Abstract
The most active anticancer component in green tea is epigallocatechin-3-gallate (EGCG). The human peptidyl prolyl cis/trans isomerase (Pin1) plays a critical role in oncogenic signaling. Herein, we report the X-ray crystal structure of the Pin1/EGCG complex resolved at 1.9 Å resolution. Notably, the structure revealed the presence of EGCG in both the WW and PPIase domains of Pin1. The direct binding of EGCG with Pin1 was confirmed and the interaction inhibited Pin1 PPlase activity. In addition, proliferation of cells expressing Pin1 was inhibited and tumor growth in a xenograft mouse model was suppressed. The binding of EGCG with Arg17 in the WW domain prevented the binding of c-Jun, a well-known Pin1 substrate. EGCG treatment corresponded with a decreased abundance of cyclin D1 and diminution of TPA-induced AP-1 or NFκB promoter activity in cells expressing Pin1. Overall, these results showed that EGCG directly suppresses the tumor promoting effect of Pin1.
Keywords: EGCG, Pin1, tumorigenesis, Green Tea, Tumor
Introduction
Epigallocatechin-3-gallate (EGCG), the most potent active anticancer tea polyphenol, inhibits multiple signal transduction pathways, including activator protein-1 (AP-1), nuclear factor kappaB (NF-κB), nuclear factor of activated T-cells (NFAT) and β-catenin (1-5). The suppression of these signals is believed to contribute to the anticancer activities of EGCG because these pathways are critical in carcinogenesis. The human peptidyl prolyl cis/trans isomerase (Pin1) is composed of two distinct functional domains, the protein-protein interaction N-terminal WW domain (aa 1-39) and the isomerization catalysis PPIase domain (aa 45-168), which are connected by a flexible linker region (aa 40-44) (6). Pin1 isomerizes the peptide bond of specific phosphorylated serine or threonine residues preceding proline in several proteins involved in various events including mitosis, transcription, differentiation and the DNA damage response (7-8). The Ser/Thr-Pro motifs appear to be exclusive phosphorylation sites for many key protein kinases involved in control of cell growth and transformation (9-10). Pin1 is most likely required for the full activation of multiple signal transduction pathways including AP-1, NF-κB, NFAT and β-catenin (7, 11-12). Furthermore, Pin1 plays a key role in oncogenic signaling pathways (13-14) and is highly abundant in various tumors (11, 15) including breast (12), colorectal (16), prostate (17), and thyroid (18). Notably, inhibition of Pin1 in cancer cells triggers apoptosis or suppresses the transformed phenotype (19-20). EGCG was reported to inhibit the activation of HER-2/neu and its downstream signaling pathway in human head and neck and breast carcinoma cells (21). Pin1 is also a downstream effector of oncogenic Neu/Ras signaling (20, 22). Therefore, we hypothesized that EGCG might directly target Pin1 to suppress critical oncogenic signaling pathways and neoplastic cell transformation.
Materials and Methods
Structure determination and refinement
The crystals of the complex belong to space group P3121 with one molecule in the asymmetric unit. The crystals of the Pin1/EGCG complex were isomorphous to the crystals of Pin1 complexed with the L-peptide (PDB code 2itk) (23). Therefore the 2itk coordinates were used as the starting model for the refinement of our structure with REFMAC5 (24) coupled with automated building and updating of the solvent structure using ARP/wARP (25). Manual adjustment of the models and interpretation of extra electron densities were performed with the programs O (26) and COOT (27) using 2Fo-Fc and Fo-Fc electron density maps. The 2Fo-Fc maps for the two EGCG molecules were calculated without contribution of EGCG's coordinates (omit) with superimposed refined molecules of EGCG (Fig. S2). The structure model was refined to a final R-factor/Rfree-factor of 0.19/0.23. The final structure contains residues 6-163 of Pin1 (but without the interdomain linker region of aa 38-51 due to the lack of electron density), 2 EGCG molecules, 3 PEG400 molecules, 1 sulfate ion and 113 water molecules. The structure has good stereochemistry and no outliers were observed in the disallowed regions of the Ramachandran plot. Refinement statistics are summarized (Supplemental Table 1).
GST/Pin1 pull-down assay
To investigate the interaction of Pin1 and c-Jun, a glutathione-S-transferase (GST) pull-down assay was performed as described (12, 28). The pcDNA3.1/c-Jun plasmid was a kind gift from Dr. Michael J. Birrer (National Cancer Institute, Bethesda, MD). For expression of c-Jun, the c-Jun plasmid (pcDNA3.1/c-Jun) was transfected into HEK293T cells. For the GST/Pin1 pull-down assay, 4 μg GST and GST/Pin1 FL or GST/Pin1 WW domain fusion proteins were collected on glutathione-Sepharose beads (Amersham Biosciences) and treated with EGCG (0, 10, 20, 30, or 40 μM). The bound proteins were denatured in sample buffer, separated and analyzed by immunoblotting with anti-phospho-c-Jun (Ser73).
In vivo xenograft mouse model
Athymic nude mice [Cr:NIH(S), NIH Swiss nude, 5 weeks old] purchased from Jackson Laboratory were pretreated by intraperitoneal (i.p.) injection with vehicle (0.9% NaCl) or 50 mg/kg BW of EGCG in vehicle (3 × week for 2 weeks). At the end of 2 weeks, mice were injected subcutaneously in the right flank with 1×106 Neu/Pin1 WT or Neu/Pin1 KO MEFs in a volume of 0.5 ml. Treatment with vehicle (n = 15/group) or EGCG (n = 15/group) continued (3 × week for 32 days) until most of the vehicle-treated mice had tumors measuring 1 cm3, which is the endpoint allowed by U of M IACUC. In an additional study, mice were pretreated by i.p. injection with vehicle (10% ethanol/PBS) or 75 mg/kg of EGCG in vehicle (3 × week for 1 week). Mice were then injected subcutaneously in the right flank with 1×106 HCT116 cells. Treatment with vehicle (n = 10) or EGCG (n = 10) continued (3 × week for 27 days). For both studies, tumors were measured twice a week by caliper and volume calculated by the formula, length × width × height × 0.52.
siRNA Pin1 plasmid construction
The mU6Pro-Pin1 plasmid was constructed using the mU6Pro vector. The inserted sequence was: 5’-TTTGGTCAGGAGAGGAAGACTTTTTCAAGAGAAAAGTCTTCCTCTCCTGACTTTTT-3’. The siRNA sequence targeted against mRNA Pin1 was GUCAGGAGAGGAAGACUUU.
Construction of wildtype (WT) or mutant Pin1 expression vectors and establishing stable cells
The coding sequences of Pin1 were obtained after polymerase chain reaction (PCR) amplification (30 cycles, annealing at 65 °C) of the cDNA derived from reverse transcription of the total RNA of Neu/PIN1 WT MEFs. The amplified PCR products were ligated into the pcDNA4.0-HisMaxC plasmid (Invitrogen, Grand Island, NY) to produce an expression vector for 6xHis-and Xpress-tagged Pin1. The mutant Pin1 R17A was constructed using the Site-Directed Mutagenesis Kit II (Stratagene, La Jolla, CA). Transformants of Pin1 were obtained by transfecting 1 μg of the pcDNA4.0-WT, mutant-Pin1-HisMaxC, or control pcDNA4.0-HisMaxC vector into Neu/Pin1 KO MEFs using jetPEI (Qbiogen, Inc, Montreal, Quebec, CA) and then maintained by addition of Zeocin into the culture medium.
Protein expression and purification
E. coli BL21 (DE3) bacteria served as the host strain for the N-terminal His6-tagged Pin1 R14A mutant protein. Transformed BL21 bacteria were grown in Luria-Bertani (LB) medium with kanamicyn at 37°C. Bacteria were induced with 0.25 mM isopropyl β-D-thiogalactoside overnight at 15°C. The proteins were purified by Ni-affinity chromatography, digested with thrombin, and further purified by gel filtration chromatography (Superdex 75).
Affinity chromatography and ex vivo pull-down assay
After suspending Sepharose 4B freeze-dried powder (0.3 g) with 1 mmol/L HCl, it was mixed with EGCG (1.5 mg) in coupling solution (0.1 mol/L NaHCO3 pH 8.3 and 0.5 mol/L NaCl) and rotated at 4°C overnight. Then EGCG coupled to the CNBr-activated Sepharose 4B matrix was transferred to 0.1 mol/L Tris-HCl buffer (pH 8.0) and again rotated end over end at 4°C overnight. The mixture was washed three times with 0.1 mol/L acetate buffer (pH 4.0) containing 0.5 mol/L NaCl followed by a wash with 0.1 mol/L Tris-HCl (pH 8.0) containing 0.5 mol/L NaCl. The resulting EGCG-Sepharose 4B beads were used to assess binding of EGCG with soluble cellular proteins from Neu/Pin1 WT, Neu/Pin1 KO, Neu/Pin1 KO + Pin1, Neu/Pin1 KO + Pin1R17A or Neu/Pin1 KO + mock cell lysates in reaction buffer (50 mM Tris pH 7.5, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% NP-40, 2 μg/ml bovine serum albumin, 0.02 mM PMSF, 1X proteinase inhibitor). The beads were washed 5 times with buffer (50 mM Tris pH 7.5, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% NP-40, 0.02 mM PMSF) and proteins bound to the beads were analyzed by immunoblotting with the appropriate antibodies or by autoradiography (29).
PPIase assay and inactivation studies
The effect of EGCG on PPIase activity of purified Pin1 was determined using the proline isomerase assay based on the isomer-specific cleavage of the phosphorylated peptide WFY(pS)PR-pNA (Bachem) by trypsin as described (30) with some modifications. In brief, the isomerase activity of Pin1 was estimated by absorbance of p-nitroaniline at 390 nm measured every 10 sec for a total of 100 sec using a Beckman DU800 spectrophotometer and a temperature controlled cuvette holder. Prior to kinetic measurements, sample buffer containing Pin1 and EGCG was incubated in the spectrophotometer at 10° C for 10 min. Then trypsin (final concentration of 0.1 mg/ml from stock solution 50 mg/ml) in 1 mM HCl and 2 mM CaCl2 was added and the reaction was immediately initiated by addition of peptide dissolved in 0.47 M LiCl/TFE (trifluoroethanol, anhydrous) at a final concentration of 20 mg/ml. The total volume of reaction was 1 ml, including 35 mM Hepes (pH 7.8), 44 nM Pin1, 20 μM peptide, 0.1 mg/ml trypsin and various concentrations of EGCG (final concentrations 0-30 μM). Determination of inhibition constant (Ki) for EGCG was performed using the Dixon approach (31) with Nα-Bezoyl-DL-arginine 4-nitroanilida hydrochloride (BAPNA) as a substrate to build a calibration curve of dependence on relative absorbance of BAPNA concentration and to determine reaction velocities. From the graphics plot, the kinetic parameter (Vmax) was determined (Fig. S4A) as described previously (31). Then, the Ki was determined from the plot of the reciprocal inhibition rates as a function of concentrations of EGCG from the negative of the x-axis value at the point of intersection of the plot with the plot at V = Vmax (Fig. S4B-E). For ex vivo experiments, Pin1WT, Pin1KO, Neu/Pin1 WT, Neu/Pin1 KO, Neu/Pin1 KO + Pin1, Neu/Pin1 KO + mock, or Neu/Pin1KO + Pin1R17A MEF lysates were used. Cells were grown in 10-cm dishes for 24 h at a density of 2×105/ml and then treated with various concentrations of EGCG (0, 10, 15, 20 or 25 μM) for 24 h. Cells were disrupted in lysis buffer (35 mM Hepes pH 7.4, containing proteinase inhibitors) and equivalent concentrations of soluble cellular proteins were used in the assays. The Ki was determined as for the in vitro experiments. All Ki values obtained from the graphical approach were confirmed by calculations made using the program GraphPad Prism (GraphPad Prism® 5.0 for Windows [GraphPad Software, San Diego, CA: www.graphpad.com]).
FKBP and cyclophilin A in vitro pull-down assay
. To detect the FK506-binding protein (FKBP) and cyclophilin A, a glutathione-S-transferase pull-down assay was performed as described for the ex vivo experiment using 100 ng of FKBP or cyclophilin A (Sigma), respectively. Proteins bound to the beads were analyzed by immunoblotting with the appropriate antibodies (FKBP12 antibody, BD Biosciences; cyclophilin A antibody, Cell Signaling Technology). The levels of FKBP and cyclophilin A in Neu/Pin1WT and KO or Pin1WT and Pin1KO MEFs were also determined by immunoblotting with the appropriate antibodies.
ITC and Data Analysis
The purified mutant human Pin1R14A protein was subjected to gel filtration using a Superdex 75 column in buffer containing 10 mM Hepes (pH 7.5) and 100 mM NaCl. The EGCG solution was prepared in the same buffer and centrifuged at 14,000 rpm for 10 min to remove any insoluble particles. Isothermal titration calorimetry (ITC) experiments were performed at 25°C using a NanoITC instrument that was coupled with NanoAnalyze v.2.0.4 calculation software (www.tainstruments.com). The titration was carried out with 25 injections of 10 μl EGCG (500 μM; spaced 300 seconds apart) into the sample cell containing a solution of 1,000 μl of 5 μM Pin1. The thermogram was plotted by the NanoAnalyze software v.2.0.4 (www.tainstruments.com) using the best-fit parameters obtained by a nonlinear least-squares fit to the independent binding model after subtracting the heats of dilution from the raw binding heats. Dilution heats were determined by a blank titration and the association constant (Ka) was obtained and the dissociation constant (Kd) was calculated from the relationship: Kd = 1/Ka. Expression, purification and crystallization of the Pin1 protein was performed as described previously (23). The complex of Pin1 and EGCG was obtained by soaking Pin1 crystals in 40% (v/v) PEG 400, 100 mM HEPES-Na+ (pH 7.5), and 200 mM EGCG for 15-20 min at room temperature. The soaking solution served as a cryoprotectant, and during X-ray exposure, crystals of the complex were picked up by a fiber loop and frozen in a stream of liquid nitrogen. Diffraction data were collected to 1.9-Å resolution at the 23 IDD NET-CAT beamline (Advanced Photons Source, Argonne National Laboratory) using a MAR-300 CCD detector. The Pin1/EGCG structure (PDB ID 3OOB) was solved by molecular replacement method. The inhibition of tumorigenesis by EGCG in Pin1 expressing cells was confirmed in a xenograft model in vivo.
RESULTS
Crystal structure of the Pin1/EGCG complex
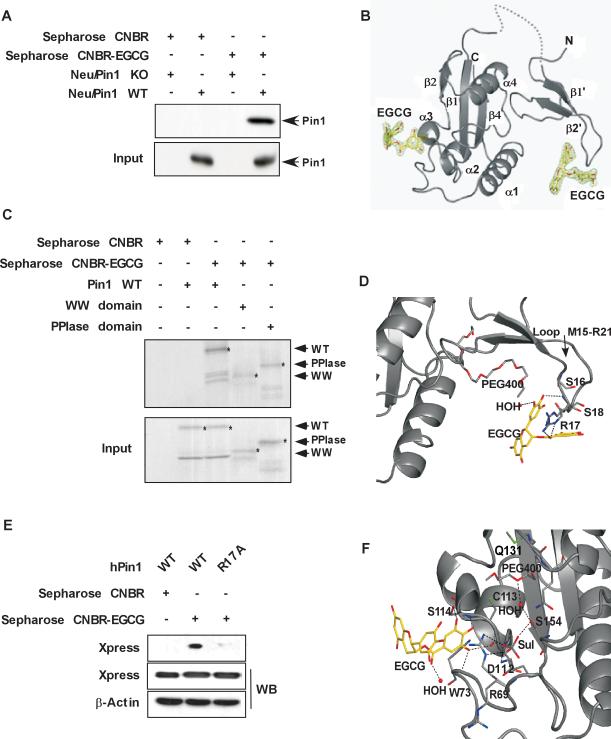
The binding of Pin1 and EGCG was confirmed in an affinity chromatography pull-down experiment and Western blotting with anti-Pin1 (Fig. 1A). We performed isothermal titration calorimetry (ITC) and calculated a dissociation constant of 21.6 μM (Fig. S1). To obtain direct information regarding the interaction between Pin1 and EGCG, the X-ray crystal structure of the Pin1/EGCG complex was determined and refined to 1.9 Å resolution (Supplemental Table 1). The analysis of the difference electron density maps revealed that two EGCG molecules (Fig. S2A) were bound to Pin1–one EGCG molecule bound the WW domain and the other to the PPIase domain (Fig. 1B, Fig. S2B, C). An additional affinity chromatography experiment using Sepharose CNBR-EGCG beads supported these findings (Fig. 1C).
Fig. 1. EGCG binds with Pin1.
(A) EGCG binds with Pin1 in Neu/Pin1 WT MEFs but not in Neu/Pin1 KO MEFs. Proteins bound to Sepharose CNBR-activated beads were analyzed by immunoblotting with the appropriate antibodies. (B) Overall structure of Pin1 complexed with EGCG. The structure of Pin1 is shown as a cartoon in gray. Secondary structure elements are marked in accordance with Ranganathan, et al. (6). Two EGCG molecules are in stick representation. The 2Fo-Fc omit electron density maps are contoured at 1.0 σ for EGCG in the complex structure. (C) Western blot analysis of Pin1 and EGCG binding in vitro using GST/Pin1 full-length (WT) protein and GST/WW and GST/PPIase domain deletion mutant proteins. (D) Close-up, enlarged view of the interaction of EGCG with the Pin1 WW domain. Residues that interact with EGCG are shown as sticks. Hydrogen bonds are indicated by dashed lines. (E) Western blot analysis of EGCG and Pin1 binding in WT and mutant Neu/Pin1 R17A MEFs. (F) Close-up, enlarged view of the interaction of EGCG with the PPIase domain of Pin1. Residues involved in hydrogen-bonding interactions are shown as sticks. Hydrogen bonds are indicated by dashed lines. Figures for the crystal structure were produced with PyMOL (http://www.pymol.org). For Western blot analyses, lysates from Neu/Pin1 WT, Neu/Pin1 KO, or mutant Neu/Pin1 R17A MEFs were used for an ex vivo assay. A GST/Pin1 full-length (WT) protein and GST/WW and GST/PPIase domain deletion mutant proteins were used for an in vitro assay. Cell lysates were applied to a Sepharose CNBR and a Sepharose CNBR-EGCG conjugated column. Pin1 was detected by Western blot using a specific Pin1 antibody or anti-Xpress.
In the WW domain, EGCG forms contacts with the pSer/Thr–Pro recognition loop of Met15–Tyr23 through hydrogen bonding with the side chain and the amide group of Arg17 (Fig. 1D). In this structure, the PEG 400 molecule (from crystallization conditions) occupies the hydrophobic pocket formed by the interface between both domains. The PEG 400 molecule might be thought to compete with EGCG for binding to the WW domain and prevent EGCG from occupying the appropriate position. To eliminate this possibility, we mutated arginine 17 to alanine (R17A) and showed that this mutant failed to bind EGCG ex vivo (Fig. 1E). This result supports the idea that EGCG binds with WW domain in the manner observed in the solved crystal structure.
We also found that a second molecule of EGCG bound to the PPIase domain of Pin1 (Fig. 1B, C). Although most of the atoms of the EGCG molecule are exposed to the surrounding solution, EGCG still creates several strong contacts with Pin1. Its gallate moiety forms bonds with the main chain carbonyl group of Asp112, the side chain of Ser114, and with the amide groups of Trp73 and Ser114 (Fig. 1F). In addition, from the crystallization conditions, the complex has a sulfate ion in the phosphate-binding cluster, which mimics the phosphorylated substrate moiety and another PEG 400 molecule in the active site (Fig. 1F).
Impact of EGCG on Pin1 activity
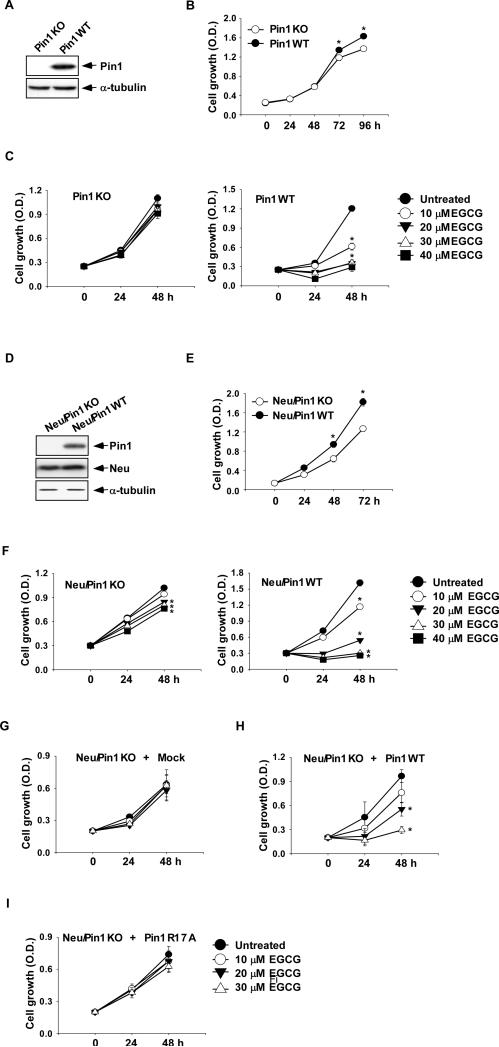
First, we showed that Pin1 is required for tumor cell proliferation in wildtype Pin1 murine embryonic fibroblasts (MEFs) (Pin1WT) and Pin1 deficient (Pin1KO) MEFs (Fig. 2A). Cell growth is decreased time dependently in Pin1WT MEFs compared to Pin1KO MEFs (Fig. 2B). To study the effect of EGCG on cell proliferation, we treated Pin1KO and Pin1WT MEFs with various concentrations of EGCG. Proliferation was assessed using the MTS assay as described in “Supplemental Methods”. Proliferation of Pin1KO MEFs was not affected by EGCG (Fig. 2C, left panel) compared to Pin1WT (Fig. 2C, right panel). These data suggest that Pin1WT MEFs are more sensitive to the anti-proliferation effects of EGCG compared to Pin1KO MEFs. Neu/Pin1 wildtype (WT) MEFs overexpress both the Neu oncogene and Pin1; and Neu/Pin1 knockout (KO) MEFs express the Neu oncogene but not Pin1 (Fig. 2D). Pin1 is also very important for cell growth in these cell lines (Fig. 2E). Treating these cells with EGCG revealed that EGCG inhibits proliferation of the Neu/Pin1 WT (Fig. 2F, right panel), but has no effect on Neu/Pin1 KO MEFs (Fig. 2F, left panel). In order to show that the resistance of Pin1 deficient MEFs to EGCG treatment is specifically due to the lack of Pin1 or the lack of Pin1 affinity for EGCG, Neu/Pin1 KO cells were transfected with the mammalian expression pcDNA4/HisMax-Pin1 plasmid, R17A-mutant-Pin1-HisMaxC plasmid, or with pcDNA4.0-HisMaxC vector as a control. These cells were then used to study the effect of EGCG on proliferation. Results indicated that only cells re-expressing Pin1 (Fig. 2H) regained the sensitivity to the anti-proliferation effects of EGCG, whereas cells transfected with vector (Fig. 2G) or the R17A mutant (Fig. 2I), which failed to bind EGCG, remained resistant to EGCG.
Fig. 2. EGCG inhibits cell growth.
The effect of EGCG on cell growth was estimated using the MTS assay kit, as described in “Supplemental Methods.” (A) The expression of Pin1 in Pin1KO and Pin1WT MEFs. (B) Growth comparison of Pin1KO and Pin1WT MEFs. Cells (1×103/well) were seeded on 96-well plates then incubated for 24, 48, 72 or 96 h and growth was estimated using the MTS assay. (C) Effect of various concentrations of EGCG (0-40 μM) on the growth of Pin1KO and Pin1WT MEFs. Cells (1×103/well) were seeded on 96-well plates then incubated in a 37°C/5% CO2 incubator for 24 h. Cells were treated with EGCG for 24 or 48 h and growth was then estimated by MTS assay. (D) Expression of Pin1 and Neu in Neu/Pin1WT and Neu/Pin1KO MEFs. (E) Growth comparison of Neu/Pin1WT and Neu/Pin1KO MEFs. Cells (1×103/well) were seeded on 96-well plates then incubated in a 37°C/5% CO2 incubator for 24, 48 or 72 h and cell growth was then estimated by MTS assay. (F) Effect of EGCG on the growth of Pin1KO and Pin1WT MEFs. Cells (1×103/well) were seeded on 96 well plates and incubated for 24 h in a 37°C/5% CO2 incubator. Cells were treated with EGCG for 24 or 48 h and then growth was estimated by MTS assay. Effect of EGCG on the growth of Neu/Pin1KO MEFs re-expressing (G) Mock, (H) Pin1WT, or (I) mutant Pin1R17A. Cells were transfected with a Mock, Pin1WT or mutant Pin1R17A plasmid. After zeocin selection, cells (1×103/well) were seeded on 96-well plates and incubated for 24 h in a 30°C/5% CO2 incubator. Cells were treated with various doses of EGCG for different times and growth was then estimated by MTS assay. Data are expressed as means ± S.D. and the asterisk (*) indicates a significant (p <0.001) decrease in proliferation of Pin1WT or Neu/Pin1WT MEFs compared to Pin1KO or Neu/Pin1KO MEFs or the untreated group compared to treated groups (20 or 30 μM EGCG).
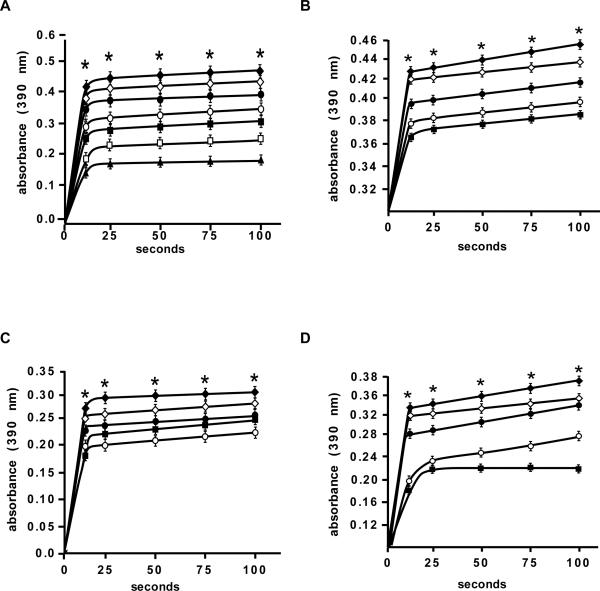
To study the functional significance of the binding of EGCG to Pin1, Pin1 PPIase activity measurements were made using an α-trypsin-coupled assay as described in Supplemental Information. For the ex vivo assays, lysates were prepared from Pin1WT and Pin1KO MEFs and Neu/Pin1 WT and Neu/Pin1 KO MEFs and treated 24 h with increasing concentrations of EGCG. The results indicated that EGCG suppressed PPIase activity in vitro (Fig. 3A), in Pin1WT (Fig. 3B), in Neu/Pin1 WT (Fig. 3C) and in Neu/Pin1 KO + WT (Fig. 3D) cells in a dose-dependent manner, but had no effect on Pin1KO (Fig. S3A) or Neu/Pin1 KO (Fig. S3B). For an ex vivo recovery experiment, Neu/Pin1 KO MEFs were transfected with a mammalian expression plasmid, pcDNA4/HisMax-Pin1, or a scrambled mammalian expression plasmid, pcDNA4/HisMax as a mock control. Also Neu/Pin1 KO MEFs were transfected with the R17A-mutant-Pin1-HisMaxC plasmid to determine whether the failure of the Pin1 mutant to bind EGCG will affect PPIase activity. The cells with re-expression of Pin1 showed a restoration of sensitivity to EGCG's inhibitory effect on PPIase activity (Fig. 3D), whereas the Neu/Pin1 KO + pcDNA4/HisMax or + R17A-mutant-Pin1-HisMaxC showed no response to EGCG (Fig. S3C, D). The inhibition constant (Ki) for EGCG in vitro was determined to be 20 μM (Kcat/Km =19.8× 106 M-s-) and for the ex vivo experiments, 45, 48 and 50 μM for PinWT, Neu/Pin1 WT, and Neu/Pin1 KO + Pin1 cells, respectively (Fig. S4B-E).
Fig. 3. EGCG inhibits Pin1 isomerase activity dose-dependently in vitro and ex vivo.
(A) In vitro measurement of Pin1 isomerase activity using increasing concentrations of EGCG. Plots reflect isomerase activity of Pin1 at a fixed concentration of purified Pin1 (44 nM), substrate (20 μM), and the indicated concentrations of EGCG in order from the top: 0, 5, 10, 15, 20, 25, and 30 μM. (B), (C) and (D), ex vivo measurement of Pin1 isomerase activity in lysates from (B) Pin1WT, (C) Neu/Pin1 WT, or (D) Neu/Pin1 KO + Pin1 MEFs treated for 24 h with the indicated concentrations of EGCG in order from the top: 0, 10, 15, 20, and 25 μM. Data are represented as means ± S.D. from 3 independent experiments. The asterisk (*) indicates a significant (p < 0.0001) inhibition of isomerase activity at all EGCG concentrations compared to the untreated control.
To investigate the specificity of EGCG for Pin1 compared to other isomerases, we determined the binding affinity of EGCG with the FK506-binding protein (FKBP) and human cyclophilin A. Western blot analysis of the application of FKBP and cyclophilin A to Sepharose 4B or a Sepharose 4B-EGCG column using a specific antibody was performed. Results indicated that both FKBP and cyclophilin A are expressed at similar levels in Neu/Pin1 WT, Neu/Pin1 KO, Pin1WT, and Pin1KO cells (Fig. S5A). However, EGCG did not bind to either protein (Fig. S5B).
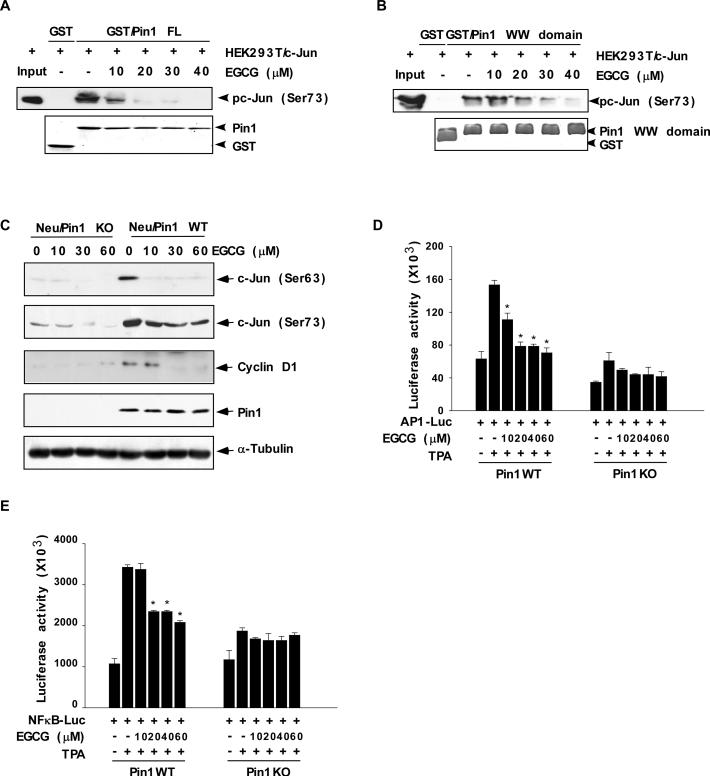
The formation of the Pin1/c-Jun complex is an important regulator of c-Jun stabilization and Pin1 has been shown to bind phosphorylated c-Jun and potentiate its transcriptional activity towards cyclin D1 (12). We therefore used a GST/Pin1 pull-down assay to determine the effect of increasing concentrations of EGCG on the formation of the Pin1 and c-Jun complex in cell lysates prepared from HEK293T cells that were transfected with c-Jun. Pin1 was phosphorylated on Ser73, as indicated by Western blot (Fig. 4A, B). No binding was observed between GST alone and phosphorylated c-Jun [pc-Jun (Ser73)], but binding did occur between phosphorylated c-Jun (Ser73) and the full-length (FL) GST/Pin1 (Fig. 4A) or the GST/Pin1 WW domain (Fig. 4B). Importantly, the binding of phosphorylated c-Jun (Ser73) with either full-length Pin1 or the WW domain of Pin1 was dramatically decreased by EGCG (Fig. 4A, B). The phosphorylation of c-Jun at Ser63 or Ser73 was examined in Neu/Pin1 WT and Neu/Pin1 KO MEFs (Fig. 4C). EGCG treatment resulted in the dose-dependent decrease in the phosphorylation of c-Jun at both sites in Neu/Pin1 WT but had no effect on Neu/Pin1 KO MEFs (Fig. 4C). Also we found that the level of cyclin D1 was decreased in EGCG-treated cells that overexpress Pin1, but was unchanged in Pin1 deficient cells treated with EGCG (Fig. 4C). The expression level of Pin1 in Neu/Pin1 WT did not change.
Fig. 4. EGCG inhibits the interaction of Pin1 with active c-Jun and attenuates TPA-induced AP-1 and TPA-induced NF-κB promoter activities.
(A) The effect of EGCG on GST/Pin1 full-length (FL) protein or (B) the GST/Pin1 WW domain and active c-Jun binding affinity. Lysates from HEK293T cells transfected with c-Jun were treated with different concentrations of EGCG and then subjected to a pull-down assay with GST, GST/Pin1 FL, or GST/Pin1 WW domain followed by immunoblotting with an antibody to detect phosphorylation of c-Jun (Ser73). Purified GST, GST/Pin1 FL, or GST/Pin WW domain abundance was visualized by Coomassie brilliant blue staining (bottom panels A, B). (C) The effect of EGCG on phosphorylation of c-Jun and cyclin D1 protein expression level in Neu/Pin1 WT and KO MEFs. Phosphorylation of c-Jun at Ser63 or Ser73 or cyclin D1 expression level was detected in lysates prepared from cells treated 24 h with various doses (0-30 μM) of EGCG. A specific Pin1 antibody was used to detect the abundance of Pin1 in the same samples. The firefly luciferase (D) AP-1- or (E) NF-κB reporter gene activity was assessed. For the reporter-gene assay, Pin1WT and Pin1KO MEFs were transfected with a plasmid mixture containing the (D) AP-1- or (E) NF-κB-luciferase reporter gene (100 ng) and the pRL-SV40 gene (10 ng) for normalization. At 24 h after transfection, cells were treated with various concentrations of EGCG (0, 10, 20, 40, or 60 μM) for 1 h before treatment with 20 ng/ml TPA for an additional 24 h. Data are shown as means ± S.D. of triplicate samples from 3 independent experiments. The asterisk (*) indicates a significant change relative to Pin1WT cells treated with only TPA (*, p < 0.005).
EGCG has been reported to inhibit AP-1 or NF-κB activity (2)'(3-4) and Pin1 is required for the full activation of signal transduction pathways including AP-1 and NF-κB (7, 12, 32). Therefore we hypothesized that the inhibitory effect of EGCG might effectively suppress AP-1 or NF-κB activity through Pin1. We examined the effect of EGCG on AP-1 and NF-κB promoter activity in Pin1WT and Pin1KO MEFs. Results indicated that EGCG significantly suppressed AP-1 (Fig. 4D) and NF-κB (Fig. 4E) reporter activity in Pin1WT cells, but had little effect on these activities in Pin1KO cells.
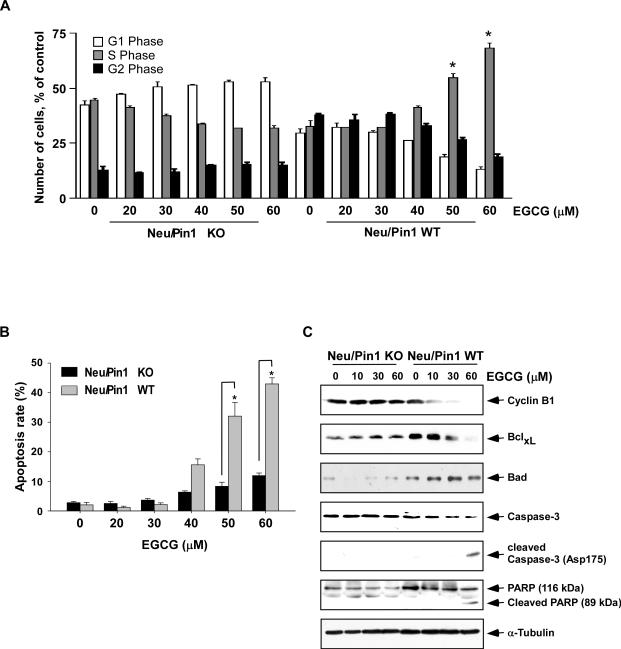
Pin1 has been shown to play an important role in cell cycle regulation (8, 33) and to trigger apoptosis with stress (11-12, 34). We compared the effect of EGCG on cell cycle and apoptosis in Pin1WT and Pin1KO MEFs. We found that EGCG induces accumulation of Pin1WT cells in S phase and apoptosis (Fig. 5A, B) and also regulates several pro- or anti-apoptotic marker genes (Fig. 5C). We observed that the number of cells in S phase was substantially increased in EGCG-treated Neu/Pin1 WT cells, but not in Neu/Pin1 KO cells (Fig. 5A). EGCG-induced more apoptosis in Neu/Pin1 WT compared to Neu/Pin1 KO cells. Additionally we found that the level of cyclin B1 and cyclin D1 was decreased in EGCG-treated cells overexpressing Pin1, but was not changed in Pin1 deficient cells treated with EGCG (Fig. 4C, 5C).
Fig. 5. EGCG induces apoptosis.
(A) Effect of EGCG on cell cycle in Neu/Pin1 WT or KO MEFs. Cells were cultured for 24 h with various doses of EGCG (0-60 μM). Cells were stained with PI and subjected to analysis by flow cytometry to assess cell distribution at each phase of the cell cycle as described in “Supplemental Methods”. All experiments were performed in duplicate and yielded similar results. Data are shown as means ± S.D. and the asterisk (*) indicates a significant increase (p < 0.001) in WT cells in S phase induced by EGCG compared to untreated control. (B) Effect of EGCG to induce apoptosis in Neu/Pin1 WT or KO MEFs. Neu Pin1 KO or WT MEFs were treated with various doses of EGCG (0-60 μM) for 24 h. Cells were then labeled with Annexin V and PI and analyzed by flow cytometry as described in ”Supplemental Methods.” The data are shown as the percentage of cells undergoing apoptosis and the asterisk (*) indicates a significant (p < 0.05) increase in apoptosis induced by EGCG in WT compared to KO cells. All experiments were performed in duplicate and yielded similar results. (C) EGCG regulates pro- or anti-apoptotic marker protein expression. Cells were treated with various doses of EGCG (0-60 μM) for 24 h and then harvested. Immunoblotting was performed with specific antibodies to detect cyclin B1, BclxL, Bad, caspase-3, cleaved caspase-3, and cleaved PARP. α-Tubulin was used as a control to verify equal loading of protein.
EGCG suppresses Pin1-induced tumorigenesis ex vivo and in vivo
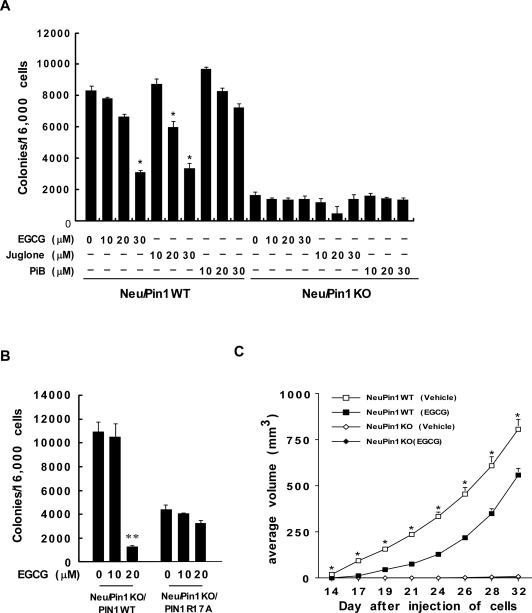
Next, we evaluated the effect of EGCG on colony formation mediated through Pin1 by performing an anchorage independent soft agar cell transformation assay using Neu/Pin1 WT and KO MEFs. Neu/Pin1 WT MEFs were treated with increasing concentrations (0-30 μM) of EGCG and results indicated that colony numbers (Fig. 6A) were significantly decreased in cells treated with EGCG compared with untreated cells. To compare the inhibitory efficiency of EGCG with that of the well known Pin1 inhibitors, Juglone (35) and PiB (30), we performed an anchorage independent soft agar cell transformation assay with Neu/Pin1 WT and Neu/Pin1 KO MEFs using the same conditions and the same concentrations (0-30 μM) of each of the inhibitors (Fig. 6A). Results indicated that EGCG was similar to Juglone in effectiveness, but more effective than PiB to inhibit colony formation of Neu/Pin1 WT MEFs. To confirm the essential role of Pin1 in colony formation, we performed an anchorage independent soft agar cell transformation assay using Neu/Pin1 KO MEFs re-expressing Pin1 WT or the R17A mutant and found that Neu/Pin1 KO MEFs re-expressing Pin1WT regained sensitivity to EGCG in contrast to the mutant (Fig. 6B).
Fig. 6. EGCG or knockout of Pin1 inhibits tumor formation and development ex vivo and in vivo.
(A) EGCG inhibits colony formation of Neu/Pin1 WT MEFs. Neu/Pin1 WT and KO MEFs were subjected to an anchorage independent soft agar assay to assess the effect of EGCG on colony formation. The assay was performed as described in “Supplemental Methods” and colonies were counted after 7 days using a microscope and the Image-Pro Plus (v.4) software program. Colonies are represented as the means ± S.D. of triplicate samples from 3 independent experiments. The asterisk (*) indicates a significant decrease (p < 0.01) in colony formation in Neu/Pin1 WT MEFs treated with EGCG compared with untreated cells. The effect of Juglone or PiB compared to EGCG is also shown. (B) Re-expression of Pin1WT, but not mutant Pin1 R17A, restores sensitivity to the inhibitory effect of EGCG on colony formation. Re-expressing Pin1WT in mutant Pin1R17A or Neu/Pin1 KO MEFs was performed as described in “Supplemental Methods” and colony formation was measured as for panel (A). (C) Athymic nude mice were treated as described in “Supplemental Methods” and tumor volume was measured and recorded 3 times a week. The asterisk (*) indicates a significantly larger volume of tumor growth (p < 0.0001) in Neu/Pin1WT mice treated with vehicle compared to Neu/Pin1WT mice treated 3 x week i.p. with 50 mg/kg BW EGCG (one way ANOVA). Two-way ANOVA indicated a significant effect of EGCG and knockout of Pin1 on tumor development (p < 0.0001).
We finally determined whether EGCG could suppress tumor growth in a xenograft study in vivo using mice injected with Neu/Pin1 WT or KO cells (1 × 106). Mice (n =15 mice/group) were either treated with vehicle or EGCG (50 mg/kg body weight) 3 × week by i.p. (intraperitoneal) injection. We found that tumor formation was significantly delayed by EGCG treatment. At day 14, 60% of WT vehicle-treated mice had visible tumors, whereas none of the EGCG-treated WT mice had tumors. At day 17, 100% of vehicle-treated WT mice had tumors and only 40% of EGCG treated mice had tumors. Notably, only 1 mouse injected with Neu/Pin1 KO cells in each of the vehicle-treated and EGCG-treated groups developed a small visible tumor (day 28) (Fig.6C). Furthermore, EGCG also suppressed tumor formation in nude mice injected with HCT116 colorectal cancer cells, which also express high levels of Pin1 (Fig. S6). The significant effect of EGCG and knockout of Pin1 on tumor development in vivo confirmed that Pin1 is an oncoprotein and the antitumor activity of EGCG is associated with Pin1 activity.
DISCUSSION
Effective inhibitors of human Pin1 activation could provide a novel clinical strategy for cancer prevention or therapy. Both the WW and PPIase domain activities of Pin1 are important for Pin1's function (6, 36-40). Thus, a highly effective inhibitor could be targeted either against the catalytic center of the PPIase domain or against the phospho-peptide-binding site of the WW domain. The only reported crystal structure of Pin1 with a ligand bound to the WW domain is the Pin1 complex with the doubly phosphorylated CTD peptide (41) (PDB code 1f8a). All other crystal structures with peptides or inhibitors of Pin1 were screened against the PPIase domain: (6) (PDB code 1pin); (23) (PDB codes 2q5a, 2itk); (42) (PDB codes 3ik8, 3ikd, 3ikg); (43) (PDB codes 3kac, 3kad, 3kaf, 3kag, 3kah, 3kai, 3kce); (44) (PDB codes 3i6c, 3jyj). Here we present the first crystal structure of the complex of Pin1 with a natural inhibitor where the inhibitor is targeted against not only the WW domain of Pin1, but also against the catalytic PPlase domain of Pin1. The binding of EGCG in both the WW domain and PPIase domain might explain why EGCG is an effective inhibitor of Pin1 and Pin1-regulated genes or signaling pathways. The crystallographic findings were supported by our studies in vitro, ex vivo, and in vivo. Previous studies demonstrated that inhibition of Pin1 in cancer cells triggers apoptosis or suppresses the transformed phenotype (19-20). We showed that the Pin1/EGCG association plays a regulatory role in cell proliferation (Fig. 2) and malignant cell transformation (Fig. 6A, B). In a xenograft athymic nude mouse model, our results indicated that EGCG significantly suppresses tumor growth of Neu/Pin1 WT MEFs (Fig.6C) as well as of HCT116 cells, which express high levels of Pin1 (Fig. S6). Pin1 has many reported protein targets and an important Pin1 substrate is the pSer63/73-Pro motif in c-Jun (12). Consistent with our data, the binding of phosphorylated c-Jun (Ser73) with either full-length Pin1 or the WW domain of Pin1 was dramatically decreased by EGCG (Fig. 4A, B). The binding of Pin1 to phosphorylated c-Jun is associated with an increased expression of cyclin D1 (12). We found that EGCG treatment corresponded with a decreased abundance of cyclin D1 in cells overexpressing Pin1, but cyclin D1 was unchanged in Pin1 deficient cells (Fig. 4C). EGCG was reported to inhibit tumor promoter-induced cell transformation mediated by AP-1 (2) and NF-κB activation (3). Pin1 was also shown to regulate NF-κB DNA binding and reporter activity after Pin1 recognition of its phosphorylated p65/RelA subunit (45). We found that EGCG effectively inhibited AP-1 or NF-κB reporter activity in Pin1 WT MEFs, but not in Pin1 deficient MEFs (Fig. 4D,E). We also showed that EGCG might effectively suppress AP-1 or NF-κB activity through Pin1, because EGCG decreased the phosphorylation of the Pin1 substrate, c-Jun at Ser63 and Ser73, in cells expressing Pin1, but not in Pin1 deficient MEFs (Fig.4C). These data also support the hypothesis that EGCG directly targets Pin1 and this association can inhibit Pin1 tumor promoter activity.
Apoptosis was markedly induced with EGCG treatment in WT compared to KO MEFs (Fig. 5B); and EGCG-induced changes in levels of various pro- or anti-apoptotic associated proteins was more apparent in Pin1 overexpressing cells (Fig. 5C). The comparison of the inhibitory effect of EGCG with other known Pin1 inhibitors, Juglone (35) and PiB (30), revealed that EGCG was comparable to these inhibitors in its ability to suppress colony formation of Neu/Pin1 WT MEFs (Fig. 6A). On the other hand, Pin1 deficient cells were unresponsive to all three inhibitors. Juglone and PiB were shown to be specific for the parvulin family of peptidyl-prolyl cis/trans isomerases and do not affect the PPIase activity of other isomerases, such as FK506 binding proteins or cyclophilins (30, 35). To determine the ability of EGCG to bind with FKBP12 or cyclophilin A, we prepared an in vitro pull-down assay with EGCG that revealed no binding between EGCG and these isomerases (Fig. S5B). In addition, our isomerase activity assays showed that Pin1-expressing and -deficient cells have a differential sensitivity to EGCG despite the same abundance of these isomerases (Fig. S5A). Overall, these results indicate a specificity of EGCG in interacting with Pin1.
The crystal structure of the Pin1/EGCG complex revealed that EGCG is bound to the Met 15-Tyr23 loop in the WW domain of Pin1 through interaction with the backbone and side chain of Arg17. The importance of this loop and especially Arg17 for the binding activity and function of Pin1 was confirmed by mutational analysis (41), which revealed that the side chain of Arg17 contributes substantially to the ligand-binding energy through the recognition of a phosphorylated serine/threonine residue of target proteins. Replacing Arg17 with Ala resulted in a 6.3-fold decrease in binding activity (41). Notably, our results indicated that mutating Arg17 to Ala resulted in an inability of Pin1 to bind EGCG ex vivo (Fig. 1E). These findings might explain why EGCG binding with WW domain can inhibit its phospho-group binding ability. The other consequence of the EGCG association with Arg17 might be its effect on Ser16 phosphorylation. The Pin1 WW domain is phosphorylated on Ser16 both in vitro and in vivo and the phosphorylation regulates the ability of the WW domain to mediate Pin1 substrate interaction and cellular localization (39). Pin1 and WW domain mutants, which cannot be phosphorylated at Ser16, were shown to act as dominant-negative mutants to induce mitotic block, apoptosis, and increase multinucleated cells (39). Even though the distances from EGCG to the hydroxyl group of Ser16 in the solved structure are too large to form hydrogen bonds, EGCG binding with Arg17 could prevent phosphate group access to this residue.
The suppressive effect of EGCG on Pin1 isomerase activity in vitro and ex vivo provides evidence that EGCG inhibits Pin1 isomerase function even after re-expression of Pin1 in Pin1 KO cells and confirms our results showing that EGCG also binds with the PPIase domain of Pin1. Cys113 was shown to be important for the isomerase activity of Pin1 because its thiol group is involved in catalysis (6). The flexibility of a main chain of residues spatially neighboring Cys113, including Trp73 and Ser114, might be critical for rotational freedom of Cys113's thiol group (46), which is necessary for the catalytic reaction (6). In the solved structure, EGCG forms hydrogen bonds with the amide groups of the backbone of Trp73 and Ser114 and the carbonyl group of the backbone of Asp112, making them more rigid. Thus, EGCG can affect the unrestricted movement of Cys113.
Finally, the changing of the surface charges around the side chain of Cys113 upon the negatively charged group binding might alter the reactivity of its thiol group (46). The apparent pKa of Cys113 increases (i.e., becomes more basic), making the residue less nucleophilic, which will inhibit the catalytic reaction (46). EGCG might possibly have an effect on changing the surface charge around Cys113 to control the isomerase activity of Pin1. Overall, our crystallographic findings and accompanying biochemical data showed that EGCG binds with both domains of Pin1 and directly inhibits the tumor-promoting activity of Pin1 and, thus, might have practical implications for cancer prevention or therapy.
Supplementary Material
Supplemental Table 1. Summary of crystallographic data collection and refinement statistics
ACKNOWLEDGMENTS
We thank Dr. Kun Ping Lu (Harvard Medical School, Boston, MA) for providing the GST/Pin1, Pin1 deficient (Pin1-/-), Neu/Pin1 KO, Pin1 expressing (Pin1+/+), and Neu/Pin1 WT cell lines; Dr. Zhiwei He for providing the siPin1 construct and assisting in the creation of the knockdown and overexpression cells. We are grateful for the gift of the Pin1 R14A mutant plasmid from Dr. Jessie Zhang (Salk Institute, University of California, CA). We acknowledge the Argonne Photon Source and Argonne National Laboratory for providing access to the X-ray facilities for data collection. We give special thanks to Dr. Valentina Terechko (University of Chicago, IL) for her assistance during the data collection and give special thanks to Dr. Svetlana Ermakova (Pacific Institute of Bioorganic Chemistry, Russia), Dr.Sergei Pletnev (National Cancer Institute ANL, IL), and Dr. Svetlana Antonyuk (University of Liverpool, UK) for their advice and assistance.
Financial Support: This work was supported by The Hormel Foundation and National Institutes of Health Grants R37 CA081064 and CA120388.
Footnotes
AUTHOR CONTRIBUTIONS
Z.D. designed research; D.V.U., J.-H. S., D.J.K., S.K.J., A.M.B., and A.C. performed research experiments; D.V.U., J.-H.S., T.A.Z., D.J.K., S.K.J, A.M.B., and A.C. analyzed data; D.V.U., D.J.K., and A.M.B. wrote the paper.
Conflicts of interest statement: Authors disclose no conflict of interest.
Accession code. Atomic coordinates and experimental structure factors of the Pin1/EGCG complex have been deposited in the Protein Data Bank under ID code 3OOB.
REFERENCES
- 1.Dashwood WM, Orner GA, Dashwood RH. Inhibition of beta-catenin/Tcf activity by white tea, green tea, and epigallocatechin-3-gallate (EGCG): minor contribution of H(2)O(2) at physiologically relevant EGCG concentrations. Biochem Biophys Res Commun. 2002;296:584–8. doi: 10.1016/s0006-291x(02)00914-2. [DOI] [PubMed] [Google Scholar]
- 2.Dong Z, Ma W, Huang C, Yang CS. Inhibition of tumor promoter-induced activator protein 1 activation and cell transformation by tea polyphenols, (-)-epigallocatechin gallate, and theaflavins. Cancer Res. 1997;57:4414–9. [PubMed] [Google Scholar]
- 3.Gupta S, Hastak K, Afaq F, Ahmad N, Mukhtar H. Essential role of caspases in epigallocatechin-3-gallate-mediated inhibition of nuclear factor kappa B and induction of apoptosis. Oncogene. 2004;23:2507–22. doi: 10.1038/sj.onc.1207353. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Jin H, Shim HE, Kim HN, Ha H, et al. Epigallocatechin-3-gallate inhibits osteoclastogenesis by down-regulating c-Fos expression and suppressing the nuclear factor-kappaB signal. Mol Pharmacol. 2010;77:17–25. doi: 10.1124/mol.109.057877. [DOI] [PubMed] [Google Scholar]
- 5.Mizuno H, Cho YY, Zhu F, Ma WY, Bode AM, et al. Theaflavin-3, 3'-digallate induces epidermal growth factor receptor downregulation. Mol Carcinog. 2006;45:204–12. doi: 10.1002/mc.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–86. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 7.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7:435–41. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- 8.Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer. 2007;7:381–8. doi: 10.1038/nrc2107. [DOI] [PubMed] [Google Scholar]
- 9.Lu KP. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem Sci. 2004;29:200–9. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 2002;12:164–72. doi: 10.1016/s0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- 11.Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 12.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. Embo J. 2001;20:3459–72. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez-Sola D, Dalla-Favera R. PINning down the c-Myc oncoprotein. Nat Cell Biol. 2004;6:288–9. doi: 10.1038/ncb0404-288. [DOI] [PubMed] [Google Scholar]
- 14.Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–7. [PubMed] [Google Scholar]
- 15.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, et al. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–37. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim CJ, Cho YG, Park YG, Nam SW, Kim SY, et al. Pin1 overexpression in colorectal cancer and its correlation with aberrant beta-catenin expression. World J Gastroenterol. 2005;11:5006–9. doi: 10.3748/wjg.v11.i32.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayala G, Wang D, Wulf G, Frolov A, Li R, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003;63:6244–51. [PubMed] [Google Scholar]
- 18.Nakashima M, Meirmanov S, Naruke Y, Kondo H, Saenko V, et al. Cyclin D1 overexpression in thyroid tumours from a radio-contaminated area and its correlation with Pin1 and aberrant beta-catenin expression. J Pathol. 2004;202:446–55. doi: 10.1002/path.1534. [DOI] [PubMed] [Google Scholar]
- 19.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–7. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 20.Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW, et al. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. 2002;22:5281–95. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda M, Suzui M, Lim JT, Weinstein IB. Epigallocatechin-3-gallate inhibits activation of HER-2/neu and downstream signaling pathways in human head and neck and breast carcinoma cells. Clin Cancer Res. 2003;9:3486–91. [PubMed] [Google Scholar]
- 22.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. Embo J. 2004;23:3397–407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Daum S, Wildemann D, Zhou X. Zh., Verdecia M, Bowman ME, Lucke Ch., Hunter T, Lu K-P, Fischer G, Noel JP. Structural Basis for High-Affinity Peptide Inhibition of Human Pin1. ACS Chemical Biology. 2007;2(5):320–8. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecule structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D. 1993;49:129–47. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 26.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Emsley PC,K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Zacchi PGM, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 2002 Oct 24;419(6909):853–7. doi: 10.1038/nature01120. [DOI] [PubMed] [Google Scholar]
- 29.Ermakova SP, Kang BS, Choi BY, Choi HS, Schuster TF, Ma WY, Bode AM, Dong Z. (-)-Epigallocatechin Gallate Overcomes Resistance to etoposide-Indused Cell death by Targeting the Molecular Chaperone Glucose-regulated Protein 78. Cancer Research. 2006;66:9260–9. doi: 10.1158/0008-5472.CAN-06-1586. [DOI] [PubMed] [Google Scholar]
- 30.Uchida T, Takamiya M, Takahashi M, Miyashita H, Ikeda H, et al. Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem Biol. 2003;10:15–24. doi: 10.1016/s1074-5521(02)00310-1. [DOI] [PubMed] [Google Scholar]
- 31.Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd ed. A John Wiley &Sons, Inc., Publication; New York/Chichester/Weinheim/Brisbane/Sigapore/Toronto: 2000. [Google Scholar]
- 32.Lu KPZX. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007 Nov;8(11):904–16. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 33.Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J Biol Chem. 2002;277:47976–9. doi: 10.1074/jbc.C200538200. [DOI] [PubMed] [Google Scholar]
- 34.Zheng H, You H, Zhou XZ, Murray SA, Uchida T, et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 2002;419:849–53. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]
- 35.Hennig L, Christner C, Kipping M, Schelbert B, Rucknagel KP, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37:5953–60. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 36.Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Structure-function-folding relationship in a WW domain. PNAS. 2006;103:10648–53. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–60. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 38.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidylprolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–20. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–4. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 40.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–8. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 41.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–43. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 42.Guo C, Hou X, Dong L, Dagostino E, Greasley S, et al. Structure-based design of novel human Pin1 inhibitors (I). Bioorg Med Chem Lett. 2009;19:5613–6. doi: 10.1016/j.bmcl.2009.08.034. [DOI] [PubMed] [Google Scholar]
- 43.Potter AJ, Ray S, Gueritz L, Nunns CL, Bryant CJ, et al. Structure-guided design of alpha-amino acid-derived Pin1 inhibitors. Bioorg Med Chem Lett. 2010;20:586–90. doi: 10.1016/j.bmcl.2009.11.090. [DOI] [PubMed] [Google Scholar]
- 44.Dong L, Marakovits J, Hou X, Guo C, Greasley S, et al. Structure-based design of novel human Pin1 inhibitors (II). Bioorg Med Chem Lett. 2010;20:2210–4. doi: 10.1016/j.bmcl.2010.02.033. [DOI] [PubMed] [Google Scholar]
- 45.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–26. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 46.Bayer E, Goettsch S, Mueller JW, Griewel B, Guiberman E, Mayr LM, Bayer P. Structural Analysis of the Mitotic Regulator hPin1 in Solution. The Journal of Biological Chemistry. 2003;278:26183–93. doi: 10.1074/jbc.M300721200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Summary of crystallographic data collection and refinement statistics