. Phys. 1970;3:1499–1504. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.