Abstract
Kinesin-5 proteins are essential for formation of a bipolar mitotic spindle in most, and perhaps all, eukaryotic cells. Several Kinesin-5 proteins, notably the human version, HsEg5, are targets of a constantly expanding group of small-molecule inhibitors, which hold promise both as tools to probe mechanochemical transduction and as anti-cancer agents. Although most such compounds are selective for HsEg5 and closely related Kinesin-5 proteins, some, such as NSC 622124, exhibit activity against at least one kinesin from outside the Kinesin-5 family. Here we show NSC 622124, despite identification in a screen that yielded inhibitors now known to target the HsEg5 monastrol-binding site, does not compete with 14C-monastrol for binding to HsEg5, and is able to inhibit the basal and microtubule-stimulated ATPase activity of the monastrol-insensitive Kinesin-5, KLP61F. NSC 622124 competes with microtubules, but not ATP, for interaction with HsEg5, and disrupts the microtubule binding of HsEg5, KLP61F and Kinesin-1. Proteolytic degradation of an HsEg5•NSC622124 complex revealed that segments of the α3 and α5 helices map to the inhibitor-binding site. Overall, our results demonstrate that NSC 622124 targets the conserved microtubule-binding site of kinesin proteins. Further, unlike compounds previously reported to target the kinesin microtubule-binding site, NSC 622124 does not produce any enhancement of basal ATPase activity, and thus acts solely as a negative regulator through interaction with a site traditionally viewed as a binding region for positive regulators (i.e., microtubules). Our work emphasizes the concept that microtubule-dependent motor proteins may be controlled at multiple sites by both positive and negative effectors.
Kinesin-5 motor proteins act to separate the spindle poles during formation of the bipolar mitotic spindle [reviewed in (1, 2)]. Representatives of this family have been identified throughout eukaryotes and may be ubiquitous. Certain Kinesin-5 family members, e.g., the human Eg5 protein (HsEg5), represent targets of an ever-expanding collection of chemically diverse, small-molecule inhibitors (3–10). The mechanism of HsEg5 inhibition, as well as the search for more potent inhibitors, is of particular interest since HsEg5 inactivation leads to cell cycle arrest, and thus inhibitors of this motor have potential as anti-cancer drugs (9, 11, 12).
Monastrol, the first recognized HsEg5 inhibitor, was so named because treatment of cultured vertebrate cells and cell extracts led to failure of spindle pole separation and subsequent formation of a monoastral spindle. Recent characterization of the monastrol:protein interaction, mostly with HsEg5, has demonstrated that monastrol binding allosterically inhibits the motor’s basal and microtubule (MT)-stimulated ATPase activities, and consequently productive mechanochemical transduction (3, 13–19). The monastrol-binding site is 12 Å from the nucleotide-binding site and is formed by elements of helix α2, insertion loop L5, and helix α3 (20). Recent characterization of other HsEg5 inhibitors suggests the L5 loop and structurally adjacent regions represent a “hot spot” that serves as a common binding site and thus modulates allosteric inhibition for many different compounds (5, 7, 10, 21, 22).
The vast majority of HsEg5 inhibitors, including monastrol, are highly specific for Kinesin-5 proteins from higher eukaryotes, and have little or no effect on many non-vertebrate Kinesin-5 motors or members of the other thirteen kinesin families. However, one recently identified inhibitor, the polyoxometalate NSC 622124 (K6Mo18O62P2), has been reported to inhibit Ncd (4), a member of the Kinesin-14 family. Since Ncd does not contain a well-defined monastrol-binding pocket (23), NSC 622124 may instead target a conserved site present in both HsEg5 and Ncd. The present study investigates the interactions between NSC 622124 and kinesin proteins in order to clarify this compound’s mechanism of action.
Materials and Methods
Reagents
14C-monastrol (specific activity: 50 mCi/mmol) was synthesized from ethyl acetoacetate, 3-hydroxybenzaldehyde and 14C-thiourea by the procedure of Kappe et al. (24). This high-yield condensation reaction of ethyl acetoacetate, 3-hydroxybenzaldehyde and 14C-thiourea (American Radiolabeled Chemicals, Inc.) resulted in radiolabeled monastrol in racemic form. HPLC analysis and UV-vis spectroscopy were employed to isolate a single chemical entity in high yield and to confirm the identity of the compound, respectively. NSC 59349, NSC 169676, and NSC 622124 were obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute. S-trityl-L-cysteine (STLC) and flexeril were obtained from Sigma-Aldrich. Inhibitors were prepared in DMSO as 50 mM solutions, with the exceptions of monastrol (100 mM in DMSO), 14C-monastrol (10 mM in DMSO), and flexeril (50 mM in H2O).
Protein Expression and Purification
The HsEg5 motor domain, composed of HsEg5 residues 1-370 and a C-terminal 6-His tag, was expressed as previously described (23). A cDNA encoding residues 1-367 of D. melanogaster KLP61F was amplified from clone LD15641 (Berkeley Drosophila Genome Project) by PCR using Pfu polymerase (Stratagene), a forward primer containing an NdeI site, and a reverse primer containing an XhoI site. The product was digested with NdeI and XhoI and inserted into pET-21a (Novagen) digested with the same restriction enzymes. Both strands of the insert were sequenced to confirm that no mutations occurred during amplification. Plasmids were transformed into BL21 Codon-plus (DE3)-RIL cells (Stratagene) for protein expression.
Overnight cultures of cells containing HsEg5 or KLP61F plasmids were diluted 1:100 into LB media supplemented with 100 µg/ml ampicillin and grown at 37°C for 2.5 hours. Protein expression was induced with 0.2 mM IPTG, and after 4 hours at room temperature, cells were pelleted, washed once with 25 mM PIPES pH 6.9, 0.25 mM MgSO4, 0.5 mM EGTA, and frozen at −80°C until use. Frozen cells were thawed in 50 mM HEPES (pH 7.5), 75 mM NaCl, 1 mM PMSF, 0.1 mM MgATP, 40 µg/mL DNAse, 0.3 mg/ml lysozyme, 10 mM MgCl2, and 1 mM DTT, and passed through a French Press three times to ensure adequate lysis. Cell lysates were then centrifuged at 13,500 × g for 30 minutes at 4°C, and the resulting supernatant was passed over a 5 ml S-Sepharose resin. After a wash with 50 mM HEPES (pH 7.5), 0.1 mM MgATP, and 1 mM DTT, the protein was eluted with 50 mM HEPES (pH 7.5), 0.2 mM MgATP, 1 mM DTT and 250 mM NaCl. The protein-containing eluate was immediately mixed with an equal volume of 50 mM HEPES (pH 7.5), supplemented with glycerol (to 10%), frozen on dry ice, and stored at −80°C until use. Protein concentration was measured by Bradford assay (Biorad) with BSA as the standard.
Full length D. melanogaster Kinesin-1 was expressed and bacterial cells were lysed and centrifuged as described for HsEg5 and KLP61F. The supernatant was then centrifuged at 100,000×g for 15 min at 4°C (25), and the resulting high speed supernatant was used directly in MT motility experiments.
14C-Monastrol Binding and Competition Experiments
Size exclusion spin columns were prepared with fine grade G25 Sephadex and Micro Bio-Spin Chromatography columns (Biorad). Sephadex was prepared per manufacturer’s instructions, exchanged into 20 mM HEPES, pH 7.2, 1 mM EDTA, and 1 mM MgCl2, and added to each column to generate a packed resin bed of 0.7 ml. Just prior to use, columns were centrifuged (1500 × g, 4 min) to remove excess liquid. To evaluate binding of 14C-monastrol to motor, 130 µl reactions containing 1 mg/ml (~24 µM) motor protein and 14C-monastrol (0.9 mM unless otherwise indicated) were prepared in HEM buffer, incubated at room temperature for 10 min, then 50 µl was applied to each of two spin columns. Columns were immediately centrifuged (1500 × g, 4 min) to separate protein with bound 14C-monastrol from unbound 14C-monastrol. Samples of the initial reaction as well as the spin column “flow-through” were analyzed by Bradford assay and liquid scintillation counting to quantify protein and 14C-monastrol, respectively. Unless otherwise indicated, binding reactions contained 29–55 mM NaCl and 24–45 µM MgATP carried over from the protein stock solution. To correct for 14C-monastrol that passed through the column in the absence of protein, duplicate reactions were prepared without motor and processed identically. Typically, < 0.24% of 14C-monastrol added to a given reaction passed through the spin column in the absence of motor protein. For competition experiments, motor was incubated with 0.5 mM inhibitor for 20 min at room temperature prior to addition of 0.9 mM 14C-monastrol, then subjected to size exclusion spin chromatography after another 10 min at room temperature. Statistical analyses (t-tests) were performed using Prism 4 software (GraphPad).
ATPase Assays
All assays were conducted at room temperature in 50 mM Tris-acetate, pH 7.4, 2 mM MgCl2. Control reactions were supplemented with DMSO to match the concentration of DMSO carried over with inhibitors.
KLP61F steady-state basal and MT-stimulated ATPase rates (Figure 2) were measured with a coupled pyruvate kinase / lactate dehydrogenase assay (26, 27) and normalized to 100% of the control rate (no inhibitor). Basal ATPase reactions contained 5 µM motor, while MT-stimulated ATPase reactions contained 200 nM motor, 20 µM paclitaxel (Calbiochem) and GTP-depleted, paclitaxel-stabilized MTs (2.3 µM bovine or bison tubulin). Inhibitor concentrations were either 200 µM (monastrol) or 100 µM (all others) in basal assay. To maintain the inhibitor-to-protein ratio in the basal assays, 4 µM inhibitor concentration was used in MT-stimulated reactions.
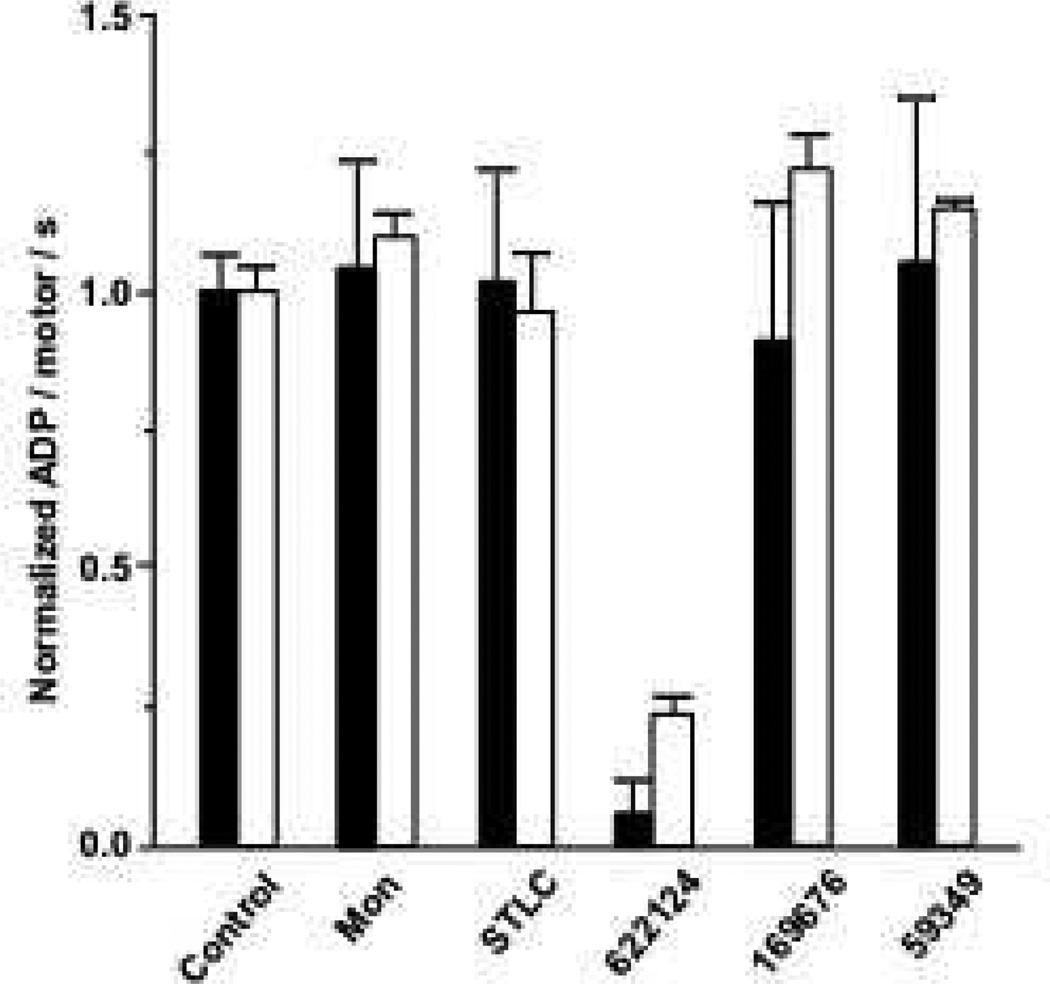
Figure 2. NSC 622124 Inhibits KLP61F Basal and MT-Stimulated ATPase Activities.
Basal (open bars) and MT-stimulated (solid bars) ATPase rates were determined for the indicated inhibitors and normalized to the appropriate control (no inhibitor) mean rate. Basal ATPase reactions contained 5 µM KLP61F and the control basal rate was 0.047 ± 0.002 ADP/motor/s. Inhibitor concentrations in the basal reactions were either 200 µM (monastrol) or 100 µM (all other inhibitors). MT-stimulated ATPase reactions contained 200 nM KLP61F and the control MT-stimulated rate was 0.63 ± 0.04 ADP/motor/s. The concentration of inhibitor was 4 µM in the MT-stimulated reactions. Each bar represents the mean ± SEM of 3-7 reactions except control basal (n=10) and MT-stimulated (n=14) values.
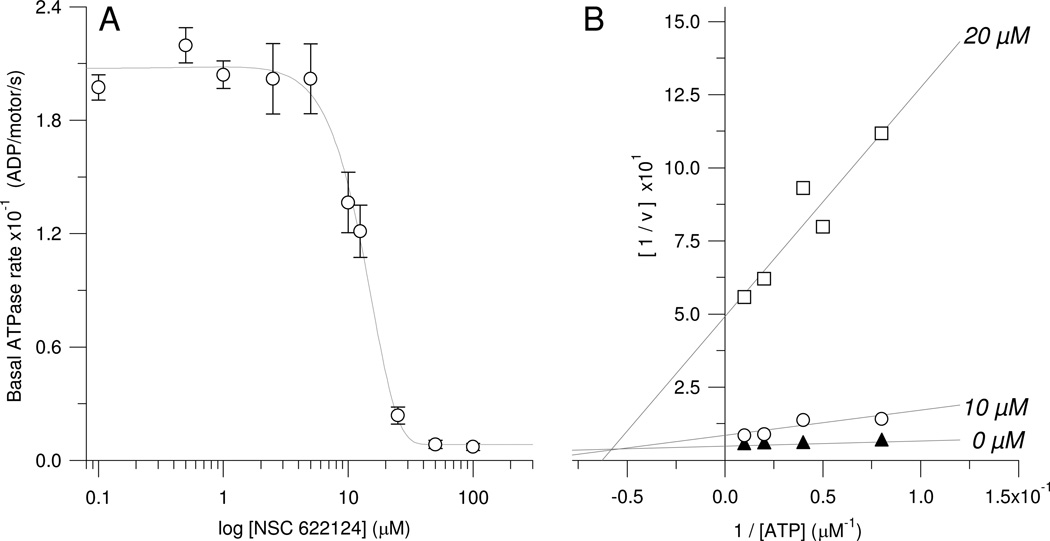
Determination of the HsEg5 basal IC50 (Figure 3A) also utilized coupled assays in which the activity of 2.5 µM HsEg5 was measured with varying NSC 622124 concentrations (0. 0.1, 0.5, 1, 2.5, 5, 10, 12.5, 25, 50, and 100 µM). Data was collected on a SpectraMax2E spectrometer (Molecular Devices). To determine the mode of basal inhibition by NSC 622124 (Figure 3B), HsEg5 activity was observed with varying NSC 622124 concentrations (0, 10, and 20 µM) and MgATP concentrations (12.5, 20, 25, 50 and 100 µM). A Lineweaver-Burk plot was graphed in Igor Pro (Wavemetrics Inc.). The x-axis intercept represents a value equal to - 1/Km. The x-coordinate and y-coordinate of the intersection from the three fitted lines, corresponding to the three concentrations of inhibitor, denotes the value of -1/αKm and 1/Vmax, respectively.
Figure 3. NSC 622124 is a Micromolar Inhibitor of HsEg5 Basal ATPase Activity and Exhibits Mixed Inhibition with MgATP in Binding to HsEg5.
(A) Inhibition of ATPase activity was tracked via NADH coupled assays containing 2.5 µM Eg5 motor protein in the presence of 0, 0.1, 0.5, 1.0, 2.5, 5.0, 10.0, 12.5, 25.0, 50.0, and 100.0 µM NSC 622124. Each data point represents the mean ± SEM of 3–4 replicates. The IC50 is 13 µM. (B) ATPase activity was measured with an array of NSC 622124 concentrations (0, 10, and 20 µM) and MgATP concentrations (12.5, 20, 25, 50 and 100 µM). Results were graphed into a Lineweaver-Burk plot. Each data point represents the average of 3–4 replicates. SEM values averaged 8% of the means (range of 0.6% to 14%, with the higher percentage corresponding to the lowest measurable rates due to inhibition).
Competition assays between NSC 622124 and MgATP or MTs for HsEg5 (Figures 4B and 4C) were measured via a malachite green ATPase assay. Briefly, 50 µl reactions containing 100 nM motor protein, 20 µM paclitaxel, GTP-depleted paclitaxel-stabilized MTs, and indicated NSC 622124 concentrations were initiated by the addition of MgATP. Aliquots (5, 10 or 15 µl) removed at 2, 3, 4 and/or 5 min were added immediately to dilute malachite green reagent (BioAssay Systems) in 96-well plates. Time-zero points were obtained by addition of MgATP after dilution of sample aliquots with malachite green reagent. After 15–30 min at room temperature, the A650 values of samples and Pi standards were measured with either a SpectraFluor Plus (Tecan) or a SpectraMax 190 (Molecular Devices) microplate reader, and rate of Pi production was calculated.
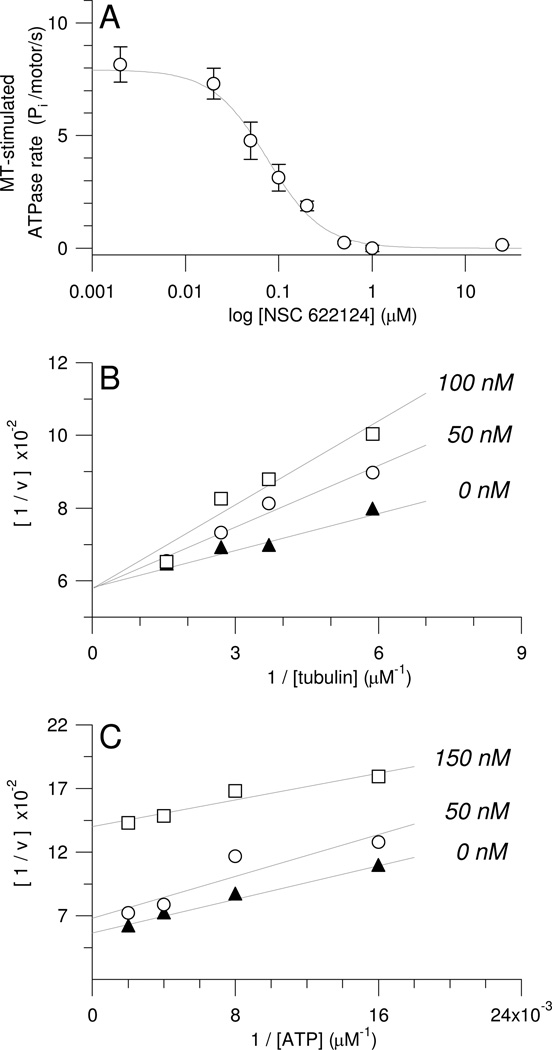
Figure 4. NSC 622124 is a Nanomolar Inhibitor of HsEg5 MT-Stimulated ATPase Activity and Competes with MTs but not ATP for Association with HsEg5.
(A) MT-stimulated ATPase activity as a function of NSC 622124 concentration. Reactions containing 100 nM HsEg5, paclitaxel-stabilized MTs (0.5 µM tubulin), and 1 mM MgATP were prepared with a range of NSC 622124 concentrations (0, 0.002, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 25, 50, and 100 µM) and ATPase rates were determined with a malachite green assay. Each point represents the mean ± SEM of 2–16 reactions. All data were used to fit the curve although only data from 0–25 µM NSC 622124 are shown. (B) MT-stimulated ATPase rates of 100 nM HsEg5 were measured in the presence of 2 mM MgATP and 0, 50 or 100 nM NSC 622124 at several tubulin concentrations (0.17, 0.27, 0.37, 0.64, and 1.08 µM), and results were graphed as a Lineweaver-Burk plot. Each data point represents the mean of 4–12 reactions. SEM values averaged 4% of the means (range = 2% to 8%). (C) MT-stimulated ATPase rates of 100 nM HsEg5 were measured in the presence of MTs (0.75 µM tubulin) and 0, 50 or 150 nM NSC 622124 over a range of MgATP concentrations (62.5, 125, 250, and 500 µM), and results were graphed as a Lineweaver-Burk plot. Each data point represents the mean of 4–8 reactions. SEM values averaged 4% of the means (range = 1% to 7%).
To determine the IC50 for NSC 622124 inhibition of HsEg5 MT-stimulated ATPase activity (Figure 4A), the malachite green assay was used to measure ATPase rates in the presence of MTs (0.5 µM tubulin) as a function of NSC 622124 concentration. The IC50 was calculated by fitting the mean values for each drug concentration as described (19). Note that, for clarity, Figure 4A shows a subset of the data points utilized for its curve fit analysis.
Trypsin Digest and Proteolytic Mapping
Four 50 µl reactions were carried out at room temperature, one with HsEg5 and NSC 622124 (10 minute preincubation) and another reaction with HsEg5 in the absence of NSC 622124. The additional two reactions consisted of a positive and negative control: HsEg5 that did not undergo digestion and a trypsin digest without HsEg5, respectively. Reactions were conducted in 50 mM Tris-acetate, pH 7.4, and 2 mM MgCl2, and contained 45 µg HsEg5 protein, 0.3 µg trypsin (Promega sequencing grade), and/or 343 µM NSC 622126. These quantities were used to ensure visualization of small peptide fragments on SDS-PAGE and to mimic molar ratios of protein to inhibitor utilized in the steady-state activity assays.
Upon addition of trypsin to the reaction, 12 µl were removed from the reaction at four time points (0, 1, 30, and 90 min) and added to an inhibitor mix that yielded final concentrations of 1.5 mM PMSF, 100 µM TLCK, and 100 µM TPCK. The proteolytic reactions were visualized on a NuPage Novex 4–12% Bis-Tris Gel with the 1X MES buffer system (Invitrogen) and stained with SYPRO Tangerine (Cambrex BioScience Rockland, Inc.).
For mass spectral analysis, bands of interest were excised from the gel under a UV-transillumination box. Gel pieces, collected in eppendorf tubes, were incubated with one round of 5% acetonitrile/25 mM ammonium bicarbonate buffer and two rounds of 0.5% acetonitrile/25 mM ammonium bicarbonate, each round requiring 20 min. Gel slices were then rinsed with 50 µL acetonitrile for 10 min and excess liquid was manually removed. Following the above treatment for the gel slices, approximately 0.1 µg trypsin in 10 µL of 25 mM ammonium bicarbonate buffer was added into each vial and incubated at 37°C overnight (~16 hours). Tryptic peptides were extracted twice with 100 µL of 50/50 0.1% trifluoroacetic acid/acetonitrile, and dried in a CentriVac. The samples were resuspended into 10 µL of 2% acetonitrile and 0.1% formic acid, prior to the LC-MS analysis.
Thermo-Fisher LTQ ion trap mass spectrometry (Waltham, MA) coupled with an Eksigent nanoLC chromatographer (Dublin, CA) was used for the LC-MS analysis. A gradient was delivered to a column/emitter (New Objective Picofrit C18 reversed phase, 75 micron ID, 30A) to elute the peptides directly into mass spectrometer. The first linear gradient was set at 200 nL/min from 5% buffer B (99.9% acetonitrile with 0.1% formic acid) and 95% buffer A (0.1% formic acid) to 40% buffer B and 60% buffer A in 20 min, and followed by the second gradient from 40% to 90% buffer B within 10 min. The mass spectrometer then scanned the ions from m/z 200 to 2000. The precursor ions scanned in MS run were subjected for MS/MS run. The data were sent to the Thermo-Fisher Sequest database search engine for database search to display the protein sequence coverage for each sample.
Co-sedimentation Assays
MT co-sedimentation assays were prepared in 50 mM PIPES, pH 6.9, 1 mM EGTA, and 0.5 mM MgCl2 and contained 20 µM paclitaxel, 10 µM tubulin (as paclitaxel-stabilized MTs), 2.5 µM HsEg5 or KLP61F, 1 mM MgAMPPNP, and 25 µM NSC 622124 (or an equivalent volume of DMSO as a control). Reactions were incubated at room temperature for 15 min and centrifuged at 110,000×g spun in a Beckman TLA 100.3 rotor at 25°C for 15 min. Supernatants and pellets were analyzed by SDS-PAGE.
MT-Motility Assays
A sample of bacterial high-speed supernatant containing full length D. melanogaster Kinesin-1 was applied to slide-coverslip chambers constructed with double-sided tape. The chamber was washed with a solution of 50 mM PIPES, pH 6.9, 1 mM EGTA, and 0.5 mM MgCl2 to remove unbound protein followed by the same buffer supplemented with paclitaxel-stabilized MTs (0.1 µM tubulin), and then either 1 mM MgATP or MgAMPPNP, was perfused into the chamber. Samples were observed at room temperature by video-enhanced differential interference contrast microscopy (28) to confirm MT attachment, and for chambers containing MgATP, MT motility. For each nucleotide condition, after 3–10 min, a solution containing the same nucleotide and 5 µM NSC 622124 was perfused into the chamber. All solutions pefused into the chamber following the addition of MTs contained 20 µM paclitaxel.
Results
To address the possibility that NSC 622124 binds HsEg5 at a site distinct from monastrol, we synthesized 14C-monastrol and utilized size exclusion spin chromatography to evaluate the binding of monastrol to HsEg5 in the absence or presence of selected HsEg5 inhibitors, including NSC 622124. Comprised of both S- and R- enantiomers, the 14C-monastrol was similar to commercially available racemic monastrol in ability to inhibit HsEg5 ATPase activity (data not shown). Binding assays consisted of HsEg5 incubation with 14C-monastrol followed by centrifugation through G25 Sephadex to separate protein with bound 14C-monastrol from unbound 14C-monastrol. In order to ensure adequate recovery of protein, 1 mg/ml (~24 µM) was selected as the default protein concentration for these assays, and typical recovery of HsEg5 protein after centrifugation was 31.4 ± 2.6%. In order to mimic typical inhibitor-to-protein ratios used in coupled assays of motor basal ATPase activity, where 5 µM protein and 200 µM monastrol were typically used (e.g., Figure 2), binding assays utilized 0.9 mM 14C-monastrol.
Under the conditions of the assay, and consistent with the moderate binding affinity (in the µM range) and specificity of monastrol (19, 29), each mole of HsEg5 that passed through the column contained 0.34 ± 0.02 mol of 14C-monastrol (Figure 1). Neither varying the duration of incubation from 10 to 70 min nor the presence of the excess nucleotide (5 mM MgATP or MgAMPPNP) had an effect on the extent of 14C-monastrol binding (data not shown). Since the 14C-monastrol was a racemic mixture of the S- and R-enantiomers and the relative proportion of each was unknown, the sub-equimolar stoichiometry was expected.
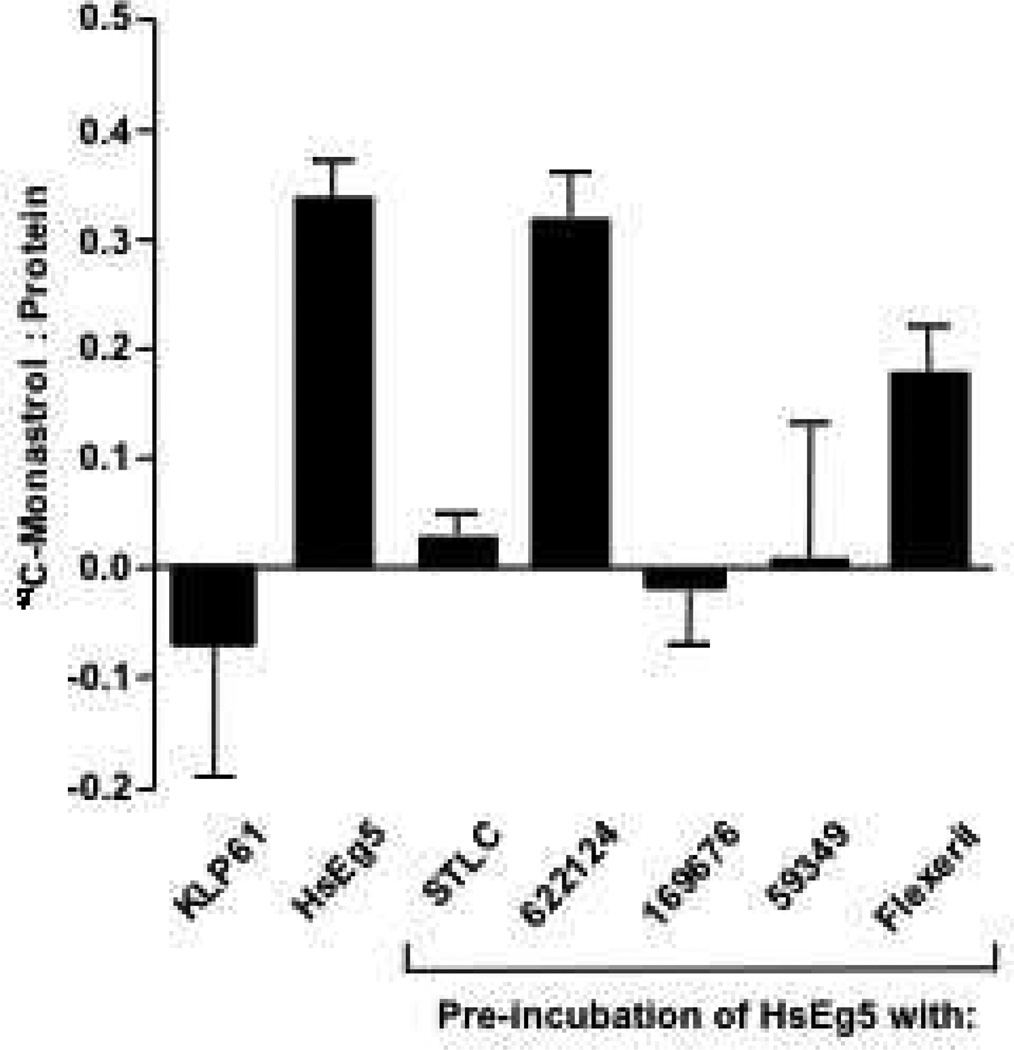
Figure 1. NSC 622124 Does Not Interfere with 14C-Monastrol Binding to HsEg5.
Reactions containing 24 µM protein (HsEg5 or KLP61F) and 0.9 mM 14C-monastrol were subjected to size exclusion spin chromatography and the amount of protein and bound 14C-monastrol was determined. Data represent the mean ± SEM of 18 reactions (36 columns) for HsEg5 and 2 reactions (4 columns) for KLP61F. To assay for competition between indicated HsEg5 inhibitors and 14C-monastrol for binding to HsEg5, reactions were prepared as above except 0.5 mM of the indicated inhibitor was incubated with the motor for 20 min at room temperature before addition of 14C-monastrol. Data represent the mean ± SEM of 4 reactions (8 columns).
The ability of the Drosophila melanogaster Kinesin-5 protein, KLP61F, to bind 14C-monastrol was evaluated next. This HsEg5 relative is not inhibited by monastrol (30), although it is unknown if this insensitivity results from an inability of KLP61F to bind monastrol, or if KLP61F binds monastrol but is unable to initiate the conformational change(s) required for inhibition. To distinguish these possibilities, KLP61F was incubated with 14C-monastrol as described for HsEg5 and subjected to size exclusion spin chromatography. The results confirmed that KLP61F does not bind 14C-monastrol (Figure 1), demonstrating that key residue differences exist in the drug-binding pocket of the two proteins.
As shown in Figure 1, pre-incubation of HsEg5 with four inhibitors previously reported to target the monastrol-binding site (22) either completely (for STLC, NSC 169676 and NSC 59349) or significantly (for flexeril, unpaired t-test, p = 0.027) reduced the binding of 14C-monastrol to HsEg5. In contrast, NSC 622124 did not significantly reduce bound 14C-monastrol (unpaired t-test). Since NSC 622124 did not appear to target the HsEg5 monastrol-binding site and has demonstrated inhibition of the Kinesin-14 motor, Ncd (4), we next investigated whether this compound affected either the basal or MT-stimulated ATPase activities of monastrol-insensitive (30) KLP61F. As expected from both previous work (29) and the inability of KLP61F to bind 14C-monastrol (Figure 1), inhibitors that target the monastrol binding-site had no effect on KLP61F ATPase activity either with or without MTs (Figure 2). In contrast, NSC 622124 significantly inhibited both basal and MT-stimulated ATPase activities of KLP61F.
Since the results from Figures 1 and 2 strongly suggested that NSC 622124 binds to HsEg5 at a site different from monastrol (and by extension, STLC, NSC 59349, NSC 169676 and flexeril), we wished to characterize further the interaction of NSC 622124 with HsEg5. The ability of NSC 622124 to inhibit both a monastrol-sensitive kinesin (HsEg5) and two monastrol-insensitive kinesins (Ncd (4) and KLP61F) suggested that NSC 622124 might bind to an orthosteric site shared by all kinesin motors, e.g., the ATP-binding site or the MT-binding site. NSC 622124 has previously been reported to inhibit HsEg5 basal ATPase activity with an IC50 of 13 µM, but no IC50 for inhibition of MT-stimulated ATPase activity was reported (4). From the data presented in Figure 3A, we confirmed that the basal IC50 is 13 µM, as previously reported (4). Also in the absence of microtubules, we examined the effects of increasing concentrations of ATP on the inhibitory activity of the small molecule, using NADH-coupled assays to monitor product formation from HsEg5 catalytic reactions. Lineweaver-Burk analysis of this data (Figure 3B) demonstrated that NSC 622124 exhibits mixed-type inhibition of this Kinesin-5 motor domain, with respect to ATP, in the absence of tubulin. Mixed-type inhibition, a form of noncompetitive inhibition, indicates that NSC622124 can bind to HsEg5 alone with micromolar affinity or bind to HsEg5•substrate binary complexes, but its affinity for the two forms of the enzyme is different. The inhibition constant (Ki) calculated for the HsEg5•NSC 622124 complex is 0.55 µM, and the α factor is 4.8, indicative that the dissociation constant for HsEg5•substrate•NSC 622124 is higher. Thus, NSC 622124 does not compete, and does not bind to, the nucleotide triphosphate site of HsEg5.
To determine the IC50 for inhibition of HsEg5 MT-stimulated ATPase activity, ATPase rates in the presence of MTs were measured as a function of NSC 622124 concentration (Figure 4A). The calculated IC50 was 69 ± 15 nM, indicating that NSC 622124 is one of the higher affinity HsEg5 inhibitors characterized to date. To determine if NSC 622124 competes with MTs for binding to HsEg5, MT-stimulated ATPase assays were conducted at different NSC 622124 concentrations for several MT (tubulin) concentrations (Figure 4B). In a Lineweaver-Burk plot of the resulting data, NSC 622124 and MTs exhibited competitive binding for HsEg5. In contrast, when MT-stimulated ATPase reactions were carried out at different NSC 622124 concentrations over a range of MgATP concentrations (Figure 4C), there was no evidence of a competitive interaction between the inhibitor and the nucleotide for binding to HsEg5.
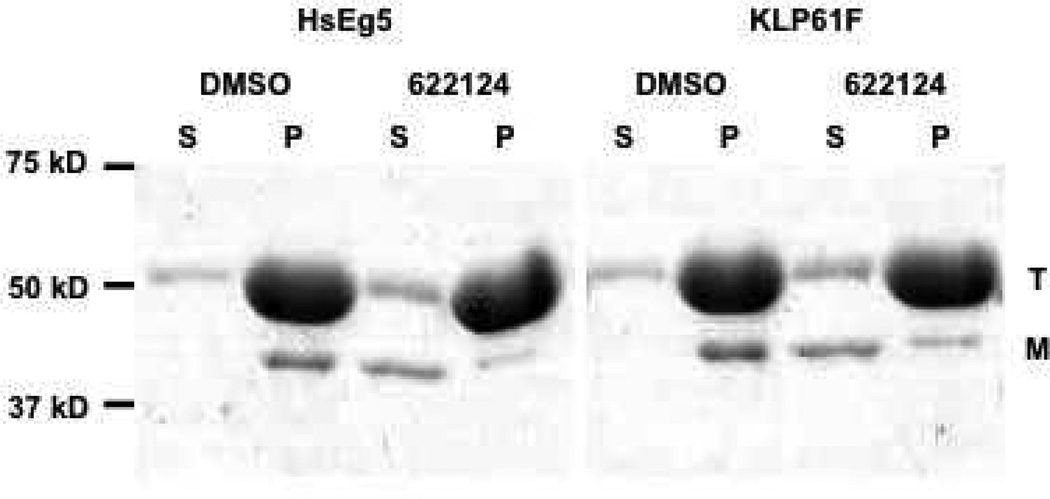
From the above series of steady-state kinetic assays, the competition between NSC 622124 and MTs for binding to HsEg5 predicted that the inhibitor should interfere with the ability of HsEg5, and perhaps other kinesins, to bind MTs. To test this possibility, three complementary approaches were used: co-sedimentation assays with two different motors proteins, MT motility assays, and proteolytic mapping of the inhibitor-binding site. In the first approach, HsEg5 and KLP61F binding to MTs was evaluated using co-sedimentation assays with and without NSC 622124, and the results (Figure 5) demonstrated that NSC 622124 substantially disrupted HsEg5 and KLP61F binding to MTs, even in the presence of rigor-inducing MgAMPPNP.
Figure 5. NSC 622124 Inhibits HsEg5 and KLP61F Binding to MTs.
Co-sedimentation assays containing HsEg5, MTs, MgAMPPNP and either DMSO or 25 µM NSC 622124 were performed as described in the Methods, and supernatant (S) and pellet (P) fractions were subjected to SDS-PAGE. The positions of molecular weight markers, tubulin (T), and relevant motor (M) are indicated.
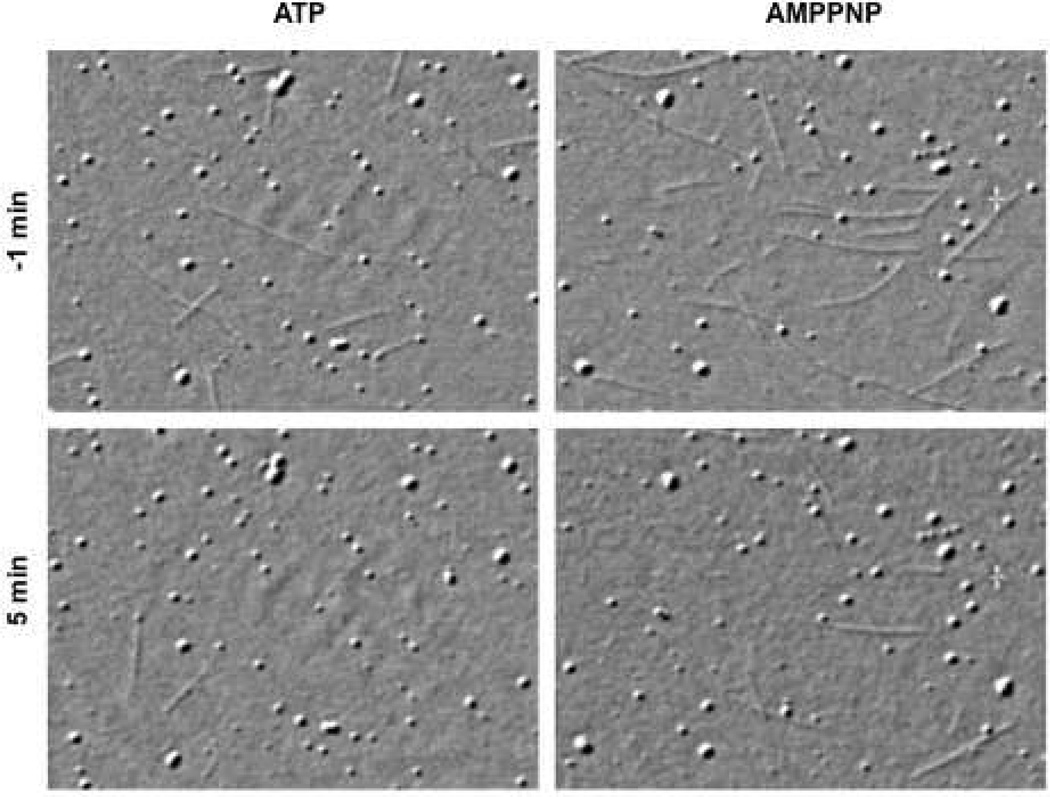
To test whether NSC 622124 would show a similar effect in MT motility assays, as well as to evaluate the compound’s effect on a kinesin motor outside the Kinesin-5 family, the effect of NSC 622124 on the D. melanogaster Kinesin-1 MT motility in the presence of either 1 mM MgATP or MgAMPPNP was observed by video-enhanced differential interference contrast microscopy (Figure 6). Without NSC 622124, kinesin in the presence of 1 mM MgATP supported MT gliding at published rates [mean rate = 0.48 ± 0.04 µm/sec, n = 10 (25)], whereas kinesin in the presence of 1 mM MgAMPPNP exhibited rigor binding to MTs. Replacement of the chamber volume with buffer containing paclitaxel and identical nucleotide had no effect on the number of MTs attached to the surface [and for MgATP, no effect on the rate of gliding (0.47 ± 0.03 µm/sec, n = 10)]. However, subsequent replacement of the chamber volume with buffer containing paclitaxel, identical nucleotide and 5 µM NSC 622124, caused a reduction in the number of MTs attached to the surface. For experiments with MgATP, the majority of MTs released from the coverslip during the time course of NSC 622124 perfusion (<25 sec) and the few MTs that remained attached showed no directed movement, and instead exhibited thermal movements consistent with single-point attachment. Experiments performed in the presence of MgAMPPNP provided similar results in terms of reduction in the number of attached microtubules and increased evidence of single-point attachment, but the time course of detachment was extended over a period of several min.
Figure 6. NSC 622124 Disrupts Kinesin-1 MT Attachment and Motility.
The effect of NSC 622124 on the interaction of D. melanogaster Kinesin-1 and MTs in the presence of either 1 mM MgATP (left column) or MgAMPPNP (right column) was observed by video-enhanced differential interference contrast microscopy. Paclitaxel-stabilized MTs and either 1 mM MgATP or MgAMPPNP were perfused into slide-coverslip chambers pre-coated with a bacterial high-speed supernatant containing Drosophila Kinesin-1. Images were collected just before (- 1 min, top row) perfusion with buffer containing 20 µM paclitaxel, 1 mM appropriate nucleotide, and 5 µM NSC 622124, as well as 5 min after initiation of perfusion (bottom row). Each panel is ~27 µm wide.
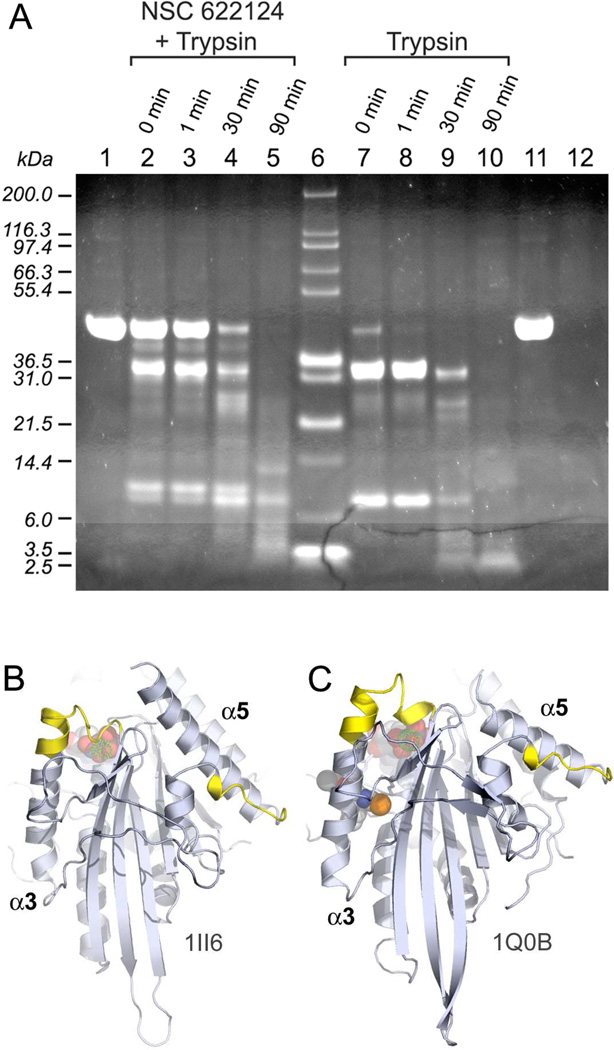
To search for a putative NSC 622124 binding site in HsEg5, purified HsEg5 motor domain samples incubated with the inhibitor were subjected to partial proteolysis by trypsin, and the resultant fragments were separated by SDS-PAGE (Figure 7). The positive control, trypsinolysis of motor alone, led to immediate digestion of the protein to yield initial 32.8 and 7.1 kD products (lanes 7 & 8) and a terminal 3.5 kD product (lane 10). Samples of HsEg5 in the presence of NSC 622124 (lanes 2–4) were not degraded as rapidly by the serine protease, and these trypsinolysis reactions yielded different digestion products: initial 34.6 and 7.9 kD products (lanes 2–4) and a terminal set of an 11 kD fragment, a 7.1 kD fragment, and a 5.1-4.0 kD smear (lane 5). Negative control reactions of HsEg5 in the absence of trypsin (lane 1& 11) over the same time course indicated no spurious degradation of the motor domain under these experimental conditions.
Figure 7. NSC 622124 Imparts Protection against Proteolytic Digestion of HsEg5.
(A) Representative samples from the trypsin digestion experiments of HsEg5, in the presence or absence of NSC 622124, were run on a 4–12% SDS-PAGE. Lanes contain: negative control HsEg5 without the small-molecule inhibitor and without trypsin at 0 min timepoint (1); reactions of HsEg5 with NSC 622124 and trypsin terminated at 0 min (2), 1 min (3), 30 min (4), and 90 min (5); Mark12 (Invitrogen) molecular weight standards (6); reactions of HsEg5 and trypsin terminated at 0 min (7), 1 min (8), 30 min (9), and 90 min (10); negative control HsEg5 without the inhibitor and without trypsin at 90 min timepoint (11); and trypsin alone at 90 min (12). The locations of peptide sequences obtained from the ~4 kD band in lane 5 are shown are labeled yellow on the X-ray structures of HsEg5 complexed with MgADP (B) and complexed with monastrol and MgADP (C).
These data show that we have generated protein fragments that are selectively protected from degradation in the presence of NSC 622124. Since these fragments may represent a subset of the minimal interaction domains of the inhibitor with HsEg5, the 5.1-4.0 kD region was excised from the SDS-gel, and subjected to an additional round of trypsin treatment before the fragments were extracted from the acrylamide matrix. Mass spectral analysis of this extraction revealed the sequence of two peptides, corresponding to residues 222–312 (TTAATLMNAYSSR) and residues 306–312 (TPHVPYR) of the HsEg5 motor domain. These peptide sequences, uniquely protected in the presence of inhibitor, may therefore represent a portion of the NSC 622124-binding site in this Kinesin-5 protein.
Discussion
Here we show that NSC 622124 does not bind to the L5 loop “hot spot” recognized by monastrol and several other HsEg5 inhibitors [Figure 1 and (21, 22)], and that NSC 622124, unlike monastrol, can inhibit KLP61F and Kinesin-1 (Figures 2, 5 and 6). Further, the mechanism of NSC 622124 inhibition is different from that of monastrol. In contrast to evidence that monastrol has little or no effect on co-sedimentation of monomeric HsEg5 (as used here) with MTs (13, 31), and also stabilizes the interaction between HsEg5 and MTs in motility assays (15), herein NSC 622124 was shown to disrupt the interaction between motor and MTs in both assays. Finally, unlike monastrol (19), NSC 622124 demonstrated direct competition with MTs for binding to HsEg5.
The simplest explanation for these results is that NSC 622124 binds at or adjacent to the conserved kinesin MT-binding site and consequently alters the interaction of the motor with MTs. This conclusion is further supported by proteolytic mapping (Figure 7), which defined two minimal HsEg5 fragments protected by NSC 622124: (a) the C-terminal residues in the L12 loop, followed by N-terminal portion of the HsEg5 α5 helix (306TPHVPYR312) and (b) the C-terminus of the α3 helix, as well as the switch I region (222TTAATLMNAYSSR234). The core of the MT-binding interface has been defined as the conserved L12 loop and subsequent helix α5 (32), and the correlation between the first fragment listed above with the alanine-scanning mutagenesis mapping of the MT-binding site (32) provides direct and strong support that NSC 622124 targets the MT-binding site of HsEg5.
How might NSC 622124 associate with the MT-binding site of kinesins? The compound is ~12 × 15 Å with a negatively charged surface and may therefore interact with the positively charged residues present in the conserved kinesin MT-binding site (32). A similar charge-dependent interaction between another polyoxometalate and the DNA-binding site of various DNA polymerases inhibits the ability of these enzymes to bind DNA (33). Binding of NSC 622124 to the MT-binding domain would clearly inhibit, through direct competition, the ability of the motor to bind MTs and to undergo MT-stimulated enhancement of ATP hydrolysis.
Two other compounds, adociasulfate-2 (AS-2) (34) and rose bengal lactone (RBL) (35), have also been reported to bind at/near the MT-binding site. Both compounds inhibit the MT-stimulated ATPase activity of Kinesin-1 and at least one other kinesin motor, and both compete with MTs but not ATP for binding to the motor. Further, AS-2 and RBL inhibit the interaction between Kinesin-1 and MTs in motility assays and in MT co-sedimentation assays (34, 35), similar to our NSC 622124 data. However, these compounds are ~100 fold less effective against HsEg5 and/or Kinesin-1 MT-stimulated ATPase activity than NSC 622124 is against HsEg5 (34–36). In fact, NSC 622124 is among the most effective inhibitors of HsEg5 MT-stimulated ATPase activity reported to date [Figure 4A and (4, 10)].
NSC 622124 also differs from AS-2 and RBL in effect on basal ATPase activity. Both AS-2 and RBL have been variously reported to either enhance or inhibit the basal ATPase activity of different kinesins (34–36). AS-2 has been proposed to act as a MT “mimic” in which negatively charged sulfate groups act analogously to the negatively charged C-termini of tubulin (34), and subsequently AS-2 has been shown to form rod-like aggregates that have been proposed to be the “active” form of AS-2 (37). Like AS-2, RBL has been reported to form aggregates, although the formation of aggregates by RBL may represent a non-specific mechanism of inhibition associated with several “promiscuous” inhibitors (38).
In contrast to AS-2 and RBL, there is no evidence that NSC 622124 can enhance the basal ATPase activity of any kinesin motor [(4), and Figures 2 and 3]. In fact, the ability of NSC 622124 to inhibit the basal ATPase activity of HsEg5 allowed the inhibitor to ”survive” a screen designed to eliminate compounds that affected MT assembly or motor binding to MTs (4). Interestingly, NSC 622124 is apparently unable to inhibit the basal ATPase activity of the A. nidulans Kinesin-5, bimC (4), although unlike most kinesins, this motor appears to contain two MT-binding sites within its motor domain (39).
Since MTs clearly influence events at the nucleotide hydrolysis site, it is perhaps not surprising that other molecules could influence nucleotide hydrolysis activity via the MT-binding site. NSC 622124 association with the MT-binding site may induce different conformation changes from those induced by MT binding, and consequently this compound may impair nucleotide hydrolysis whereas MTs enhance hydrolytic rates. The proteolytic mapping of the switch I sequence provides an explanation for the mixed-type inhibitions exhibited by NSC 622124 for HsEg5. The binding of ATP is expected to induce conformational switching of the switch I sequence, implicated in interactions with the γ-phosphate moiety of the substrate, and thereby alter the inhibition constant of the polyoxometalate for the motor protein. Conversely, as our kinetic data indicate that NSC 622124 can bind to HsEg5 in the absence of substrate, binding of the small-molecule inhibitor may alter the conformation of switch I and directly affect substrate binding. Examination of topological representations of these proteolytic fragments in HsEg5 in the absence (Figure 7B) or presence (Figure 7C) of an L5-directed allosteric inhibitor permits appreciation of the different conformational changes recognized in this Kinesin-5 protein and the outcome of these structural alterations on the NSC 622124 binding site. However, atomic resolution of the exact NSC 622124 binding site and its allosteric regulation of ATP hydrolysis will likely depend on co-crystallization of the compound and motor protein.
It is relevant to note that NSC 622124 was recently found to inhibit protein kinase CK2 (40). This work demonstrated that NSC 622124 is a nanomolar inhibitor of CK2 and, similar to our results, that the compound did not target the enzyme’s ATP- binding site. However, unlike our results in which the inhibitor targeted the MT-binding site, NSC 622124 did not compete with a substrate peptide for the substrate-binding site of the kinase, and thus appears to interact with CK2 via a distinct binding site. Further, unlike our results in which NSC 622124 affected multiple kinesins, the inhibitor was specific for CK2 in a screen of 29 kinases. Thus, NSC 622124 is not specific for kinesin proteins, but is clearly able to target different proteins through different mechanisms.
Overall, our data reinforce the concept that small molecules can control kinesins through sites other than the L5 loop specific to Kinesin-5 motors. Although a pan-kinesin inhibitor targeting a site shared by multiple proteins may not initially appear promising for therapeutic uses, recent work has identified a novel class of HsEg5 specific, ATP-competitive inhibitors that interact either directly with the nucleotide-binding site, or through allosteric interactions (41, 42). The ability of such compounds to target a conserved binding site shared by all kinesins yet still retain specificity to a select few suggests that it may be possible to generate NSC 622124 derivatives that show specificity for certain kinesins and thereby selectively interfere with cell processes that depend on those motors.
Acknowledgments
The Proteomics Core Facility at LSU Health Sciences Center is acknowledged for their contribution to this work. We also thank Jessica Richard for assistance with ATPase assays.
This work was supported by NSF MCB-0130910 (R.A.W.), NIH GM066328 (E.W.), the Virginia Tech ASPIRES program (S.K., R.A.W., and E.W.), the OSER/Carillion Biomedical Institute (E.W. and S.K.), Sigma Xi (S.S.L), and the Graduate School and the Virginia Tech Graduate Student Association (S.S.L)
Abbreviations
- ADP
adenosine 5’-diphosphate
- AMPPNP
adenosine 5’-(β,γ-imido)triphosphate
- ATP
adenosine 5’-triphosphate
- AS-2
adociasulfate-2
- DMSO
dimethyl sulphoxide
- HsEg5
Homo sapiens Kinesin-5
- HEPES
4-(2-hydroxylethyl)piperazine-1-ethanesulfonic acid
- IC50
median inhibitory concentration
- MT
microtubule
- monastrol
4-(3-hydroxyphenyl)-6-methyl-2-thioxo-1,2,4-trihydro-pyridimine-5-carboxylic acid ethyl ester
- STLC
S-trityl-L-cysteine
References
- 1.Kashina AS, Rogers GC, Scholey JM. The BimC Family of Kinesins: Essential Bipolar Mitotic Motors Driving Centrosome Separation. Biochim Biophys Acta. 1997;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- 2.Sharp DJ, Rogers GC, Scholey JM. Microtubule Motors in Mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 3.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small Molecule Inhibitor of Mitotic Spindle Bipolarity Identified in a Phenotype-Based Screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 4.DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. In Vitro Screening for Inhibitors of the Human Mitotic Kinesin Eg5 with Antimitotic and Antitumor Activities. Mol Cancer Ther. 2004;3:1079–1090. [PubMed] [Google Scholar]
- 5.Cox CD, Breslin MJ, Mariano BJ, Coleman PJ, Buser CA, Walsh ES, Hamilton K, Huber HE, Kohl NE, Torrent M, Yan Y, Kuo LC, Hartman GD. Kinesin Spindle Protein (KSP) Inhibitors. Part 1: The Discovery of 3,5-Diaryl-4,5-Dihydropyrazoles as Potent and Selective Inhibitors of the Mitotic Kinesin KSP. Bioorg Med Chem Lett. 2005;15:2041–2045. doi: 10.1016/j.bmcl.2005.02.055. [DOI] [PubMed] [Google Scholar]
- 6.Hotha S, Yarrow JC, Yang JG, Garrett S, Renduchintala KV, Mayer TU, Kapoor TM. HR22C16: A Potent Small-Molecule Probe for the Dynamics of Cell Division. Angew Chem Int Ed Engl. 2003;42:2379–2382. doi: 10.1002/anie.200351173. [DOI] [PubMed] [Google Scholar]
- 7.Kim KS, Lu S, Cornelius LA, Lombardo LJ, Borzilleri RM, Schroeder GM, Sheng C, Rovnyak G, Crews D, Schmidt RJ, Williams DK, Bhide RS, Traeger SC, McDonnell PA, Mueller L, Sheriff S, Newitt JA, Pudzianowski AT, Yang Z, Wild R, Lee FY, Batorsky R, Ryder JS, Ortega-Nanos M, Shen H, Gottardis M, Roussell DL. Synthesis and SAR of Pyrrolotriazine-4-One Based Eg5 Inhibitors. Bioorg Med Chem Lett. 2006;16:3937–3942. doi: 10.1016/j.bmcl.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 8.Nakazawa J, Yajima J, Usui T, Ueki M, Takatsuki A, Imoto M, Toyoshima YY, Osada H. A Novel Action of Terpendole E on the Motor Activity of Mitotic Kinesin Eg5. Chem Biol. 2003;10:131–137. doi: 10.1016/s1074-5521(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 9.Sakowicz R, Finer JT, Beraud C, Crompton A, Lewis E, Fritsch A, Lee Y, Mak J, Moody R, Turincio R, Chabala JC, Gonzales P, Roth S, Weitman S, Wood KW. Antitumor Activity of a Kinesin Inhibitor. Cancer Res. 2004;64:3276–3280. doi: 10.1158/0008-5472.can-03-3839. [DOI] [PubMed] [Google Scholar]
- 10.Tarby CM, Kaltenbach RF, 3rd, Huynh T, Pudzianowski A, Shen H, Ortega-Nanos M, Sheriff S, Newitt JA, McDonnell PA, Burford N, Fairchild CR, Vaccaro W, Chen Z, Borzilleri RM, Naglich J, Lombardo LJ, Gottardis M, Trainor GL, Roussell DL. Inhibitors of Human Mitotic Kinesin Eg5: Characterization of the 4-Phenyl-Tetrahydroisoquinoline Lead Series. Bioorg Med Chem Lett. 2006;16:2095–2100. doi: 10.1016/j.bmcl.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto DT, Perlman ZE, Mitchison TJ, Shirasu-Hiza M. Dynamics of the Mitotic Spindle--Potential Therapeutic Targets. Prog Cell Cycle Res. 2003;5:349–360. [PubMed] [Google Scholar]
- 12.Bergnes G, Brejc K, Belmont L. Mitotic Kinesins: Prospects for Antimitotic Drug Discovery. Curr Top Med Chem. 2005;5:127–145. doi: 10.2174/1568026053507697. [DOI] [PubMed] [Google Scholar]
- 13.Cochran JC, Gatial JE, 3rd, Kapoor TM, Gilbert SP. Monastrol Inhibition of the Mitotic Kinesin Eg5. J Biol Chem. 2005;280:12658–12667. doi: 10.1074/jbc.M413140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochran JC, Gilbert SP. ATPase Mechanism of Eg5 in the Absence of Microtubules: Insight into Microtubule Activation and Allosteric Inhibition by Monastrol. Biochemistry. 2005;44:16633–16648. doi: 10.1021/bi051724w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crevel IM, Alonso MC, Cross RA. Monastrol Stabilises an Attached Low-Friction Mode of Eg5. Curr Biol. 2004;14:R411–R412. doi: 10.1016/j.cub.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 16.DeBonis S, Simorre JP, Crevel I, Lebeau L, Skoufias DA, Blangy A, Ebel C, Gans P, Cross R, Hackney DD, Wade RH, Kozielski F. Interaction of the Mitotic Inhibitor Monastrol with Human Kinesin Eg5. Biochemistry. 2003;42:338–349. doi: 10.1021/bi026716j. [DOI] [PubMed] [Google Scholar]
- 17.Krzysiak TC, Gilbert SP. Dimeric Eg5 Maintains Processivity through Alternating-Site Catalysis with Rate-Limiting ATP Hydrolysis. J Biol Chem. 2006;281:39444–39454. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric Inhibition of Kinesin-5 Modulates Its Processive Directional Motility. Nat Chem Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- 19.Maliga Z, Kapoor TM, Mitchison TJ. Evidence That Monastrol Is an Allosteric Inhibitor of the Mitotic Kinesin Eg5. Chem Biol. 2002;9:989–996. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Sardana V, Xu B, Homnick C, Halczenko W, Buser CA, Schaber M, Hartman GD, Huber HE, Kuo LC. Inhibition of a Mitotic Motor Protein: Where, How, Conformational Consequences. J Mol Biol. 2004;335:547–554. doi: 10.1016/j.jmb.2003.10.074. [DOI] [PubMed] [Google Scholar]
- 21.Brier S, Lemaire D, Debonis S, Forest E, Kozielski F. Identification of the Protein Binding Region of S-Trityl-L-Cysteine, a New Potent Inhibitor of the Mitotic Kinesin Eg5. Biochemistry. 2004;43:13072–13082. doi: 10.1021/bi049264e. [DOI] [PubMed] [Google Scholar]
- 22.Brier S, Lemaire D, DeBonis S, Kozielski F, Forest E. Use of Hydrogen/Deuterium Exchange Mass Spectrometry and Mutagenesis as a Tool to Identify the Binding Region of Inhibitors Targeting the Human Mitotic Kinesin Eg5. Rapid Commun Mass Spectrom. 2006;20:456–462. doi: 10.1002/rcm.2329. [DOI] [PubMed] [Google Scholar]
- 23.Wojcik EJ, Dalrymple NA, Alford SR, Walker RA, Kim S. Disparity in Allosteric Interactions of Monastrol with Eg5 in the Presence of ADP and ATP: A Difference FT-IR Investigation. Biochemistry. 2004;43:9939–9949. doi: 10.1021/bi048982y. [DOI] [PubMed] [Google Scholar]
- 24.Kappe CO, Shishkin OV, Uray G, Verdino P. X-Ray Structure, Conformational Analysis, Enantioseparation and Determination of Absolute Configuration of the Mitotic Kinesin Eg5 Inhibitor Monastrol. Tetrahedron. 2000;56 1859-1832. [Google Scholar]
- 25.Yang JT, Saxton WM, Stewart RJ, Raff EC, Goldstein LS. Evidence That the Head of Kinesin Is Sufficient for Force Generation and Motility in Vitro. Science. 1990;249:42–47. doi: 10.1126/science.2142332. [DOI] [PubMed] [Google Scholar]
- 26.Moore JD, Song H, Endow SA. A Point Mutation in the Microtubule Binding Region of the Ncd Motor Protein Reduces Motor Velocity. Embo J. 1996;15:3306–3314. [PMC free article] [PubMed] [Google Scholar]
- 27.Deavours BE, Reddy AS, Walker RA. Ca2+/Calmodulin Regulation of the Arabidopsis Kinesin-Like Calmodulin-Binding Protein. Cell Motil Cytoskeleton. 1998;40:408–416. doi: 10.1002/(SICI)1097-0169(1998)40:4<408::AID-CM8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 28.Karabay A, Walker RA. Identification of Microtubule Binding Sites in the Ncd Tail Domain. Biochemistry. 1999;38:1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- 29.Maliga Z, Xing J, Cheung H, Juszczak LJ, Friedman JM, Rosenfeld SS. A Pathway of Structural Changes Produced by Monastrol Binding to Eg5. J Biol Chem. 2006;281:7977–7982. doi: 10.1074/jbc.M511955200. [DOI] [PubMed] [Google Scholar]
- 30.Maliga Z, Mitchison TJ. Small-Molecule and Mutational Analysis of Allosteric Eg5 Inhibition by Monastrol. BMC Chem Biol. 2006;6:2. doi: 10.1186/1472-6769-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, Gilbert SP, Hoenger A. A Structural Model for Monastrol Inhibition of Dimeric Kinesin Eg5. Embo J. 2006;25:2263–2273. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule Interaction Site of the Kinesin Motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 33.Sarafianos SG, Kortz U, Pope MT, Modak MJ. Mechanism of Polyoxometalate-Mediated Inactivation of DNA Polymerases: An Analysis with HIV-1 Reverse Transcriptase Indicates Specificity for the DNA-Binding Cleft. Biochem J. 1996;319:619–626. doi: 10.1042/bj3190619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakowicz R, Berdelis MS, Ray K, Blackburn CL, Hopmann C, Faulkner DJ, Goldstein LS. A Marine Natural Product Inhibitor of Kinesin Motors. Science. 1998;280:292–295. doi: 10.1126/science.280.5361.292. [DOI] [PubMed] [Google Scholar]
- 35.Hopkins SC, Vale RD, Kuntz ID. Inhibitors of Kinesin Activity from Structure-Based Computer Screening. Biochemistry. 2000;39:2805–2814. doi: 10.1021/bi992474k. [DOI] [PubMed] [Google Scholar]
- 36.Brier S, Carletti E, Debonis S, Hewat E, Lemaire D, Kozielski F. The Marine Natural Product Adociasulfate-2 as a Tool to Identify the Microtubule-Binding Region of Kinesins. Biochemistry. 2006;45:15644–15653. doi: 10.1021/bi061395n. [DOI] [PubMed] [Google Scholar]
- 37.Reddie KG, Roberts DR, Dore TM. Inhibition of Kinesin Motor Proteins by Adociasulfate-2. J Med Chem. 2006;49:4857–4860. doi: 10.1021/jm060115z. [DOI] [PubMed] [Google Scholar]
- 38.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 39.Stock MF, Chu J, Hackney DD. The Kinesin Family Member BimC Contains a Second Microtubule Binding Region Attached to the N Terminus of the Motor Domain. J Biol Chem. 2003;278:52315–52322. doi: 10.1074/jbc.M309419200. [DOI] [PubMed] [Google Scholar]
- 40.Prudent R, Moucadel V, Laudet B, Barette C, Lafanechere L, Hasenknopf B, Li J, Bareyt S, Lacote E, Thorimbert S, Malacria M, Gouzerh P, Cochet C. Identification of Polyoxometalates as Nanomolar Noncompetitive Inhibitors of Protein Kinase Ck2. Chem Biol. 2008;15:683–692. doi: 10.1016/j.chembiol.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Parrish CA, Adams ND, Auger KR, Burgess JL, Carson JD, Chaudhari AM, Copeland RA, Diamond MA, Donatelli CA, Duffy KJ, Faucette LF, Finer JT, Huffman WF, Hugger ED, Jackson JR, Knight SD, Luo L, Moore ML, Newlander KA, Ridgers LH, Sakowicz R, Shaw AN, Sung CM, Sutton D, Wood KW, Zhang SY, Zimmerman MN, Dhanak D. Novel ATP-Competitive Kinesin Spindle Protein Inhibitors. J Med Chem. 2007;50:4939–4952. doi: 10.1021/jm070435y. [DOI] [PubMed] [Google Scholar]
- 42.Rickert KW, Schaber M, Torrent M, Neilson LA, Tasber ES, Garbaccio R, Coleman PJ, Harvey D, Zhang Y, Yang Y, Marshall G, Lee L, Walsh ES, Hamilton K, Buser CA. Discovery and Biochemical Characterization of Selective ATP Competitive Inhibitors of the Human Mitotic Kinesin KSP. Arch Biochem Biophys. 2008;469:220–231. doi: 10.1016/j.abb.2007.10.016. [DOI] [PubMed] [Google Scholar]