Abstract
Peroxisome proliferator-activated receptors (PPARs) are nuclear hormone receptors that regulate genes involved in energy metabolism and inflammation. For biological activity, PPARs require cognate lipid ligands, heterodimerization with retinoic × receptors, and coactivation by PPAR-γ coactivator-1α or PPAR-γ coactivator-1β (PGC-1α or PGC-1β, encoded by Ppargc1a and Ppargc1b, respectively). Here we show that lipolysis of cellular triglycerides by adipose triglyceride lipase (patatin-like phospholipase domain containing protein 2, encoded by Pnpla2; hereafter referred to as Atgl) generates essential mediator(s) involved in the generation of lipid ligands for PPAR activation. Atgl deficiency in mice decreases mRNA levels of PPAR-α and PPAR-δ target genes. In the heart, this leads to decreased PGC-1α and PGC-1β expression and severely disrupted mitochondrial substrate oxidation and respiration; this is followed by excessive lipid accumulation, cardiac insufficiency and lethal cardiomyopathy. Reconstituting normal PPAR target gene expression by pharmacological treatment of Atgl-deficient mice with PPAR-α agonists completely reverses the mitochondrial defects, restores normal heart function and prevents premature death. These findings reveal a potential treatment for the excessive cardiac lipid accumulation and often-lethal cardiomyopathy in people with neutral lipid storage disease, a disease marked by reduced or absent ATGL activity.
PPARs belong to a superfamily of nuclear hormone receptors. Ligand-induced activation of PPARs controls the expression of innumerable genes involved in energy homeostasis, lipid and lipoprotein metabolism, and inflammation. The PPAR family consists of PPAR-α, PPAR-δ (also known as PPAR-β), and three isoforms of PPAR-γ (ref. 1). PPAR-α is found mainly in oxidative tissues like cardiac muscle, skeletal muscle and liver, where it activates the transcription of genes stimulating fatty acid transport and oxidation, ketogenesis and gluconeogenesis2. PPAR-δ is expressed ubiquitously and activates genes for fatty acid and glucose utilization3. PPAR-γ is more common in cells involved in fat storage, and it induces the expression of lipogenic genes4. Activation of all PPAR isoforms involves the binding of lipid ligands, dimerization with retinoid × receptor (R×R), and coactivation by PGC-1α or PGC-1β (refs. 5–7). Because both PPAR-α and PPAR-δ are crucial for the coordinated regulation of processes involved in the uptake, transport and oxidation of energy substrates2,3, their dysregulation causes major metabolic derangements, thereby resulting in ectopic lipid accumulation in cardiac and skeletal muscle as well as in the liver. This lipid overflow in nonadi-pose tissues leads to pronounced lipotoxicity, which is characterized by defective insulin signaling, insulin resistance and cellular dysfunction that may eventually lead to cell death.
Although it is currently not clear how specific endogenous lipid ligands activate the PPARs and whether ligand preference varies in a tissue-specific manner, it is generally accepted that fatty acids directly (as PPAR ligands) or indirectly (as precursors for other lipid ligands of PPARs) activate target gene expression. Established ligands include mono- and polyunsaturated fatty acids, as well as fatty acid derivatives such as fatty acid-Coenzyme A (CoA), eicosanoids and phospholipids8–13. Cellular fatty acids originate from the uptake of unesterified plasma fatty acids, the uptake of fatty acids generated by lipoprotein lipase (LPL)-mediated hydrolysis of triglyceride-rich lipoproteins in tissue capillaries or de novo synthesis. Fatty acids from all these sources can activate PPAR-mediated gene expression13,14; however, it is currently unknown whether fatty acids require esterification to triglycerides and subsequent rehydrolysis (lipolysis) before signal transduction to the nucleus. If lipolysis is required, then the activity of ATGL and/or hormone-sensitive lipase (encoded by LIPE, referred to hereafter as HSL)15 would be an expected step in the activation of PPARs. The relatively benign phenotype of Hsl deficiency in mice (Hsl-knockout mice, hereafter referred to as HslKO) argues against a crucial role of this enzyme in the regulation of PPARs and energy homeostasis16,17. In contrast, Atgl deficiency in mice (Atgl-knockout mice, referred to hereafter as AtglKO; synonymous with Pnpla2–/–) is associated with reduced expression of genes responsible for oxidative phosphorylation (OXPHOS)18, massive lipid accumulation in multiple tissues, severe skeletal- and cardiomyopathies, and premature death19. Similar defects in lipid- and energy homeostasis are observed in humans with mutations in the ATGL gene20,21. All affected individuals suffer from neutral lipid storage disease (NLSD) and many people develop severe skeletal myopathies, as well as cardiomyopathies that often necessitate heart transplantation21.
These findings inspired the hypothesis that the provision of fatty acids for PPAR activation in oxidative organs such as the heart requires ATGL-mediated lipolysis. To test this hypothesis, we assessed the expression of established PPAR-α and PPAR-δ target genes in relation to mitochondrial morphology, OXPHOS and heart function in normal and mutant mice lacking either Hsl or Atgl. We show that Atgl-mediated lipolysis in cardiac muscle is essential for the biological activity of the PPAR-α–PGC-1 complex and normal heart function.
Results
Atgl deficiency impairs PPAR target gene expression
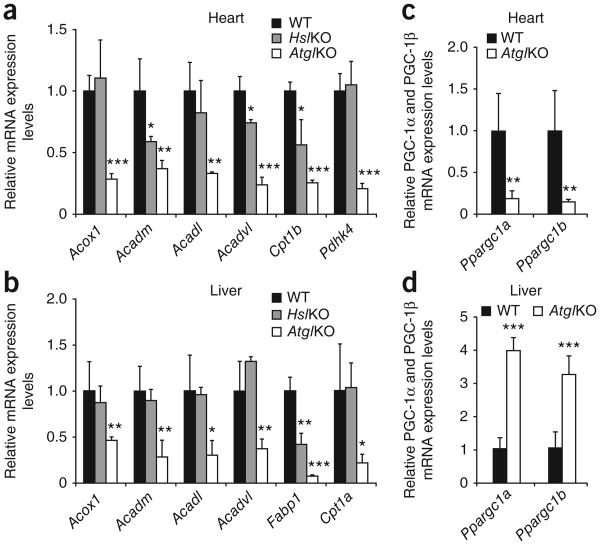
Our analysis of mRNA expression in cardiac muscle of AtglKO mice revealed markedly reduced expression of PPAR-α and PPAR-δ target genes, including acyl-CoA oxidase (Acox1, −72%), medium-chain acyl-CoA dehydrogenase (Acadm, −64%), long-chain acyl-CoA dehydrogenase (Acadl, −67%), very long–chain acyl-CoA dehydrogenase (Acadvl, −77%), carnitine palmitoyltransferase 1b (Cpt1b, −75%) and pyruvate dehydrogenase kinase 4 (Pdhk4, −79%) compared to the levels found in wild-type mice (Fig. 1a). Reduction in the expression of these genes in HslKO mice was less pronounced (Acadm, −42%; Acadvl, −26%; Cpt1b, −44%) or nonexistent (Acox1, Acadl, Pdhk4). Our analysis of liver samples showed a similar mRNA expression profile characterized by marked reduction of the expression of all PPAR-α and PPAR-δ target genes examined (−50% to −90%) in AtglKO mice compared to wild-type mice (Fig. 1b), whereas only gene expression of liver-specific fatty acid–binding protein (L-Fabp, encoded by Fabp1) expression was reduced in HslKO mice (−58%). We found that measurements of PPAR proteins in AtglKO mice showed reduced concentrations of PPAR-α in the heart (−37%) and liver (−33%) (Supplementary Fig. 1a), and of PPAR-δ in the heart (−19%) compared with wild-type mice (Supplementary Fig. 1b). As the main regulatory step for efficient transactivation of gene transcription involves the hetero dimerization of PPAR proteins with R×R-α in a ligand-dependent manner, the moderate reduction in PPAR protein concentrations is unlikely to mediate the drastic decrease of PPAR target gene expression in Atgl-deficient cardiac muscle or liver. Notably, we found that PGC-1α (−80%) and PGC-1β (−85%) mRNA levels were also drastically decreased in cardiac muscle of AtglKO mice compared to wild-type mice (Fig. 1c). This decrease was specific to cardiac muscle because mRNA levels of PGC-1α and PGC-1β were higher in the liver of AtglKO mice than in wild-type mice (Fig. 1d), thus arguing for a tissue-specific difference in the regulation of PGC-1 expression. In cardiac muscle, we found that Atgl-deficient mice had normal PGC-1α and PGC-1β mRNA levels at birth (Supplementary Fig. 1c). At weaning (14 ± 1 d after birth), however, we found that the concentration of PGC-1α mRNA decreased relative to wild-type mice by 64%, and that PGC-1β tended to decrease (−18%). This decrease in mRNA levels in both types of PGC-1 typically precedes pronounced cardiac lipid accumulation and defective heart function.
Figure 1.
Expression of PPAR-α and PPAR-δ target genes and PGC-1α and PGC-1β in Atg/KO, Hs/KO, and wild-type tissues. mRNA expression levels for selected PPAR-α and PPAR-δ target genes and PGC-1α and PGC-1β were determined by RT-qPCR analysis. (a,b) Cardiac (a) and hepatic (b) mRNA expression of PPAR-α and PPAR-δ target genes were markedly decreased in fasted 8- to 10-week-old female Atg/KO mice compared to age-matched Hs/KO and wild-type mice. (c,d) mRNA levels of genes encoding PGC-1α and PGC-1β mRNA were also reduced in cardiac muscle (c) but increased in the liver (d) of fasted Atg/KO mice compared to wild-type mice. n = 4. Error bars show means ± s.d. *P < 0.05, **P < 0.01 and ***P < 0.001.
Impaired cardiac mitochondrial respiration in AtglKO mice
We carried out biochemical analyses that confirmed the previously observed increased cardiac triglyceride content in AtglKO mice (20-fold; ref. 19) and additionally found increased glycogen concentrations (+70%) in cardiac muscle of the mutant mice compared with wild-type mice (Fig. 2a). Our histomorphological analysis showed that these increases were a result of increased size and number of lipid droplets and glycogen granules within cardiomyocytes (Fig. 2b). Furthermore, although the overall morphology of mitochondria and the structure of mitochondrial cristae appeared normal in electron microscopical analyses of AtglKO cardiac muscle, mitochondrial size compared to that of wild-type mice was increased when we applied higher-resolution morphometric (+18%) and cytofluorimetric (+45%) analyses (Fig. 2c,d). Concomitantly, we found that mitochondrial DNA content in cardiac muscle was 32% lower in AtglKO mice than in wild-type mice, thus indicating a decreased number of mitochondria in the hearts of AtglKO mice (Fig. 2e).
Figure 2.

Morphology, glycogen content, mitochondria size and mitochondrial DNA content in cardiac muscle of wild-type and Atg/KO mice. (a) Cardiac muscle glycogen content (measured as glucose after hydrolysis) of 10-week-old female wild-type and Atg/KO mice (n = 9). (b) Transmission electron microscopy of cardiac muscle sections from 10-weeks old female mice. Top images, wild-type cardiac muscle sections show a typical intermyofibrillar network containing mitochondria (M), glycogen (*Gly) and lipid droplets (LD). In Atg/KO cardiac muscle (lower panels) lipid droplet size and the number of glycogen granules embedded within the intermyofibrillar network are increased. VE, vessel. Scale bars, 1 μm for upper and lower left images; 0.5 μm for upper and lower right images. (c,d) Morphometric (c) and cytofluorimetric (d) analyses of mitochondria from cardiac muscle of wild-type and Atg/KO mice. Size was either determined from sections of 100 randomly selected mitochondria per genotype or from isolated mitochondria (fluorescence-activated cell sorting (FACS) analysis, n = 4). AU, arbitrary units. (e) Relative mitochondrial DNA (mtDNA) content (normalized to the single-copy nuclear gene Ndufv1) in cardiac muscle of 10-week-old female wild-type and Atg/KO mice (n = 5). Error bars are means ± s.d. **P < 0.01.
We also found that Atgl deficiency prompted severe impairment of cardiac mitochondrial function. We prepared heart homogenates from 4-week-old (Fig. 3a) and 8-week-old (Fig. 3b) AtglKO mice and found drastically reduced oxygen consumption under both basal and succinate-stimulated conditions. We also found that reduced oxygen consumption was irrespective of the presence of glucose (Fig. 3a,b) or palmitate (Supplementary Fig. 2a), thereby suggesting that the substrate energy source did not affect the mitochondrial defect. Cyanide (1 mM) stopped both basal and succinate-stimulated cardiac respiration, excluding non-mitochondrial mechanisms of oxygen consumption (data not shown). We showed that the reduction of mitochondrial oxygen consumption was similar in 4-week-old and 8-week-old AtglKO mice (between −63% and −74%) and did not correlate with cardiac triglyceride content (Fig. 3c), which was 4.6-fold higher in older than in younger AtglKO mice.
Figure 3.

Mitochondrial OXPHOS function and oxidative stress in cardiac muscle of wild-type and Atg/KO mice. (a,b) Oxygen consumption, an indicator for mitochondrial respiration, in Atg/KO cardiac homogenates of 4-week-old (a) and 8-week-old (b) male mice in the presence of glucose (n = 6). (c) Triglyceride (TG) content in cardiac muscle of wild-type and Atg/KO mice. (d,e) Oxygen flux of mitochondria isolated from cardiac tissue of 8- to 9-week-old male wild-type and Atg/KO mice. ADP-driven (state 3) and uncoupled (state U) oxygen flow was measured in the presence of pyruvate (d) and palmitoyl-CoA (e) in subsarcolemmal (SS) and in intramyofibrillar (IMF) mitochondria (n = 6). (f) Western blotting analysis of mitochondrial respiratory chain proteins NDUFA9 of complex I and SDHA of complex II in mitochondrial preparations of Atg/KO mice and wild-type mice. MTCO1, a marker of complex IV, served as loading control. (g) Mitochondrial membrane potential (tetramethyl-rhodaminemethylester perchlorate (TMRM) staining) in isolated cardiac mitochondria of 8- to 9-week-old female Atg/KO compared to wild-type mice (n = 4). (h) Relative concentrations of non-oxidized (free) thiol groups in isolated mitochondria of 8- to 9-week-old female Atg/KO mice compared to wild-type mice (n = 4). Error bars are means ± s.d. *P < 0.05, **P < 0.01 and ***P < 0.001.
Our analysis of respiration in isolated cardiac mitochondria revealed a markedly decreased oxygen flux specifically to the metabolically more flexible subsarcolemmal mitochondria22,23 (Fig. 3d,e). We found that both ADP-driven (state 3) and uncoupled (state U) oxygen flow was decreased in AtglKO subsarcolemmal mitochondria in the presence of pyruvate (−49% and −43%, respectively) and palmitoyl-CoA (−79% and −78%, respectively). The decrease in oxygen flux in the presence of both glycolytic and lipolytic substrates associated with marked derangements in the mitochondrial respiratory chain (Fig. 3f). Furthermore, protein quantities for NDUFA9 (complex I; NADH dehydrogenase [ubiquinone] 1α subcomplex subunit 9) and SDHA (complex II; succinate dehydrogenase complex subunit A) were 75% and 29% lower, respectively, in AtglKO mitochondria than in wild-type mitochondria (Fig. 3f). In contrast, MTCO1 (complex IV; mitochondrial cytochrome c oxidase 1) protein quantity remained unchanged (Fig. 3f). We showed that significantly reduced mito-chondrial membrane potential (−41%) (Fig. 3g), reduction of non-esterified free thiol groups (−51%) (Fig. 3h) and decreased fatty acid oxidation rates (Supplementary Fig. 2b) added to the mitochondrial dysfunction in AtglKO cardiomyocytes. Thus, we suggest that reduced PPAR target gene and PGC-1α and PGC-1β expression in response to Atgl deficiency leads to defective OXPHOS in the hearts of AtglKO mice. This reduced oxidative capacity may contribute to decreased substrate utilization and increased cardiac deposition of neutral lipids and glycogen in the cardiomyocytes of AtglKO mice.
PPAR activation and OXPHOS depend on cardiac-specific ATGL
To examine whether the observed defects in PPAR signaling and substrate oxidation are specific to the absence of cardiac Atgl, we pursued two strategies. First, we analyzed conditional knockout mice lacking Atgl only in muscle by generating a mouse line with a loxP-flanked Atgl gene (referred to hereafter as AtglFLOX mice) and breeding it with transgenic mice expressing the Cre recombinase under the control of the muscle-specific creatine kinase promoter. Muscle-specific recombination generated mice, referred to here-after as muscleAtglKO mice, with extremely low Atgl mRNA levels in cardiac (−95% compared to wild-type mice) (Supplementary Fig. 3a) and skeletal muscle (data not shown). We found that Atgl mRNA levels in other tissues (for example, liver) were comparable to those in wild-type mice (data not shown). Muscle-specific Atgl deletion increased heart weight 1.5-fold (127 ± 5 mg for muscleAtglKO mice versus 84 ± 12 mg for controls, P < 0.001, n = 7) and cardiac triglyceride content 10.7-fold (Fig. 4a). mRNA expression for PPAR-α and PPAR-δ target genes in cardiac muscle of muscleAtglKO mice decreased by 50–80%, and PGC-1α mRNA levels decreased by 85% (Fig. 4b). This cardiac phenotype of muscleAtg1KO mice is indistinguishable from the one observed in conventional AtglKO mice, and it suggests a specific role for cardiac Atgl in PPAR-regulated energy metabolism. Expectedly, and in contrast to AtglKO mice, we found that muscleAtglKO mice had normal hepatic mRNA levels for the PPAR-α and PPAR-δ target genes we studied (Supplementary Fig. 3b).
Figure 4.

Changes in PPAR-α and PPAR-δ activated gene expression and OXPHOS in mice lacking or overexpressing Atgl in cardiac muscle. (a) Cardiac triglyceride content in wild-type and conditional knockout mice lacking Atgl in cardiac and skeletal muscle (muscleAtg/KO mice) demonstrating a drastic cardiac steatosis in muscleAtg/KO mice (n = 5). Scale bars, 5 mm. (b) mRNA expression levels of PPAR-α and PPAR-δ target genes and of the gene encoding PGC-1α in cardiac muscle of muscleAtg/KO mice compared to wild-type mice (n = 5). (c–e) Heart weight (c), cardiac muscle triglyceride (TG) content (d), and white and brown adipose tissue (WAT and BAT) weight (e) of wild-type, Atg/KO and Atg/KO-cmAtg/TG mice expressing an Atgl transgene on an Atg/KO background (n = 6). (f) mRNA expression levels of PPAR-α and PPAR-δ target genes and genes encoding PGC-1α and PGC-1β in cardiac muscle of wild-type, Atg/KO and Atg/KO-cmAtg/TG mice (n = 4). (g) Oxygen consumption in cardiac homogenates prepared from 8- to 9-week-old female wild-type and Atg/KO-cmAtg/TG mice (n = 6). (h) Relative luciferase activities in lysates of HepG2 cells transfected with a PPRE-luciferase reporter plasmid and a PPAR-α expression vector. The additional expression of Atgl increases luciferase activity in the absence or presence of exogenously added linoleic acid (LA). Transfection of the bacterial β-galactosidase gene (lacZ)-containing plasmid and colorimetric determination of β-galactosidase (β-gal) enzyme activity was used for normalization of transfection efficiency. Error bars show means ± s.d. *P < 0.05, **P < 0.01 and ***P < 0.001.
Our second strategy tested whether Atgl in the heart alone is sufficient to rescue the cardiac phenotype of AtglKO mice by generating transgenic mice with cardiomyocyte-specific overexpression of Atgl (hereafter referred to as cmAtglTG mice) (Supplementary Fig. 4a). We subsequently backcrossed cmAtglTG mice onto an AtglKO background, yielding mice with Atgl present exclusively in cardiac muscle (designated AtglKO-cmAtglTG mice). We found that Atgl enzyme activity in cardiac muscle of AtglKO-cmAtglTG mice was five- to sevenfold higher than in wild-type mice (Supplementary Fig. 4b). Cardiac-exclusive Atgl expression entirely rescued the lethal heart phenotype of conventional AtglKO mice and prolonged life span to periods comparable to those in wild-type mice (>2 years) (Supplementary Fig. 4c). Cardiac hypertrophy and massive triglyceride accumulation, characteristics of the Atgl-deficient heart, reverted to normal in AtglKO-cmAtglTG mice (Fig. 4c,d), whereas adipose tissue mass remained high (Fig. 4e). Restoration of Atgl enzyme activities in cardiac muscle caused a substantial increase in PPAR-α and PPAR-δ target gene and PGC-1α and PGC-1β mRNA expression (Fig. 4f), and it increased oxidative capacity to values found in wild-type mice (Fig. 4g).
Together, these results assign a crucial and specific role to cardiac Atgl in the activation of PPAR target gene expression and lipid oxidation. To determine whether these parameters could be attributed to differences in availability of extracellular lipid substrates, we measured plasma concentrations of lipid substrates (fatty acids and triglycerides) and cardiac LPL enzyme activities in all mouse models and correlated these with PPAR target gene expression and cardiac phenotype. We found that fasted AtglFLOX and conditional muscleAtglKO mice showed normal plasma fatty acid, triglyceride and total cholesterol concentrations, similar to those observed in fasted wild-type mice (Supplementary Table 1). In contrast, plasma fatty acid and triglyceride concentrations were much lower in fasted AtglKO (−67% and −66%, respectively) and AtglKO-cmAtglTG mice (−74% and −69%, respectively) than in wild-type mice. Cardiac-specific LPL enzyme activities varied among different genotypes, with the highest values observed in AtglKO and AtglKO-cmAtglTG mice (Supplementary Fig. 5a). In AtglKO mouse hearts, this led to a 53% increase in the total intracellular concentration of unesterified fatty acids (Supplementary Fig. 5b). In summary, these results indicate that both plasma lipid concentrations and cardiac LPL activities did not correlate with PPAR target gene expression or the severity of the cardiac phenotype.
To test whether overexpression of Atgl in an in vitro cell culture system enhances PPAR-α target gene expression, we performed reporter gene transactivation assays. We co-transfected HepG2 cells with a plasmid expressing a luciferase reporter gene under the control of multiple PPAR-responsive elements (PPREs) and an expression vector for PPAR-α. This co-transfection strongly induced luciferase reporter expression compared to cells transfected with only the PPRE-luciferase vector (Fig. 4h). On addition of exogenous linoleic acid (18:2), we found that reporter activity increased further (by 38%) (Fig. 4h), confirming previous reports that fatty acids induce PPAR-α target gene expression24. We found that additional overexpression of Atgl via an Atgl expression plasmid further augmented luciferase activity in the presence or absence of exogenous fatty acids (44% and 61%, respectively) (Fig. 4h). Thus, we suggest that the overexpression of Atgl stimulates PPAR-α–mediated reporter gene transcription in vitro.
PPAR-α agonist restores cardiac respiration and function
Next, we assessed whether the severe lipid and respiratory defects in Atgl-deficient mice can be bypassed by lipolysis-independent PPAR activation via PPAR agonists. We found that feeding 6-week-old female AtglKO mice a chow diet containing 0.1% (wt/wt) of the PPAR-α agonist Wy14643 for 3 weeks decreased circulating triglycerides and fatty acids (Supplementary Table 2) and substantially reduced triglyceride content in heart (−52%) and liver (−50%) compared to AtglKO mice fed a normal chow diet (Fig. 5a). Similarly, feeding AtglKO mice a chow diet containing 0.2% (wt/wt) of the alternative PPAR-α agonist fenofibrate for 10 weeks reduced the cardiac and hepatic triglyceride contents by 62% and 49%, respectively (Fig. 5b). In addition, we found that Wy14643 treatment caused a substantial increase in PPAR-α target gene expression (Fig. 5c). Notably, we found that PGC-1α and β mRNA levels also reverted to wild-type values after Wy14643 treatment (Fig. 5c). Agonist treatment also improved basal and succinate-stimulated respiratory rates of AtglKO hearts to wild-type values (Fig. 5d), normalized free thiol concentrations (Supplementary Fig. 6a), and partially restored mitochondrial size (Supplementary Fig. 6b) and membrane potential (Supplementary Fig. 6c). We found that the beneficial effects of Wy14643 were not caused by an induction of cardiac triglyceride hydrolase activity (Supplementary Fig. 6d).
Figure 5.

Triglyceride content, oxygen consumption and cardiac function in Atg/KO mice treated with PPAR-α agonists. (a) Cardiac and hepatic triglyceride content in 6-week-old female Atg/KO mice on chow diet with or without 0.1% Wy14643 for 3 weeks (n = 5). (b) Cardiac and hepatic triglyceride content in 6-week-old female Atg/KO mice on chow diet with or without 0.2% fenofibrate for 10 weeks (n = 4–5). (c) mRNA expression levels of PPAR-α and PPAR-δ target genes and genes encoding PGC-1α and PGC-1β in cardiac muscle of female wild-type and AtglKO mice fed a chow diet with or without 0.1% Wy14643 for 3 weeks (n = 5). (d) Oxygen consumption in cardiac muscle preparations under both basal conditions and succinate-stimulated conditions of 9-week-old male wild-type, Atg/KO mice and Atg/KO mice fed a chow diet with 0.1% (wt/wt) Wy14643 for 3 weeks (n = 5). (e) Representative echocardiographic images (M- and B-Mode) of a 9-week-old female wild-type and Atg/KO mouse on chow diet and a 9-week-old female Atg/KO mouse fed a chow diet containing 0.1% (wt/wt) Wy14643 for 3 weeks. We measured interventricular septum (IVS) and posterior wall (PW) thickness from original tracings. We measured left ventricular end-systolic dimensions (ESD) and left ventricular end-diastolic dimensions (EDD) from original tracings according to the leading edge convention of the American Society of Echocardiography. (f,g) Left ventricular fractional shortening (LVFS) (f) and left ventricular (LV) mass (g), calculated from the echocardiographic tracings as previously described54 (n = 5). Error bars show means ± s.d. *P < 0.05, **P < 0.01 and ***P < 0.001.
PPAR-α agonist treatment substantially improved heart function in AtglKO mice. In echocardiography (Fig. 5e and Supplementary Table 3), AtglKO mice showed a massive increase in left ventricular mass index. This was largely the result of a thickening of the septal and posterior walls (without substantial change in the left ventricular end-diastolic dimension), thus resulting in a concentric hypertrophic phenotype (Fig. 5e and Supplementary Table 3). Left ventricular systolic function was significantly impaired in AtglKO mice, as indicated by an increase in left ventricular end-systolic dimensions and a robust decrease in left ventricular fractional shortening (Fig. 5e and Supplementary Table 3). A 3-week treatment with the PPAR-α agonist Wy14643 completely reversed this phenotype (Fig. 5e and Supplementary Table 3). We showed that treated AtglKO mice had normal ventricular systolic function and a left ventricular mass index comparable to wild-type animals (Fig. 5f,g). In fact, left ventricular end-diastolic dimensions of treated AtglKO mice were even lower than in wild-type mice (Supplementary Table 3). Thus, we suggest that exogenous PPAR-α stimulation is sufficient to markedly improve mitochondrial function and to fully restore the cardiac performance of AtglKO mice.
Wy14643 prevents cardiac death and improves fatty acid utilization
To investigate whether PPAR-α agonist-mediated improvement of respiratory function and cardiac performance prevents the early death of AtglKO mice, we kept 8-week-old AtglKO males on a chow diet with and without Wy14643 for 12 weeks. Although all vehicle-treated knockout mice died within the first 4 weeks of the treatment period, 4 out of 5 Wy14643-treated knockout mice survived the complete duration of the experiment (Fig. 6a). One animal of the Wy14643 group was bitten to death during the second week of treatment. Notably, we found that on this long-term treatment regimen with the PPAR-α agonist, cardiac muscle and liver triglyceride content in AtglKO mice was equivalent to that seen in wild-type mice (Fig. 6b). Triglyceride content also was low in other Atgl-deficient organs including kidney, testis and ileum (Fig. 6b).
Figure 6.

Life span, tissue triglyceride content and energy substrate utilization in wild-type and Atg/KO mice treated with the PPAR-α agonist Wy14643. (a,b) Treatment of 8-week-old Atg/KO mice on chow diet containing 0.1% WY14643 for 12 weeks prevented cardiac death (a) and lowered tissue triglyceride (TG) content (b), including in cardiac muscle and liver, compared to that observed in wild-type animals (n = 4). (c) Relative whole-body oxygen consumption of 8- to 9-week-old female wild-type and Atg/KO mice housed in metabolic cages (n = 5). (d) Respiratory quotients (calculated from the ratio of carbon dioxide elimination versus oxygen consumption) in Atg/KO mice compared to wild-type during the light period and in the fasted state indicating preferential glucose utilization as oxidative fuel (n = 5). Error bars show means ± s.d. *P < 0.05, **P < 0.01 and ***P < 0.001. (e) Scheme of the integration of Atgl-mediated lipolysis in PPAR signaling. Fatty acids from exogenous or endogenous sources are not available as ligands for nuclear receptor signaling but instead are activated to acyl-CoAs and subsequently oxidized or esterified to triglycerides. Atgl-mediated lipolysis of triglyceride stores preferentially generates ligands or precursors of ligands for nuclear receptors controlling mitochondrial function and OXPHOS. CD36, cluster of differentiation 36; Fatp, fatty acid transport protein; FFA, free fatty acid; TGRLP, triglyceride-rich lipoproteins.
At the same time, the physical performance of AtglKO mice and their ‘metabolic flexibility’ in terms of energy substrate utilization improved after a 7-d treatment with Wy14643. We found that during the dark cycle when the mice are more active, oxygen consumption in metabolic chambers was 12% lower in AtglKO mice than in wild-type mice (Fig. 6c). This difference decreased to 9% when the mice received Wy14643. To test for metabolic flexibility, we assessed mice fed a chow diet with or without Wy14643 by indirect calorimetry (Fig. 6d). During the dark period (when mice eat), we found that values for the respiratory quotient were ∼1.0, suggesting that glucose was the only energy substrate used in both genotypes (Fig. 6d). During the light period (when mice typically decrease their food intake) or during food deprivation 3 h before the experiment, the respiratory quotient values of wild-type animals dropped to 0.89 and 0.80, respectively, indicating a switch from glucose to fatty acid utilization (Fig. 6d). These decreases were less pronounced in AtglKO mice (0.92 in animals during the light cycle and 0.83 after fasting), suggesting that the animals were less efficient in switching the energy substrate from glucose to fatty acids (Fig. 6d). In addition, we found that Wy14643 administration significantly decreased the respiratory quotient values in both wild-type and AtglKO mice (Fig. 6d). However, respiratory quotients remained higher in AtglKO mice than in wild-type mice (Fig. 6d). Thus, we suggest that PPAR agonist treatment increases fatty acid utilization in both genotypes but is unable to fully restore fatty acid oxidation in AtglKO mice to wild-type values. Presumably, this results from the restricted fatty acid availability in fasted AtglKO mice due to lipolytic defects in white adipose tissue.
PPAR-δ agonists do not improve the cardiometabolic defects
In addition to PPAR-α, PPAR-δ has a prominent role in the transcriptional regulation of genes involved in fatty acid uptake and oxidation in many tissues, including cardiac and skeletal muscle25–27. To differentiate between PPAR-α– and PPAR-δ–mediated effects, we investigated whether the administration of the well-established selective PPAR-δ agonists GW501516 (ref. 27) and GW0742 (ref. 28) can also improve the cardiometabolic defects in AtglKO mice. Consistent with previous observations27, we found that daily intraperitoneal administration of GW501516 at 5 μg per g body weight to 7-week-old mice for 2 weeks decreased hepatic triglyceride content in wild-type and AtglKO mice (−37% and −29%, P < 0.08, respectively) (Supplementary Fig. 7a) and increased the expression of PPAR-α and PPAR-δ target genes to wild-type values (Supplementary Fig. 7b). Unexpectedly, however, we found that PPAR-δ agonist treatment did not improve the cardiac phenotype of AtglKO mice (Supplementary Fig. 7c-e). GW501516 increased rather than decreased the cardiac triglyceride content (Supplementary Fig. 7c) in both wild-type (2.2-fold) and AtglKO mice (1.3-fold). PPAR target gene expression (Supplementary Fig. 7d) and cardiac oxygen consumption (Supplementary Fig. 7e) remained unaffected. We found that application of GW501516 by oral gavage (5 μg per g body weight) twice a day for 4 d yielded similar results (Fig. 7f–h). Whereas we found that hepatic mRNA levels for PPAR-δ targets in AtglKO mice increased to normal (Supplementary Fig. 7f), cardiac PPAR-δ target gene expression (Supplementary Fig. 7g) and triglyceride content (Supplementary Fig. 7h) remained abnormal. We also found that cardiac PGC-1α and PGC-1β mRNA concentrations slightly increased in wild-type and AtglKO mice in response to GW501516; however, in treated AtglKO mice they remained 60% below those observed in untreated wild-type mice (Supplementary Fig. 7i). In contrast, hepatic PGC-1α and PGC-1β mRNA levels were consistently higher in AtglKO than in wild-type mice independently of PPAR-δ agonist treatment (Fig. 1d and Supplementary Fig. 7j). In addition, oral administration of the alternative PPAR-δ agonist GW0742 also had no effect on cardiac PPAR target gene expression, tissue triglyceride content, plasma lipid parameters or body weight (data not shown). Thus, treatment with the PPAR-δ agonists GW501516 and GW0742 did not rescue metabolic and oxidative defects in the cardiac muscle of AtglKO mice.
Discussion
This study shows that the activation of cardiac and hepatic gene transcription by the PPAR-α–PGC-1 complex depends on the lipolytic catabolism of cellular triglyceride depots. This process requires the hydrolytic activity of Atgl. The absence of this enzyme in cardiac muscle of conventional AtglKO mice causes a lipolytic defect that results in massive lipid accumulation, drastic reduction of PPAR-α–regulated gene expression, defective mitochondrial substrate oxidation, severe cardiac dysfunction and premature death. A similar phenotype is observed in conditional knockout mice lacking Atgl only in cardiac and skeletal muscle (muscleAtglKO), indicating that the defects are specific to the absence of Atgl in cardiomyocytes. Concurrent analysis of the liver of AtglKO mice confirmed previous observations that showed that Atgl regulates triglyceride turnover and the expression of PPAR-α target genes29–31. In humans, ATGL deficiency provokes a similar clinical phenotype; affected individuals develop NLSD with severe cardiomyopathy that requires cardiac transplantation in a subgroup of patients. The pronounced phenotype with respect to energy homeostasis and cardiac function is specific for Atgl deficiency because Hsl or other potential lipid hydrolases cannot compensate for the absence of this enzyme, and also because all metabolic and functional derangements in the heart can be corrected by the cardiac-specific expression of an Atgl transgene in mice that lack Atgl in all other tissues. Thus, the Atgl-mediated generation of lipolytic products is an indispensable requirement for the regulation of PPAR-α target genes and represents a previously undescribed mechanism for gene regulation. It supports the concept that lipid droplets are not only cellular compartments for energy storage but also serve as reservoirs for specific lipid mediators, which require lipolysis for their spatio-temporally coordinated release and subsequent action.
Fatty acids as potential ligands for PPAR-α activity originate from two major sources. Endogenously synthesized fatty acids (that is, ‘new fat’)14 predominantly regulate hepatic PPAR-α activity. In contrast, exogenous fatty acids (that is, ‘old fat’)32 derived from adipose tissue (fatty acid–albumin complexes) or from the hydrolysis of hepatic very-low-density lipoprotein particles via LPL regulate cardiac PPAR-α activity. Changes in liver-specific fatty acid synthase or cardiac-specific LPL activities in mutant mouse models strongly affect energy substrate usage, PPAR signaling and cellular lipid homeostasis14,32–34, proving the crucial role of these enzymes in the delivery of PPAR ligands. Our data add an additional aspect to these findings: functional PPAR signaling requires Atgl-catalyzed hydrolysis of intracellular triglyceride stores generated from endogenous or exogenous fatty acids. Apparently, fatty acids must first be esterified to triglycerides and rehydrolyzed by Atgl before they become active signaling lipids. Ligand delivery via LPL and fatty acid uptake is indispensable but by itself not sufficient for normal PPAR signaling and cardiometabolic function. Consistent with this conclusion, conventional AtglKO mice (with decreased plasma fatty acid and increased cardiac LPL activity) and conditional muscle Atgl KO mice (with normal plasma fatty acid and decreased cardiac LPL activity) showed impaired PPAR target gene expression and lethal cardiomyopathy, whereas AtglKO-cmAtglTG mice (with decreased plasma fatty acid and increased cardiac LPL activity) showed improved target gene expression and no apparent cardiomyopathy. Whether enhanced ligand delivery to the heart by the tissue-specific overexpression of LPL33 or the fatty acid transporter CD36 (ref. 35) can overcome the cardiometabolic phenotype associated with Atgl deficiency remains to be assessed.
Our current concept can be summarized as follows: exogenous fatty acids from hydrolyzed fatty acid–albumin complexes or triglyceride-rich lipoproteins enter the cell and are coenzyme-A activated by cell membrane–bound acyl-CoA synthetases and/or fatty acid transport proteins (Fig. 6e). Depending on energy demand, these fatty acid–CoAs either enter mitochondria for oxidation or are re-esterified and stored in lipid droplet triglycerides. The generation of lipolytic products such as fatty acids or diglycerides by Atgl provides ligands or ligand precursors for functional signaling by the PPAR-α–PGC-1 complex, which, in turn, activates mitochondrial biogenesis and OXPHOS and directs fatty acid–CoAs to oxidation. Conversely, downregulation or absence of Atgl leads to decreased mitochondrial biogenesis and function and directs fatty acid–CoAs toward triglyceride synthesis and accumulation.
This study did not address the molecular nature of mediators that regulate the PPAR-α–PGC-1 complex after lipolysis. Accordingly, any considerations of why lipolysis is required for the production of these mediators remain speculative. The molecular identity of PPAR ligands and the mechanisms of their transduction from the cytoplasma to the nucleus are still a matter of intense debate. A widely accepted model involves the binding of mono- and polyunsaturated fatty acids to cytoplasmic fatty acid–binding proteins (FABPs), translocation into the nucleus and binding to PPAR-R×R heterodimers, thereby accomplishing transcriptional activation24,36. Alternatively, it has been proposed that fatty acid or diglyceride derivatives such as prostaglandins J2 (refs. 9,11) or 1-palmitoyl-2-oleyl-sn-glycerol-3-phosphocholine12, respectively, act as PPAR ligands. Atgl-mediated hydrolysis of triglycerides is compatible with both mechanisms because it generates unesterified fatty acids and diglyceride as potential lipid ligands or ligand precursors. Finally, it is even conceivable that components of lipid droplets, such as lipid droplet-associated proteins, dissociate during lipolysis and regulate PPAR signaling.
Atgl deficiency results in decreased expression of PGC-1α and PGC-1β in cardiac muscle. Consistent with the established roles of PGC-1α and PGC-1β in cardiac mitochondrial biogenesis, substrate oxidation and prevention of oxidative stress35,37, low cardiac PGC-1 concentrations may contribute to decreased numbers of mitochondria, decreased amounts of respiratory complex I and II proteins, accumulation of reactive oxygen species and impaired OXPHOS in cardiac muscle of AtglKO mice. Decreased mitochondrial oxygen consumption was specific to subsarcolemmal mitochondria, which are highly sensitive to cellular PGC-1α concentrations22,23,38.
The molecular mechanism whereby defective lipolysis suppresses cardiac PGC-1α and PGC-1β expression is currently unknown. Multiple physiological stimuli, including fatty acids, regulate PGC-1α expression and activity via numerous signaling pathways and post-translational modifications7,39–41. A plausible mechanism suggests a direct regulation of PGC-1α by lipolysis-dependent PPAR-α activation. A previous study42 characterized a functional PPRE within the PGC-1α promoter that is regulated by PPAR-α agonists, whereas another43 showed that the upregulation of mitochondrial biogenesis and function in a mouse model with insulin resistance depends on the PPAR-α–mediated upregulation of PGC-1α. Our data are consistent with these observations and suggest that lipolysis regulates PGC-1α via PPAR-α signaling. Therefore, the cardiac phenotype we found in AtglKO was more severe than in mouse lines with individual deficiencies for PGC-1α or PGC-1β, but less severe than that observed in a mouse model with combined PGC-1α and PGC-1β deficiencies7,44. The latter animals die within 24 h after birth from cardiac mitochondrial dysfunction and heart failure44. Whether alternative mechanisms involving additional nuclear receptors (for example, the estrogen-related receptor or the retinoid-related orphan receptors) or non-nuclear receptors, such as the nuclear respiratory factors, also contribute to decreased cardiac PGC-1 expression requires clarification. Notably, our finding that PGC-1α and PGC-1β expression in the liver of AtglKO mice was higher than in wild-type mice despite decreased PPAR-α and PPAR-δ target gene expression suggests basic tissue-specific differences in the regulation of PGC-1 expression in the heart and the liver that remain to be elucidated.
Treatment of AtglKO mice with PPAR-α agonists bypassed the lipolytic defect and prevented the adverse metabolic alterations and oxidative dysfunction in the heart. Wy14643 treatment of mice was previously shown to stimulate PPAR-α–activated gene expression and mitochondrial oxidation, thereby reducing cellular triglyceride content in cardiac muscle and liver45–47. In AtglKO mice, Wy14643 treatment restored the cardiac expression of PPAR-α target genes and PGC-1α and PGC-1β, reversed excessive systemic lipid accumulation, normalized oxidative function and oxidative stress in mitochondria, improved cardiac performance and prevented lethal cardiomyopathy. The normalization of cardiac steatosis in response to the PPAR-α agonist occurred without increasing cellular triglyceride hydrolysis, thus arguing against an induction of alternative lipase(s). The reduction in triglyceride content may be mostly secondary to increased oxidation of exogenous fatty acids and altered kinetics of the intramyocellular triglyceride pool.
In contrast, we show that PPAR-δ agonist treatment of AtglKO mice via intraperitoneal injection or oral gavage neither reduces cardiac lipid content nor normalized PGC-1α and PGC-1β and PPAR target gene expression. This was unexpected as PPAR-δ activation is associated with increased fatty acid oxidation in muscle and liver27,48. PPAR-δ agonist effects on cardiac triglyceride content have not been reported, but mice lacking PPAR-δ suffer from mild cardiac steatosis (normal cardiac triglycerides in 2-month-old mice and a twofold increase when they are 9 months old) and decreased OXPHOS in cardiac muscle25. Our results indicate that defective PPAR-α activation and decreased PGC-1α and PGC-1δ expression account for most of the cardiometabolic derangements observed in Atgl-deficient mice and that PPAR-δ activation alone is not sufficient to overcome these defects in AtglKO mice. In the liver, administration of PPAR-δ agonists normalized PPAR target gene expression. The agonist-mediated induction of target mRNAs was independent of PGC-1α and PGC-1β expression, which was consistently higher in the liver of AtglKO mice than in wild-type mice. These findings suggest remarkable tissue-specific differences in cardiac muscle and liver with respect to the regulation of the PPAR–PGC-1 complex by lipolysis.
The possibility that the pharmacological activation of PPAR-α can bypass the ATGL pathway offers a potential treatment for people with NLSD. Two distinct genetic defects are known to cause NLSD49: Mutations in ATGL cause NLSD with myopathy (NLSDM), whereas mutations in the ATGL activator CGI-58 cause NLSD with ichthyosis (NLSDI). Both conditions cause ectopic systemic lipid storage, although not to the same extent50. People with NLSDM accumulate large amounts of triglycerides, particularly in cardiac and skeletal muscle, and they develop only mild hepatosteatosis. People with NLSDI suffer from more severe hepatosteatosis and accrue fewer triglycerides in muscle. If our results in AtglKO mice also apply to humans, treatment with PPAR-α agonists should reduce ectopic lipid accumulation and improve organ dysfunction in these individuals. Fenofibrate, an approved and well-established activator of PPAR-α (ref. 51), may be particularly beneficial for the treatment of liver steatosis in NLSDI. Currently, there is no clinical evidence that PPAR-α agonists also affect cardiac lipid metabolism and heart function in humans (unlike in the mouse52). Instead, human skeletal muscle and cardiac energy metabolism may rely more on the activity of PPAR-δ (refs. 3,53). Accordingly, administration of recently available PPAR-δ or nonselective PPAR agonists may be an effective measure to combat excessive skeletal and cardiac lipid deposition and heart failure in people with NLSDM.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
Acknowledgments
This research was supported by the following grants: GOLD - Genomics of Lipid-Associated Disorders as part of the Austrian Genome Project (GEN-AU) funded by the Forschungsförderungsgesellschaft (FFG) and the Bundesministerium für Wissenschaft und Forschung (BMWF); Spezialforschungsbereich (SFB) LIPOTOX F30, Doktoratskolleg: Molecular Enzymology W901, the Wittgenstein Award 2007 Z136 and research grant P20602 funded by the Austrian Science Fund (FWF); Targeting Obesity-driven Inflammation (TOBI) contract no. 201608 and LipidomicNet contract no. 202272 funded by the European Commission. Additional funding for the SFB LIPOTOX was granted by the County of Styria and the City of Graz. P.S. is supported by the Research Grant for Innovative Research from the Netherlands Organization for Scientific Research (Grant 918.96.618). T.v.d.W. was supported by the Center for Translational Molecular Medicine (CTMM) project PREDICCt (Grant 01C-104) and the Netherlands Heart Foundation, the Dutch Diabetes Research Foundation and the Dutch Kidney Foundation. We thank E. Zechner and C. Schober-Trummler for proofreading the manuscript and S. Lang for the preparation of the cartoon. The pBS II SK+ vector containing the α-MHC promoter region (Genbank: U71441) was provided by J. Robbins (University of Cincinnati). The expression plasmids for (PPRE)6-tk-luciferase and human PPAR-α were provided by B. Staels (University of Lille).
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
Author Contributions: G.H. and R.Z. designed the study, were involved in all aspects of the experiments and wrote the manuscript. T.M. and D.J. were responsible for quantitative RT-qPCR–based gene expression analyses and luciferase assays. G.W. and B.M. were responsible for the measurements of tissue oxygen consumption. P. K., D.K. and S.C. were responsible for electron microscopy. S.B., F.M., N.W., T.v.d.W., M.H. and P.S. were responsible for mitochondrial analyses. P.C.K., T.K. and T.R. generated the transgenic mouse strains. K.Z., F.P.W.R., R.S., T.E., M.S., M.K., S.E., G.S. and N.M.P. were responsible for agonist application, dietary studies, plasma and tissue parameter analyses and enzymatic assays. A.S. and B.P. were responsible for echocardiography. E.E.K. generated Atgl-floxed mice. K.P.-L., M.T., A.L., R.Z. and G.H. discussed the results and commented on the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR-α in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barish GD, Narkar VA, Evans RM. PPAR-δ: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPAR-γ. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 5.Lehman JJ, et al. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 7.Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kliewer SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krey G, et al. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 11.Yu K, et al. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarthy MV, et al. Identification of a physiologically relevant endogenous ligand for PPAR-α in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziouzenkova O, et al. Lipolysis of triglyceride-rich lipoproteins generates PPAR ligands: evidence for an antiinflammatory role for lipoprotein lipase. Proc Natl Acad Sci USA. 2003;100:2730–2735. doi: 10.1073/pnas.0538015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarthy MV, et al. “New” hepatic fat activates PPAR-α to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Osuga J, et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci USA. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haemmerle G, et al. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 18.Pinent M, et al. Differential transcriptional modulation of biological processes in adipocyte triglyceride lipase and hormone-sensitive lipase-deficient mice. Genomics. 2008;92:26–32. doi: 10.1016/j.ygeno.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Haemmerle G, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 20.Fischer J, et al. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 21.Hirano K, Ikeda Y, Zaima N, Sakata Y, Matsumiya G. Triglyceride deposit cardiomyovasculopathy. N Engl J Med. 2008;359:2396–2398. doi: 10.1056/NEJMc0805305. [DOI] [PubMed] [Google Scholar]
- 22.Benton CR, et al. Modest PGC-1α overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem. 2008;283:4228–4240. doi: 10.1074/jbc.M704332200. [DOI] [PubMed] [Google Scholar]
- 23.Holloway GP, Gurd BJ, Snook LA, Lally J, Bonen A. Compensatory increases in nuclear PGC-1α protein are primarily associated with subsarcolemmal mitochondrial adaptations in ZDF rats. Diabetes. 2010;59:819–828. doi: 10.2337/db09-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfrum C, Borrmann CM, Borchers T, Spener F. Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors alpha- and gamma-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus. Proc Natl Acad Sci USA. 2001;98:2323–2328. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng L, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-δ deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–1250. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 26.Lee CH, et al. PPAR-δ regulates glucose metabolism and insulin sensitivity. Proc Natl Acad Sci USA. 2006;103:3444–3449. doi: 10.1073/pnas.0511253103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka T, et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci USA. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sznaidman ML, et al. Novel selective small molecule agonists for peroxisome proliferator-activated receptor delta (PPAR-δ)–synthesis and biological activity. Bioorg Med Chem Lett. 2003;13:1517–1521. doi: 10.1016/s0960-894x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 29.Reid BN, et al. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J Biol Chem. 2008;283:13087–13099. doi: 10.1074/jbc.M800533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 2011;53:1116–1126. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sapiro JM, Mashek MT, Greenberg AS, Mashek DG. Hepatic triacylglycerol hydrolysis regulates peroxisome proliferator-activated receptor alpha activity. J Lipid Res. 2009;50:1621–1629. doi: 10.1194/jlr.M800614-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Augustus A, et al. Cardiac-specific knock-out of lipoprotein lipase alters plasma lipoprotein triglyceride metabolism and cardiac gene expression. J Biol Chem. 2004;279:25050–25057. doi: 10.1074/jbc.M401028200. [DOI] [PubMed] [Google Scholar]
- 33.Yagyu H, et al. Lipoprotein lipase (LpL) on the surface of cardiomyocytes increases lipid uptake and produces a cardiomyopathy. J Clin Invest. 2003;111:419–426. doi: 10.1172/JCI16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan JG, et al. Rescue of cardiomyopathy in peroxisome proliferator-activated receptor-alpha transgenic mice by deletion of lipoprotein lipase identifies sources of cardiac lipids and peroxisome proliferator-activated receptor-α activators. Circulation. 2010;121:426–435. doi: 10.1161/CIRCULATIONAHA.109.888735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Tan NS, et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valle I, Alvarez-Barrientos A, Arza E, Lamas S, Monsalve M. PGC-1α regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 38.Hood DA. Invited Review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90:1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- 39.Sugden MC, Caton PW, Holness MJ. PPAR control: it's SIRTainly as easy as PGC. J Endocrinol. 2010;204:93–104. doi: 10.1677/JOE-09-0359. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 42.Hondares E, et al. Thiazolidinediones and rexinoids induce peroxisome proliferator-activated receptor-coactivator (PGC)-1α gene transcription: an autoregulatory loop controls PGC-1α expression in adipocytes via peroxisome proliferator-activated receptor-γ coactivation. Endocrinology. 2006;147:2829–2838. doi: 10.1210/en.2006-0070. [DOI] [PubMed] [Google Scholar]
- 43.Duncan JG, Fong JL, Medeiros DM, Finck BN, Kelly DP. Insulin-resistant heart exhibits a mitochondrial biogenic response driven by the peroxisome proliferator-activated receptor-α/PGC-1α gene regulatory pathway. Circulation. 2007;115:909–917. doi: 10.1161/CIRCULATIONAHA.106.662296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai L, et al. Transcriptional coactivators PGC-1α and PGC-lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoyama T, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARα) J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 46.Chou CJ, et al. WY14,643, a peroxisome proliferator-activated receptor alpha (PPARα) agonist, improves hepatic and muscle steatosis and reverses insulin resistance in lipoatrophic A-ZIP/F-1 mice. J Biol Chem. 2002;277:24484–24489. doi: 10.1074/jbc.M202449200. [DOI] [PubMed] [Google Scholar]
- 47.Ye JM, et al. Peroxisome proliferator-activated receptor (PPAR)-α activation lowers muscle lipids and improves insulin sensitivity in high fat-fed rats: comparison with PPAR-γ activation. Diabetes. 2001;50:411–417. doi: 10.2337/diabetes.50.2.411. [DOI] [PubMed] [Google Scholar]
- 48.Cheng L, et al. Peroxisome proliferator-activated receptor gamma activates fatty acid oxidation in cultured neonatal and adult cardiomyocytes. Biochem Biophys Res Commun. 2004;313:277–286. doi: 10.1016/j.bbrc.2003.11.127. [DOI] [PubMed] [Google Scholar]
- 49.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 50.Igal RA, Rhoads JM, Coleman RA. Neutral lipid storage disease with fatty liver and cholestasis. J Pediatr Gastroenterol Nutr. 1997;25:541–547. doi: 10.1097/00005176-199711000-00011. [DOI] [PubMed] [Google Scholar]
- 51.Rosenson RS. Fenofibrate: treatment of hyperlipidemia and beyond. Expert Rev Cardiovasc Ther. 2008;6:1319–1330. doi: 10.1586/14779072.6.10.1319. [DOI] [PubMed] [Google Scholar]
- 52.Vikramadithyan RK, et al. Peroxisome proliferator-activated receptor agonists modulate heart function in transgenic mice with lipotoxic cardiomyopathy. J Pharmacol Exp Ther. 2005;313:586–593. doi: 10.1124/jpet.104.080259. [DOI] [PubMed] [Google Scholar]
- 53.Wang YX, et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 54.Song Q, et al. Rescue of cardiomyocyte dysfunction by phospholamban ablation does not prevent ventricular failure in genetic hypertrophy. J Clin Invest. 2003;111:859–867. doi: 10.1172/JCI16738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.