Summary
In a forward genetic screen in Drosophila, we have isolated insomniac, a mutant that severely reduces the duration and consolidation of sleep. Anatomically-restricted genetic manipulations indicate that insomniac functions within neurons to regulate sleep. insomniac expression does not oscillate in a circadian manner, and conversely, the circadian clock is intact in insomniac mutants, suggesting that insomniac regulates sleep by pathways distinct from the circadian clock. The protein encoded by insomniac is a member of the BTB/POZ superfamily, which includes many proteins that function as adaptors for the Cullin-3 (Cul3) ubiquitin ligase complex. We show that Insomniac can physically associate with Cul3, and that reduction of Cul3 activity in neurons recapitulates the insomniac phenotype. The extensive evolutionary conservation of insomniac and Cul3 suggests that protein degradation pathways may have a general role in governing the sleep and wakefulness of animals.
Introduction
Sleep is an essential and conserved animal behavior. In humans, sleep occupies approximately one third of life, and its importance is underscored by the overpowering drive to obtain sleep after a period of sleep deprivation. Intensive study over several decades has elucidated many neuroanatomical, neurochemical, and electrophysiological aspects of sleep (Dement, 2005), and the circadian clock that controls the phase of sleep is now understood in considerable molecular detail (Zhang and Kay, 2010). Yet despite this progress, major gaps remain in our understanding of sleep. The purpose of sleep is still not well understood, and the molecular pathways that regulate sleep, particularly those that control sleep duration and homeostasis, are poorly characterized.
Although sleep has been studied most extensively in mammals, various invertebrates of the arthropod phylum, including the honeybee, cockroach, scorpion, and fruitfly, among others, exhibit behavioral states whose attributes fulfill the criteria for sleep (Kaiser and Steiner-Kaiser, 1983; Tobler, 1983; Campbell and Tobler, 1984; Kaiser, 1988; Tobler and Stalder, 1988; Tobler and Neuner-Jehle, 1992; Hendricks et al., 2000; Shaw et al., 2000; Sauer et al., 2004; Ramón et al., 2004). These attributes include behavioral immobility associated with an increased arousal threshold, a homeostatic drive to increase the amount or depth of sleep after deprivation, and altered postures specific to sleep. Although invertebrate brains lack cortical and thalamic structures that give rise to the characteristic electroencephalographic attributes of sleep in mammals, activity within the central nervous system has been correlated with arousal states in several cases where invertebrate sleep has been examined electrophysiologically (Kaiser and Steiner-Kaiser, 1983; Nitz et al., 2002; Vanswinderen et al., 2004; Ramón et al., 2004). In addition, the circadian clock regulating the timing of sleep onset is composed of genes and molecular networks that are, to a remarkable degree, shared by vertebrates and invertebrates (Zhang and Kay, 2010). These lines of evidence therefore suggest that sleep is an evolutionarily ancient behavior not unique to vertebrates (Allada and Siegel, 2008), and that the study of invertebrate model systems is likely to elucidate fundamental principles of sleep regulation.
In particular, the finding that Drosophila melanogaster exhibits a sleep state (Hendricks et al., 2000; Shaw et al., 2000) has enabled powerful genetic tools to be applied to understand the regulation and function of sleep (Hendricks, 2003; Ho and Sehgal, 2005). The relevance of Drosophila for studying sleep has been reinforced by pharmacological and candidate gene approaches, in which manipulations of molecules and pathways implicated in the regulation of sleep in vertebrates have demonstrated similar functions in Drosophila. Alteration of conserved neurotransmitter systems including GABA (Agosto et al., 2008; Parisky et al., 2008; Chung et al., 2009), serotonin (Yuan et al., 2006), and dopamine (Andretic et al., 2005; Kume et al., 2005; Lebestky et al., 2009), as well as the cAMP pathway (Hendricks et al., 2001), elicit effects on sleep and arousal that largely parallel analogous manipulations in mammalian systems (reviewed in Saper et al., 2005; Andretic et al., 2008; Crocker and Sehgal, 2010). Perhaps the greatest potential of Drosophila for understanding the regulation and function of sleep, however, resides in employing forward genetic screens to identify genes that regulate sleep and wakefulness.
Previous screens have led to the isolation of mutations in the voltage-gated potassium channel encoded by Shaker (Cirelli et al., 2005), and in sleepless (Koh et al., 2008), which encodes an extracellular membrane-linked peptide that physically associates with the Shaker channel and regulates its abundance and activity (Koh et al., 2008; Wu et al., 2010). Hyperkinetic, which encodes the cytoplasmic beta-subunit of the Shaker channel, has also been shown to regulate sleep (Bushey et al., 2007). In addition to sharply reducing sleep, loss-of-function mutations in each of these genes are associated with reduced longevity, suggesting a link between decreased sleep and lifespan (Cirelli et al., 2005; Koh et al., 2008; Bushey et al., 2010).
Here we describe the molecular cloning and characterization of insomniac, a mutant isolated in a forward genetic screen for altered sleep-wake behavior. insomniac animals exhibit severely reduced sleep, shortened sleep bouts, and decreased sleep consolidation. insomniac expression does not oscillate in a circadian manner, and the circadian clock is intact in insomniac animals, suggesting a function in pathways distinct from the circadian clock. Neuronally-restricted depletion of insomniac mimics the phenotype of insomniac mutants, indicating that insomniac is required in the nervous system for the proper regulation of sleep and wakefulness. Conversely, restoration of insomniac expression to the brains of insomniac animals is largely sufficient to rescue normal sleep-wake behavior.
insomniac encodes a protein of the BTB/POZ superfamily. Closely related members of this superfamily function as adaptors for the Cullin-3 (Cul3) ubiquitin ligase complex and thus contribute to protein degradation pathways. Consistent with the hypothesis that Insomniac may function as a Cul3 adaptor, we show that Insomniac can physically interact with Cul3, and that neuronal RNAi directed against Cul3 recapitulates the insomniac phenotype.
Results
Isolation and Molecular Cloning of insomniac
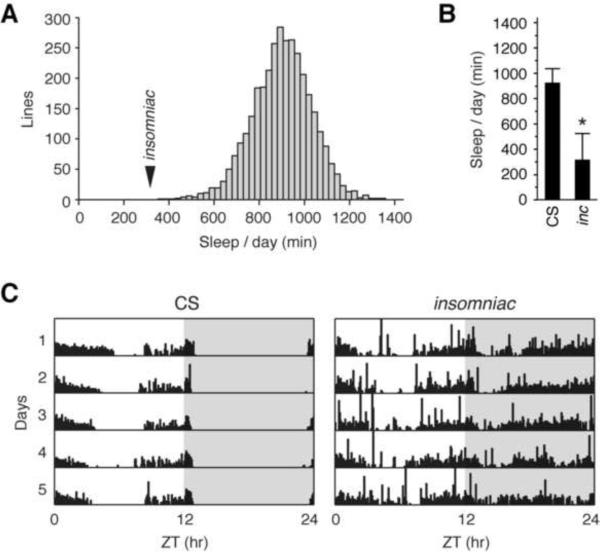
To identify mutations altering sleep, we selected a Canton-S (CS) strain exhibiting well consolidated sleep and subjected it to chemical mutagenesis with ethyl methanesulfonate. The screening regimen we employed, in which F2 males are screened, enriches for X-chromosome mutations. Over 20,800 animals, representing 3550 lines, were screened in alternating 12 hour light-12 hour dark (LD) cycles using an automated locomotor activity monitoring system. Three mutant lines exhibiting severe X-linked sleep defects were characterized further. Two of the lines shake under ether anesthesia and fail to complement the sleep defect of Shaker mutants (Cirelli et al., 2005), indicating that they are alleles of Shaker (data not shown). The third mutation does not shake under ether anesthesia and complements Shaker. This mutation, insomniac (inc), causes a severe reduction of sleep to an average of 317 minutes per day, over four standard deviations from the mean of all screened lines (Figure 1A) and a >65% reduction from that of wild-type CS control animals, which average 927 minutes of sleep per day (Figure 1B). Individual mutant animals exhibit strikingly reduced sleep during both day and night (Figure 1C), but do not display other obvious behavioral (including geotaxis and phototaxis) or morphological abnormalities.
Figure 1.
Isolation of insomniac
(A) Histogram of average daily sleep for screened lines. Mean ± SD across all lines is 901 ± 131 min. Arrowhead indicates mean sleep for insomniac. (B) Average daily sleep for wild-type CS (n=95) and insomniac (n=90) males. Mean ± SD is shown. * p < .0001. (C) Five day locomotor traces of the median individual CS and insomniac animals analyzed in (B) are shown. Shading within plots indicates night.
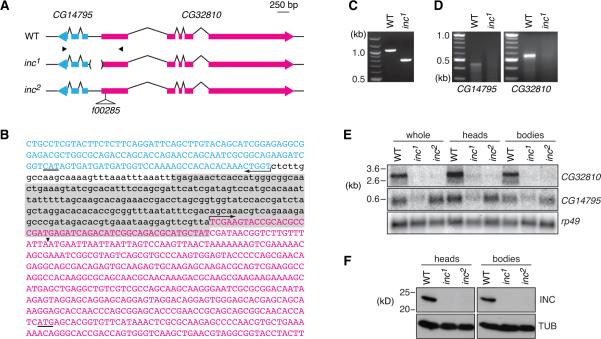
We mapped insomniac to a region of 250kb–1Mb near the tip of the X-chromosome (Figure S1A and Experimental Procedures), and further analysis identified a deficiency, removing 190kb and nine annotated genes, that failed to complement insomniac (Figure S1B). Coding regions of these candidate genes, as well as some introns and intergenic regions, were amplified from insomniac and CS animals and sequenced. A single mutation was identified, a 257 bp deletion between two divergently transcribed genes, CG14795 and CG32810 (Figure 2A–2C). This deletion is not present in other wild-type strains, and complete sequencing of CG32810 and CG14795 revealed no other alterations in either gene (data not shown).
Figure 2.
Molecular Cloning of insomniac
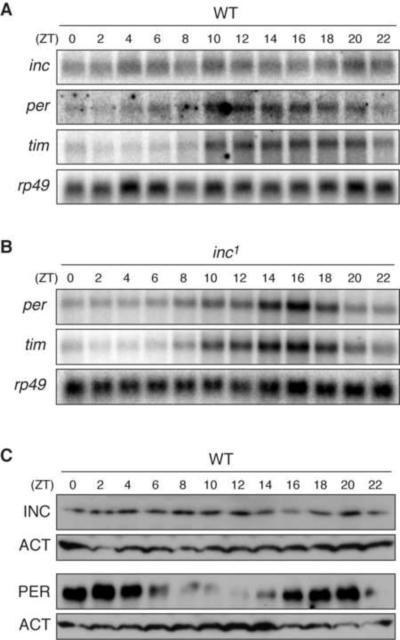
(A) Schematic of CG14795 and CG32810 loci and inc mutants. Exons are shown as thicker bars, introns by connecting lines. Arrowheads below wild-type locus denote location of primers used for PCR amplification and 5' RACE. (B) Sequence of CG14795 first exon (cyan), promoter region, and portion of CG32810 first exon (magenta). Uppercase, transcribed sequence; arrows indicate putative transcription initiation sites of longest 5' RACE products; start codons are underlined. Sequence removed by the inc1 deletion is shaded. Arrowhead indicates PBac{WH}CG32810f00285 insertion site in the inc2 mutant. (C) PCR amplification of wild-type and inc1 genomic DNA with primers shown in (A). (D) 5' RACE analysis of CG32810 and CG14795. (E) Northern analysis of CG32810 and CG14795 transcripts. (F) Western blot analysis for Insomniac protein. See also Figures S1 and S4.
To map the 5' termini of CG32810 and CG14795 with respect to the deletion, we performed 5' RACE. This analysis indicated that the deletion removes the transcriptional initiation site of CG32810 and 50 bp of its 5'UTR, but leaves intact the 5' terminus of CG14795 and a small amount of upstream sequence (Figure 2C). In insomniac animals, discrete 5' RACE products were reduced or absent for both CG32810 and CG14795, indicating that the transcription of both genes is affected by the deletion (Figure 2D). To further assess whether disruption of one or both genes causes the insomniac phenotype, we obtained a transposon insertion located in the 5'UTR of CG32810 (CG32810f00285; Figures 2A and 2B). As described below, molecular and behavioral analysis of these mutations indicates that CG32810 corresponds to insomniac, and we therefore refer to the 257 bp deletion as inc1 and the transposon insertion as inc2.
To quantitatively compare the effects of these mutant alleles on transcript levels of CG32810 and CG14795, we prepared RNA from whole animals, as well as from isolated heads and bodies, and performed Northern blotting analysis using probes specific for each transcript and for rp49, a control gene. The inc1 deletion is associated with a severe (>90%) decrease in CG32810 transcript levels and a substantial (~60%) reduction in those of CG14795 (Figure 2E and data not shown), consistent with the reduced transcript levels observed in 5' RACE analysis (Figure 2D). The inc2 transposon insertion similarly reduces CG32810 transcript levels (>90% decrease), but in contrast, has no detectable effect on levels of CG14795 RNA, indicating that it specifically disrupts the expression of CG32810. As indicated by Western blotting with an antibody raised against the protein product of CG32810, both inc1 and inc2 are null alleles at the level of protein (Figure 2F).
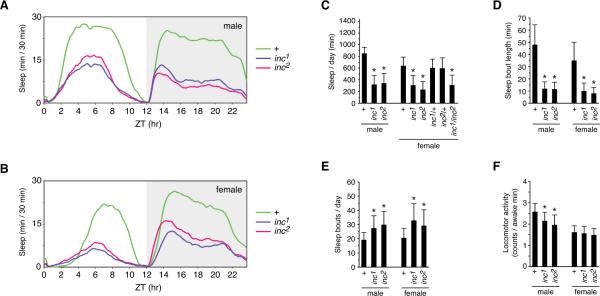
To assess the behavioral consequences of specifically disrupting CG32810 expression, we backcrossed the transposon insertion and the 257 bp deletion to an isogenic w1118 strain for eight generations, enabling phenotypic comparisons in the same genetic background. Animals bearing the deletion or the transposon insertion exhibit the same severe sleep defect, as indicated by average sleep traces (Figures 3A and 3B) and locomotor records of individual animals (Figure S2). Quantitative analysis indicates that the mutations are essentially indistinguishable in their phenotypes. inc1 and inc2 males obtain an average of 320 and 342 minutes of daily sleep respectively, a >60% reduction in comparison to control animals that sleep an average of 852 minutes per day (Figure 3C). Female inc1 and inc2 siblings respectively exhibit an average of 308 and 231 minutes of sleep per day, a ~50–60% decrease with respect to control females, which average 639 minutes daily. The sex-specific differences in total sleep have been observed previously and reflect the increased daytime sleep of males (Figures 3A and 3B, and Figure S3). For both male and female mutant animals, sleep was reduced during the day as well as during the night (Figures 3A and 3B, and Figure S3). The indistinguishable phenotypes of inc1 and inc2 animals indicate that the disruption of CG32810 causes the insomniac phenotype. Indeed, inc1 and inc2 fail to complement each other, indicating that the mutations are allelic (Figure 3C). inc1/+ and inc2/+ heterozygotes are wild-type, demonstrating that both mutants are recessive (Figure 3C).
Figure 3.
Phenotypes of inc1 and inc2 Mutants
(A and B) Average sleep traces for populations of male (A) and female (B) inc1, inc2, and control w1118 animals, plotted as a 30-minute moving average. (C) Average total sleep per day. (D) Average sleep bout length. (E) Average number of sleep bouts per day. (F) Average activity per waking minute. For all panels, n=154–155 for males, n=142–152 for homozygous females, n=63–72 for heterozygous females. For (C–F), mean ± SD is shown; * p < .0001, for comparisons against same-sex control animals. See also Figures S2 and S3.
In insomniac animals the length of sleep bouts was sharply reduced, with males averaging less than 12 minutes per sleep bout, in contrast to 48 minutes for control animals, a 75% reduction (Figure 3D). Female siblings display a similarly sharp decrease in average sleep bout length, from 35 minutes in control animals to 8 or 10 minutes for inc mutants (Figure 3D). The number of daily sleep bouts was elevated for all classes of mutant animals (Figures 3E). The substantially shorter sleep bouts and more frequent awakenings of insomniac animals reflect poorly consolidated sleep. The locomotor activity per awake minute of inc mutants was modestly reduced with respect to control animals, indicating that prolonged wakefulness is not associated with a general increase in activity level (Figure 3F).
insomniac Functions within Neurons to Govern Sleep and Wakefulness
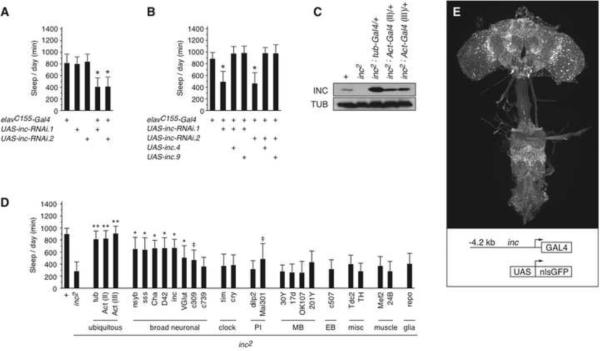
insomniac mRNA and protein are expressed in both the head and body of adult animals (Figures 2E and 2F), and our examination of transcriptome profiling databases (Chintapalli et al., 2007; Graveley et al., 2011) indicates expression in many adult tissues and during embryonic, larval, and pupal development. The sleep defect of insomniac mutants could therefore originate in a number of tissues. To test the anatomical requirements for insomniac function, we used the Gal4/UAS system (Brand and Perrimon, 1993) to direct RNAi against insomniac. More than thirty Gal4 driver lines were tested, including those driving expression in muscle, the eye, glia, and various regions of nervous system (Supplemental Experimental Procedures and data not shown). Among the lines tested, only the pan-neuronal elavC155-Gal4 driver (Lin and Goodman, 1994) was able to recapitulate the sleep defect of insomniac null mutants. Both male and female animals bearing elavC155-Gal4 and a UAS-inc-RNAi transgene integrated at either of two independent sites exhibited sharply reduced sleep (Figure 4A and data not shown). Control animals lacking either elavC155-Gal4 or the UAS-inc-RNAi transgene exhibited wild-type sleep patterns (Figure 4A). Furthermore, the RNAi phenotype induced by elavC155-Gal4 is suppressed by the co-expression of insomniac from a UAS-inc transgene (Figure 4B), indicating that the sleep defects elicited by neuronally-restricted RNAi arise from the specific depletion of insomniac and not from off-target effects (Scacheri et al., 2004; Ma et al., 2006). Thus, we conclude that insomniac is required in neurons for the normal regulation of sleep and wakefulness.
Figure 4.
Anatomically-restricted Depletion and Rescue of insomniac
(A) Neuronal depletion of insomniac. n=22–145 (mean n=69). (B) Suppression of neuronally-restricted RNAi by co-expression of UAS-inc transgenes. UAS-inc.4 and UAS-inc.9 are independent transgene insertions. n=29–36. For (A and B) * p < .0001 for comparison against elavC155-Gal4 control animals. (C) Gal4-mediated restoration of inc2 expression. Western blot of whole animal extracts is shown for males of indicated genotypes. (D) Behavioral rescue of inc2 using anatomically-restricted isogenic Gal4 drivers. PI, pars intercerebralis; MB, mushroom bodies; EB, ellipsoid body. Animals bear one copy of indicated drivers. n=9–41 (mean n=23). ** complete rescue; means are not statistically distinguishable from those of control (+) animals. * partial rescue; means are significantly different (p < .001) from those of control (+) and inc2 animals. ‡ partial rescue; means are significantly different (p < .001) from those of control (+) animals, and inc2 animals (p < .05). For (A, B, and D), total daily sleep (mean ± SD) of male animals is shown. (E) Maximal Z-projection of whole mount preparation of inc-Gal4.1; UAS-nlsGFP/+ brain stained with anti-GFP. See also Figure S4A.
In a second series of experiments, we employed the Gal4/UAS system to restore insomniac expression to inc1 and inc2 mutants. The inc2 transposon (Figure 2A) contains a UAS/TATA element within its downstream-facing terminus, juxtaposed in the correct orientation for Gal4 to drive transcription through the insomniac locus. Introduction of one copy of actin-Gal4 or tubulin-Gal4 restores insomniac expression from the inc2 allele (Figure 4C), but not from inc1 (data not shown), indicating that inc2 functions as a null allele that can be reverted in the presence of Gal4. The restoration of insomniac expression by these ubiquitous drivers rescues the sleep defect of inc2 animals completely (Figure 4D).
To further map the anatomical requirements for insomniac, we used a panel of isogenic Gal4 drivers to restore insomniac expression in restricted patterns, particularly within the brain. We were unable to assess rescue using elavC155-Gal4, as this driver is closely linked to insomniac and recombinants containing both alleles could not be isolated (data not shown). Drivers expressed broadly within the nervous system, including nsyb-Gal4, sss-Gal4, and D42-Gal4, restored the sleep of inc2 animals to 70% of wild-type levels or greater (Figure 4D). A similarly strong rescue was observed with the Cha-Gal4 driver (Figure 4D) expressed in abundant cholinergic neurons. Two drivers with relatively broad patterns of neuronal expression, glutamatergic neuron-specific VGlut-Gal4, and c309-Gal4, rescued the sleep defect of inc2 animals weakly, as did the pars intercerebralis-specific driver Mai301-Gal4 (Figure 4D). In each of these three cases however, a considerable sleep defect persists, with animals sleeping an average of ~500 minutes or less daily. We failed to obtain rescue with numerous drivers with more restricted neuronal expression (Figure 4D), including: tim-Gal4 and cry-Gal4 expressed in circadian clock cells; drivers representing various brain regions implicated in regulating sleep, including the mushroom bodies (Joiner et al., 2006; Pitman et al., 2006), pars intercerebralis (Foltenyi et al., 2007), and Tdc2-Gal4-expressing tyraminergic/octopaminergic neurons (Crocker and Sehgal, 2008; Crocker et al., 2010); c507-Gal4 expressed in the ellipsoid-body, a brain structure contributing to locomotor control; TH-Gal4 expressed in dopaminergic cells; and with glial- and muscle-specific drivers. As an independent test of these results, we performed rescue experiments in inc1 animals, using the same panel of Gal4 drivers to express a UAS-inc transgene, and obtained a pattern of rescue identical to that observed for inc2 (data not shown).
These results indicate that broad neuronal expression of insomniac is sufficient to restore sleep to near wild-type levels. Together with the consequences of depleting insomniac from neurons (Figure 4A), we conclude that insomniac functions within neurons to govern sleep. The inability of more restricted neuronal drivers to rescue insomniac is consistent with a generalized neuronal requirement, or with a requirement in dispersed neuronal subpopulations that are not represented effectively by individual Gal4 drivers we have assayed.
In a third experiment, we tested whether insomniac might regulate sleep in a dose-dependent manner, by overexpressing insomniac in a wild-type background using the pan-neuronal elavC155-Gal4 driver. For multiple insertion sites of a UAS-inc transgene, this manipulation did not increase sleep above wild-type levels (data not shown). Consistent with this finding, the levels of Insomniac protein in inc2 animals bearing tubulin-Gal4 or actin-Gal4 drivers, which exceed those of wild-type animals (Figure 4C), are not associated with an increase in sleep above wild-type levels (Figure 4D). Conversely, heterozygous inc1/+ and inc2/+ females obtain a similar amount of sleep to control animals (Figure 3C). Together, these results indicate that above a certain level, the abundance of Insomniac does not appear to regulate sleep in a dose-dependent manner.
Distribution of insomniac Expression within the Nervous System
Although antibodies raised against Insomniac specifically recognize an antigen of the appropriate size in Western blots (Figure 2F), we were unable to obtain specific staining of endogenous Insomniac protein in whole mount brain preparations (data not shown). To assess the pattern of insomniac expression, we generated transgenic animals in which insomniac genomic sequences, extending from −4.2 kb to the endogenous start codon, direct the expression of Gal4. The inc-Gal4 driver is expressed broadly in the central brain and ventral nerve cord of adult animals, including within the mushroom bodies and pars intercerebralis among other regions, as assessed by the expression of nuclear GFP from a UAS-nlsGFP reporter (Figure 4E). The inc-Gal4 driver rescues the sleep defect of inc2 animals strongly (Figure 4D), suggesting that it recapitulates endogenous insomniac expression in functionally relevant neuronal populations. Three independent insertion sites of the inc-Gal4 transgene behave similarly with respect to neuronal expression and rescue (data not shown), further supporting the notion that it provides a faithful proxy for insomniac expression. In situ hybridization experiments confirm that insomniac is expressed broadly within the brain (Figure S4A).
insomniac Functions in Pathways Distinct from the Circadian Clock
In prevailing models for how sleep is governed, the timing and amount of sleep are governed by the interaction of circadian and homeostatic mechanisms (Borbely, 1982). The increased wakefulness of insomniac mutants could in principle reflect an alteration in either mechanism. In constant darkness, inc mutants exhibit a sleep phenotype similar to that observed in LD cycles (Figures S5A and S5B). In contrast to control animals that display uniformly strong behavioral rhythms in constant darkness (100% rhythmic, τ=23.7±0.4 h, n=16), the behavioral rhythms of inc animals are weak and are observed in fewer than half of mutant animals (45% rhythmic, n=29). Nevertheless, rhythmic inc animals exhibit behavioral periods indistinguishable from those of wild-type flies (τ=23.6±0.7 h).
To further test whether the circadian clock is altered in insomniac mutants, and conversely, whether insomniac expression is regulated by the circadian clock, we performed Northern blot analysis. In the heads of wild-type animals, the levels of insomniac transcripts do not oscillate throughout the day, in contrast to those of the core clock genes period (per) and timeless (tim) (Figure 5A and Figure S5C). Similarly, there is no detectable oscillation in the abundance of Insomniac protein, in contrast to that of Period (Figure 5C). Thus, the expression of insomniac does not oscillate in a circadian fashion. In insomniac mutant heads, per and tim transcripts oscillate in a manner indistinguishable from that observed in wild-type controls (Figure 5B and Figure S5C). The circadian clock is therefore intact in insomniac mutants, suggesting that the prolonged wakefulness of insomniac animals reflects alterations in distinct molecular pathways, possibly in those that govern the homeostatic control of sleep.
Figure 5.
Relationship of insomniac and the Circadian Clock
(A and B) Northern blots of inc, per, tim, and rp49 transcripts from wild-type (A) and inc1 (B) heads. (C) Western blots of wild-type head extracts. The same extract preparation was run on 12% and 6% gels respectively, for blots against INC and PER. See also Figure S5.
Reduced Longevity of insomniac Mutants Can Be Uncoupled from Reduced Sleep
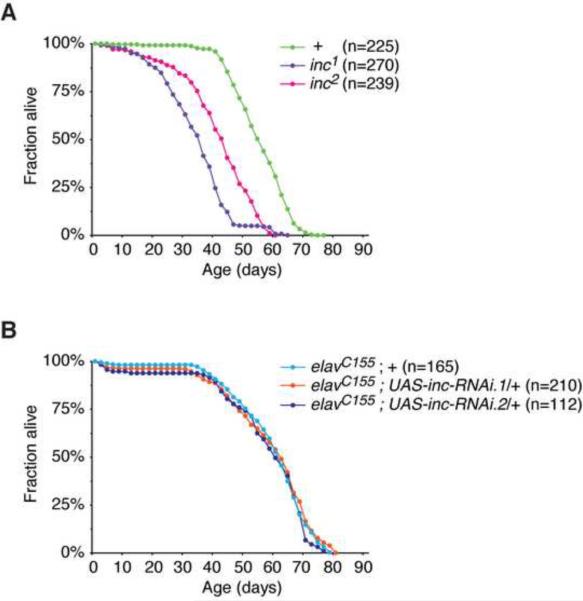
Long-term sleep deprivation leads to decreased longevity and lethality in rats (Rechtschaffen et al., 1983) and Drosophila (Shaw et al., 2002). Mutations that strongly reduce sleep in Drosophila, including Shaker, sleepless, and Hyperkinetic, are associated with decreased longevity (Cirelli et al., 2005; Koh et al., 2008; Bushey et al., 2010). As is the case for these mutations, inc1 and inc2 animals exhibit significantly reduced longevity compared to control animals (Figure 6A). To test whether the reduced longevity of insomniac mutants has a neuronal origin, we assessed the longevity of animals in which insomniac is depleted from neurons, using the elavC155-Gal4 driver and UAS-inc-RNAi transgenes. In contrast to inc null mutations, neuronally-restricted RNAi against insomniac is not associated with a decrease in longevity (Figure 6B) despite a strong reduction in sleep (Figure 4A). Although neuronal insomniac depletion reduces sleep less severely than the inc1 or inc2 mutations (Figures 3C and 4A), and does not completely eliminate Insomniac protein expressed in the head (data not shown), these results nonetheless indicate that an acute reduction in sleep can be uncoupled from reduced longevity.
Figure 6.
insomniac Longevity Phenotypes
(A) Longevity of inc1, inc2, and control w1118 males. Longevity of inc1 and inc2 animals is reduced with respect to control animals (p < .0001). Lifespan of inc1 animals is reduced with respect to inc2 animals (p < .0001), and is likely to reflect the consequences of altered CG14795 expression caused by the inc1 deletion. (B) Neuronal depletion of insomniac does not alter longevity. For males of indicated genotypes, lifespan is not significantly different (p = 0.1 to 0.4).
insomniac Encodes a Highly Conserved Protein that is a Putative Adaptor for the Cul3 Ubiquitin Ligase
The predominant insomniac transcript contains five exons and a large 3'UTR (Figure 2A and Figure S4), and encodes a 211 amino acid protein. We also detected a rare transcript variant in which the last intron is retained (Figure S4C), yielding a predicted protein three residues shorter and with a different terminal residue (Figure S4E). The mobility of endogenous Insomniac protein is slightly less than 25 kD in Western blots (Figure 2F), consistent with the 24.3 kD predicted molecular weight.
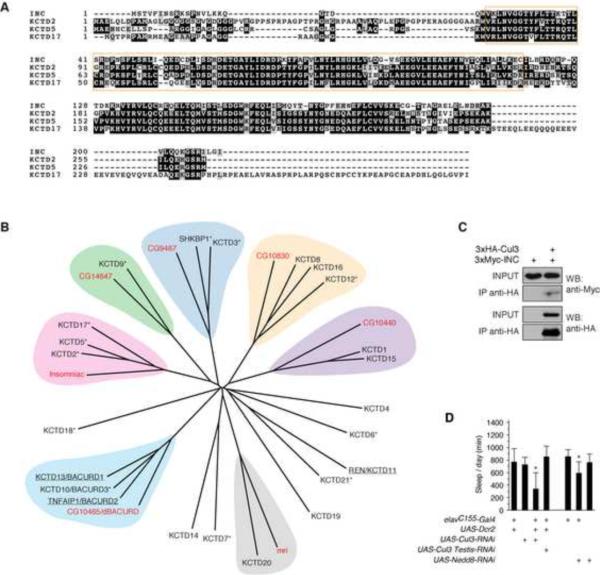
The only characterized motif within Insomniac is an N-terminal Bric-à-brac, Tramtrack and Broad / Pox virus and Zinc finger (BTB/POZ) domain, a ~95 residue motif that mediates self-association as well as interaction with heterologous proteins (Stogios et al., 2005). Three highly conserved orthologs of Insomniac, KCTD2, KCTD5, and KCTD17, are present in humans and other vertebrates (Figure 7A and data not shown). Sequence alignment of Insomniac and these orthologs reveals divergent N- and C-termini flanking the shared BTB domain and a C-terminal block of homology that corresponds to a globular domain of unknown function (Dementieva et al., 2009). Insomniac exhibits >60% identity and >75% similarity to each of its orthologs.
Figure 7.
Conservation, Phylogenetic Relationships, and Function of insomniac
(A) Alignment of Insomniac and human KCTD2, KCTD5, and KCTD17. Black and grey shading respectively indicate identical amino acids or conservative substitutions. The BTB/KCTD motif is boxed in orange. (B) Phylogenetic tree of Drosophila and human non-channel KCTD proteins. Drosophila orthologs are labeled in red. Known Cul3 adaptors are underlined; putative adaptors that co-purify with Cul3 (Bennett et al., 2010) are marked with an asterisk. This figure draws in part from a similar tree in Bayón et al., (2008). (C) Western blot analysis of Cul3 and Insomniac co-immunoprecipitation. Input is 3% of total protein used for immunoprecipitation. (D) Protein degradation pathways regulate sleep. Daily sleep (mean ± SD) is shown for male animals bearing one copy of indicated transgenes. n=15–36. * p < .001. See also Figures S6 and S7.
BTB proteins are classified on the basis of their sequence and structure into several distinct subfamilies (Stogios et al., 2005), with Insomniac and its orthologs grouped into the potassium channel tetramerization domain (KCTD) subfamily. Although the KCTD is named for its initial characterization as a sequence promoting the assembly of voltage-gated potassium (Kv) channels (Li et al., 1992; Shen et al., 1993), Insomniac and its orthologs are shorter in size and structurally distinct from Kv channels, as they lack characteristic transmembrane and ion transport domains. Including Insomniac, seven non-channel KCTD proteins are present in Drosophila. Twenty-three evolutionarily related, non-channel KCTD proteins are found in humans (Figure 7B).
Many BTB proteins function as adaptors for Cul3 ubiquitin ligase complexes (Xu et al., 2003; Pintard et al., 2003; Geyer et al., 2003), including several well-characterized proteins of the non-channel KCTD subfamily. KCTD13/BACURD1 and TNFAIP1/BACURD2 (Chen et al., 2009), and KCTD11 (Canettieri et al., 2010) associate with Cul3 and recruit specific target substrates to Cul3 complexes for ubiquitination and degradation. An additional thirteen KCTD proteins, including KCTD2, KCTD5, and KCTD17, are putative Cul3 adaptors, as they co-purify with Cul3 but not with other cullins (Bennett et al., 2010) (Figure 7B). Furthermore, both KCTD5 (Bayón et al., 2008) and TAG-303 (Xu et al., 2003), the C. elegans ortholog of Insomniac, physically interact with Cul3 in co-precipitation studies. Given the ability of highly conserved Insomniac orthologs to interact physically with Cul3, we tested whether Insomniac is able to associate with Cul3. We performed co-immunoprecipitations from Schneider S2 cells transfected with HA-tagged Cul3 and Myc-tagged Insomniac, and observed physical association of the two proteins (Figure 7C). This association is consistent with the possibility that Insomniac may serve as an adaptor for the Cul3 ubiquitin ligase complex.
Protein Degradation Pathways Regulate Sleep and Wakefulness in Drosophila
The ability of Insomniac to associate with Cul3 suggests that Insomniac may engage protein degradation pathways to regulate sleep. Cul3 null alleles are lethal (Mistry et al., 2004). To test whether Cul3 regulates sleep, we directed RNAi against Cul3 using the pan-neuronal elavC155-Gal4 driver. Animals bearing elavC155-Gal4 and a UAS-Cul3-RNAi transgene exhibited a small decrease in sleep duration (data not shown). To enhance the strength of RNAi, we co-expressed the Dicer-2 ribonuclease using a UAS-Dcr2 transgene (Dietzl et al., 2007). Animals bearing elavC155-Gal4, UAS-Dcr2, and a UAS-Cul3-RNAi transgene displayed a severe decrease in sleep duration and bout length, similar to that of insomniac animals (Figure 7D and Figure S6). Control animals bearing elavC155-Gal4 and UAS-Dcr2, or UAS-Cul3-RNAi alone, exhibited wild-type sleep (Figure 7D). Importantly, RNAi targeting a testes-specific exon of Cul3 (Arama et al., 2007) had no effect on sleep (Figure 7D). Neuronal RNAi directed against Cul1 (D. Rogulja and M.W.Y., unpublished data) or Cul2 (Figure S7) does not alter sleep significantly, suggesting that the alteration in sleep elicited by RNAi against Cul3 reflects the regulation of specific target substrates, rather than global alterations in protein degradation pathways.
We next extended our study to Nedd8, a ubiquitin-like protein whose covalent conjugation to Cul3 and other cullins is required for their activity (Petroski and Deshaies, 2005). Neuron-specific RNAi against Nedd8 elicited a significant decrease in sleep (Figure 7D). We note that a recently conducted neuronal RNAi screen for sleep defects involving over 4,000 UAS-RNAi lines also led to our identification of Nedd8 as a gene regulating sleep (D. Rogulja and M.W.Y., unpublished data). Nedd8 is essential (Ou et al., 2002), and augmenting the strength of Nedd8 RNAi by UAS-Dcr2 co-expression results in lethality (data not shown). We conclude from these studies that cullin-dependent protein degradation pathways, and particularly Cul3, are required in neurons for normal sleep and wakefulness in Drosophila.
Discussion
Emerging evidence has suggested that the sleep states of diverse animals may be regulated by conserved molecular mechanisms, although many of these mechanisms remain undefined. Here we have isolated and characterized insomniac, a gene that governs the duration of sleep and wakefulness in Drosophila, and we have shown that insomniac is likely to engage protein degradation pathways to regulate sleep. Both insomniac and these pathways are well conserved, suggesting that they may be employed generally to regulate sleep in animals.
Sleep and Longevity
In rats and Drosophila, chronic sleep deprivation leads to reduced lifespan and lethality (Rechtschaffen et al., 1983; Shaw et al., 2002). Mutations in Shaker, sleepless, and Hyperkinetic that strongly reduce sleep in Drosophila are also associated with decreased longevity (Cirelli et al., 2005; Koh et al., 2008; Bushey et al., 2010). In each case, longevity has been assessed for classical mutants in which gene function is reduced or absent in all tissues. We have shown that two independent insomniac mutants exhibit similarly decreased longevity. However, neuronally-restricted depletion of insomniac, which sharply reduces the duration of sleep, has no measurable effect on longevity, demonstrating that the two attributes can be uncoupled. Similarly, fumin mutants affecting the Drosophila dopamine transporter gene display a strong decrease in sleep but normal longevity (Kume et al., 2005).
These results do not contradict the notion that sleep has critical physiological functions or that sleep deprivation leads to deficits in waking performance, although they do suggest that certain disruptions of sleep can be tolerated without impacting lifespan. Reductions in sleep duration may need to exceed a certain threshold to affect longevity, and the lethality elicited by chronic sleep deprivation regimens, as well as that of especially severe sleep mutants (Koh et al., 2008), may reflect the reduction of sleep to extremely low levels. For mutations with more modest effects on sleep, interpretations that attribute a causal relationship between altered sleep and reduced longevity may be problematic, particularly for those genes that are broadly expressed and whose loss-of-function is likely to have numerous pathological consequences. Additional genetic manipulations that perturb sleep in increasingly specific ways are required to further assess the relationship between sleep and longevity in both Drosophila and vertebrates.
A Non-circadian Neuronal Requirement for insomniac
Our anatomically-restricted manipulations of insomniac indicate that its expression within neurons is essential for normal sleep and wakefulness. The neuronal requirement for insomniac appears to be broad, as drivers that provide pan-neuronal or broad neuronal expression alter sleep most strongly in depletion and rescue experiments. We cannot however exclude the possibility that insomniac regulates sleep by functioning in a smaller number of neurons that are dispersed within the brain and not effectively represented by individual drivers we have assayed. In particular, an insomniac-Gal4 reporter is expressed in regions of the Drosophila brain that are implicated in regulating sleep, including the mushroom bodies and the pars intercerebralis (Pitman et al., 2006; Joiner et al., 2006; Foltenyi et al., 2007), although driving insomniac expression in these areas individually does not rescue the sleep defect of insomniac mutants, with the exception of a weak rescue provided by the pars-intercerebralis-specific Mai301-Gal4 driver. Further manipulations of insomniac within the nervous system are necessary to understand the neuroanatomical basis by which it regulates sleep.
Several lines of evidence indicate that insomniac exerts its effects on sleep by a mechanism functionally distinct from the circadian clock. The circadian clock is intact in insomniac mutants, and insomniac expression is not regulated in a circadian fashion. Furthermore, the expression of insomniac in clock neurons is unable to restore normal sleep patterns in insomniac mutant backgrounds. Consistent with these data, daily sleep profiles indicate that the circadian control of sleep is intact in insomniac mutants. As is the case for wild-type animals, the highest probability of sleep during the dark phase is observed soon after the onset of darkness, with a decreasing sleep drive as the dark phase proceeds. The profile of sleep probability during the light phase is similarly intact. The principal alteration of sleep in insomniac animals is a reduced likelihood of sleeping throughout the day and night, consistent with the inference that insomniac may contribute to homeostatic mechanisms that regulate sleep need.
Insomniac as a Putative Adaptor for the Cul3 Ubiquitin Ligase Complex and the Role of Protein Degradation Pathways in Regulating Sleep
Cullins are scaffold proteins that assemble multisubunit E3 ubiquitin ligase complexes that ubiquitinate and degrade a variety of protein substrates in diverse biological contexts (Petroski and Deshaies, 2005). The C-termini of cullins interact with RING-domain ubiquitin ligases, while the N-termini interact with adaptor proteins that recruit substrates for ubiquitination. Cul3 complexes utilize proteins of the BTB superfamily as their adaptors (Pintard et al., 2003; Xu et al., 2003; Geyer et al., 2003), including members of the non-channel KCTD subfamily (Chen et al., 2009; Canettieri et al., 2010). In addition to the KCTD proteins that are known to function as Cul3 adaptors, more than half of the non-channel KCTD proteins, including the three vertebrate orthologs of Insomniac, are candidate Cul3 adaptors, as they co-purify specifically with Cul3, but not with other cullins (Bennett et al., 2010). For several of these candidate adaptors, including KCTD5 and TAG-303, the C. elegans ortholog of Insomniac, independent biochemical evidence confirms their ability to associate physically with Cul3 (Xu et al., 2003; Bayón et al., 2008; De Smaele et al., 2011). Our finding that Insomniac is able to physically interact with Cul3 indicates that this interaction is evolutionarily conserved, and supports the hypothesis that Insomniac serves as a Cul3 adaptor.
The reductions in sleep elicited by neuronal depletion of Cul3, and of its activator Nedd8, show that protein degradation pathways have a vital role in regulating sleep in Drosophila. Although we cannot exclude alternative mechanisms, the simplest hypothesis consistent with our data is that Insomniac engages the Cul3 protein degradation pathway to regulate sleep. One clear implication of this hypothesis is that the increased wakefulness of insomniac and/or Cul3 mutants may result from the inappropriate accumulation of substrates whose degradation is normally mediated by these proteins. Our results suggest that such target substrates promote wakefulness and inhibit sleep, but they do not distinguish the neuronal function of these substrates. Target substrates regulated by Insomniac and/or Cul3 might function in a developmental manner, for example, in the elaboration of neural circuits that regulate sleep. Indeed, Cul3 has been implicated in regulating axonal and dendritic branching (Zhu et al., 2005). Alternatively, such substrates might actively promote waking in adult animals, such that their ongoing degradation is part of the homeostatic mechanism contributing to the regulation of sleep-wake cycles.
Experimental Procedures
Genetics and Screening
CS males were mutagenized with 25 to 40 mM ethylmethane sulfonate and crossed en masse to virgins from an isogenic CS/FM7 stock. F1 FM7 virgins were backcrossed individually to CS males to establish lines. Four F2 males from each line were screened. Putative mutants were bred to isogenic CS/^XX females and 8–24 males were rescreened.
inc1 was mapped by crossing to y1 v1 f1 malF1 virgins and backcrossing F1 virgins to CS males. Analysis of male F2 recombinants placed the inc1 mutation proximal to y. For further mapping, 11 polymorphisms were developed by amplifying and sequencing ~1kb regions from the CS and mapping stocks at selected chromosomal positions. Mapping of inc1 with these polymorphisms and subsequent deficiency noncomplementation analysis is described in Figure S1.
Animals were backcrossed eight generations to an isogenic w1118 stock wild-type for circadian rhythms and other behaviors (Bloomington #5905, referred to elsewhere as iso31)(Ryder et al., 2004). The inc2 transposon (CG32810f00285) contains w+mC and was backcrossed by selecting w+ female offspring. The inc1 mutation induced in the CS stock is closely linked to w+ and was backcrossed similarly; the regime was carried out for several independent vials in parallel and the presence of inc1 monitored by PCR every few generations. After eight generations, w+ males exhibiting the inc phenotype were crossed to isogenic w1118/FM7c females to generate homozygous stocks; the presence of the inc1 deletion was confirmed by PCR.
Stocks and Transgenes
sss-Gal4, Cha-Gal4, D42-Gal4, VGlut-Gal4, c309-Gal4, c739-Gal4, tim-Gal4, cry-Gal4, 30Y-Gal4, 17d-Gal4, OK107-Gal4, 201Y-Gal4, c507-Gal4, Tdc2-Gal4, TH-Gal4, 24BGal4, and repo-Gal4 drivers backcrossed to the w1118 iso31 background have been described previously (Wu et al., 2010; Crocker et al., 2010). actin-Gal4 (#3954 and 4414) and tubulin-Gal4 (#5138) drivers were obtained from the Bloomington Stock Center; nsyb-Gal4 was a gift from J. Simpson; Mef2-Gal4 was a gift from R. Galindo; all were backcrossed six to eight generations to the iso31 background. UAS-inc-RNAi.1, UAS-inc-RNAi.2, and UAS-Nedd8-RNAi are in the iso31 background and correspond to VDRC stocks 18225, 18226, and 28444 respectively (Dietzl et al., 2007). UAS-Cul2-RNAi, UASCul3-RNAi, and UAS-Cul3 Testis-RNAi correspond to NIG-Fly stocks 1512R-3, 11861R-2, and 31829R-2 respectively. UAS-inc and inc-Gal4 stocks were generated in the iso31 background (Bestgene). UAS-inc.4 and UAS-inc.9 are third chromosome insertions. inc-Gal4.1 is an X-chromosome insert; inc-Gal4.2 and inc-Gal4.3 are second chromosome insertions.
As noted in the text, mutants in the CS and w1118 iso31 backgrounds were compared to their respective matched genetic backgrounds. For crosses involving transgenes, control animals were obtained by crossing transgenes to the appropriate isogenic background (e.g. for elav-Gal4 x w1118; UAS-RNAi, control crosses of elav-Gal4 x w1118 were performed). For X-linked transgenes, progeny from reciprocal crosses provided an additional control.
Sleep and Circadian Analysis
1–5 day old animals eclosing from LD-entrained cultures were loaded into glass tubes, and assayed for 5–7 days at 25°C in LD cycles on cornmeal, agar, and molasses food using DAM5 monitors (Trikinetics). Animals were allowed to acclimate after loading for 1–2 days before data collection was initiated. For females, virgins were assayed. Locomotor data was collected in 1 minute bins, and a five minute period of inactivity (Shaw et al., 2000; Huber et al., 2004) was used to define sleep. Sleep parameters were analyzed with custom software written in MATLAB (Mathworks). Dead animals were excluded from analysis by a combination of automated filtering and visual inspection of locomotor traces.
For statistical analysis of all sleep parameters that approximate normal distributions, unpaired Student's t-tests were used when comparing two genotypes; for comparisons of more than two genotypes, one-way ANOVA followed by Tukey-Kramer post-hoc tests were used. For comparisons of sleep bout length, nonparametric Kruskal-Wallis tests followed by Bonferroni-corrected Mann-Whitney post-hoc tests were used.
For analysis in constant darkness, LD-entrained animals were placed in darkness and assayed otherwise as above. To assess rhythmicity and period length, data were binned at 30 min and analyzed with chi-square periodograms (p=.01); autocorrelation analysis yielded essentially identical results.
Longevity Assay
Fifteen animals eclosing from LD-entrained cultures were placed into each individual vial on the day of eclosion, and the vials subsequently placed in LD cycles at 25°C. The number of dead animals was recorded every two days and animals transferred in parallel to new vials every two to four days. Logrank tests were used to determine statistical significance; flies lost during transfer (<4%) were included in the analysis as right-censored events.
Molecular Biology
Total RNA was isolated using TRIZOL (Invitrogen). For 5' RACE analysis of inc, RNA isolated from CS (2.5 μg) or inc1 (5 μg) adult male heads was reverse transcribed using primer ns189 and SuperScript II enzyme. cDNA was column purified and TdT tailed as per the manufacturer's protocol (Invitrogen 5' RACE System 2.0), and amplified with primers ns190 and AAP and Taq polymerase (95°C, 2 min; 30× (95°C, 30 sec; 55°C, 1 min; 72°C, 1 min); 72°C, 5 min). KpnI/SpeI-digested PCR products were cloned into pBSSK- (Stratagene) and twenty four clones sequenced. 5' RACE analysis of CG14795 was performed similarly, using primer ns195 to reverse transcribe 2.5 μg of adult male head CS or inc1 RNA; primers ns197 and AAP were used for 35 cycles of amplification as above. XhoI/SpeI-digested PCR products were cloned into pBSSK- and twenty three clones sequenced. For 3' RACE analysis of inc, 5 μg RNA was reverse transcribed using primer 3'AP and amplified using primers ns187 and 3'UAP.
Northern analysis was performed using standard methods with random-labeled hybridization probes. Templates for probes were derived by PCR amplification of cDNA using primers ns180 and ns181 for inc, and by amplification of genomic DNA with primers ns142 and ns196 for CG14795; DJS35 and DJS22 for per; DJS18 and DJS32 for tim; and DJS44 and DJS45 for rp49.
Genomic DNA was isolated with standard methods. PCR flanking the inc1 deletion (Figure 2A) was performed with primers ns198 and ns119.
Plasmids
UAS-inc expresses Insomniac under UAS control. inc-Gal4 contains inc sequences, extending from −4.2 kb upstream of the transcription start site to the endogenous start codon, fused to GAL4. Actin-3xHA-Cul3 and Actin-3xMyc-Insomniac express N-terminally tagged Cul3 and Insomniac respectively, under the control of an Actin promoter. tac-GST-Insomniac expresses a GST-Insomniac fusion protein under the control of a bacterial tac promoter. Construction details are in Supplemental Experimental Procedures.
Antibodies and Western blots
GST-Insomniac was expressed in E. coli and purified as described previously (Frangioni and Neel, 1993). GST-Insomniac bound to agarose-glutathione was cleaved with PreScission Protease (GE Healthcare) and Insomniac protein was eluted, concentrated, flash frozen, and injected into rats (Covance).
Protein extracts were prepared from whole animals, fractionated heads, and fractionated bodies by homogenization in ice-cold lysis buffer (20 mM HEPES pH 7.5, 100 mM KCl, 10 mM EDTA, 50 mM NaF, 0.1% Triton X-100, 10% glycerol) supplemented with 1mM DTT, protease inhibitors (Calbiochem), and phosphatase inhibitors (Sigma) before use. The lysate was centrifuged at 4°C at 15,000g for 15 min, quantitated, and resolved by SDS-PAGE.
For probing of Western blots, rat anti-Insomniac was used at 1:1000 to 1:2000; goat anti-Per (Santa Cruz Biotechnology) at 1:100; rabbit anti-actin (Sigma) at 1:10,000; and mouse anti-tubulin (DM1A, Sigma) at 1:200,000 to 1:1,000,000. HRP-conjugated secondary antibodies (Jackson Immunoresearch) were used at 1:10,000 and visualized with ECL plus substrate (GE Healthcare).
Immunoprecipitation
Schneider S2 cells were grown under standard conditions. 1–2×106 cells were plated in each well of 6 well plates 24 h prior to transfection, and transfected for 24 h with 400 ng of DNA using Effectene (Qiagen) according to the manufacturer's protocol. An equimolar ratio of plasmids encoding Insomniac and Cul3 was typically used. Cells were resuspended 48 h post-transfection, washed twice in PBS, and lysed in ice cold lysis buffer (50 mM Tris, 150 mM NaCl, and 0.5% NP40) containing protease and phosphatase inhibitors. In some experiments 2mM orthophenanthroline, an inhibitor of cullin deneddylation (Bennett et al., 2010), was added to the lysis buffer. 700–1000 μg total protein was incubated overnight at 4°C with 1:100 anti-HA antibody (3F10). Complexes were precipitated by incubation with Gammabind G sepharose beads (GE Healthcare) for 1 hr at room temperature on a nutator, washed with lysis buffer (4×10 min), resolved by SDS-PAGE, and subjected to Western blotting.
Immunohistochemistry
Whole animals were fixed with 4% paraformaldehyde in PBST (1× PBS, 0.2% Triton X-100) for 3h at 4°C, and washed four times in PBST (2×1 min, 2×30min) at room temperature. Brains were dissected in PBST, blocked in PBST containing 5% normal donkey serum for 30 min at room temperature, and stained for 2 days at 4°C in a cocktail containing PBST, 5% donkey serum, 1:1000 rabbit anti-GFP (Invitrogen), and 1:40 mouse anti-nc82 (DSHB). Washing in PBST (4×15min) at room temperature was followed by staining with Alexa 488 anti-rabbit (Invitrogen) and Cy3 anti-mouse (Jackson ImmunoResearch) secondary antibodies, both at 1:500, for 2 days at 4°C followed by washes as above. Brains were mounted in Vectashield (Vector Labs) and imaged on a LSM 510 confocal microscope (Zeiss).
Alignments
Protein sequences (see Supplemental Experimental Procedures) were aligned and plotted with ClustalW2, PHYLIP, and BOXSHADE.
Supplementary Material
Highlights
Mutations in insomniac reduce sleep duration and consolidation in Drosophila
insomniac regulates sleep by a mechanism distinct from the circadian clock
insomniac encodes a putative Cul3 adaptor and is highly conserved in vertebrates
insomniac and Cul3 function within neurons to control sleep
Acknowledgements
We thank Lino Saez for his advice and guidance throughout the course of these experiments, and Dragana Rogulja for communicating her independent isolation of Nedd8 from a sleep screen. We also thank J. Stieglitz and A. Sarma for technical assistance; F. Lam and S. Syed for advice on MATLAB coding; P. Kidd for assistance with circadian analysis; D. Seay for primers; M. Crickmore, R. Galindo, R. Jackson, W. Joiner, K. Koh, H. Kramer, A. Sehgal, J. Simpson, G. Tononi, L. Vosshall, and the Bloomington, NIG-Fly, and VDRC stock centers for stocks; A. Sehgal and DSHB for antibodies; and the RU Bio-Imaging Resource Center for use of microscopes. We thank C. Bargmann, L.Vosshall, and members of the Young Lab for critical comments on the manuscript and discussions. This research was supported by NIH grants NS053087, GM054339, and MH015125 to M.W.Y., and by NIH Ruth L. Kirchstein postdoctoral fellowship GM080934 to N.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–R679. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, Franken P, Tafti M. Genetics of sleep. Annu Rev Genet. 2008;42:361–388. doi: 10.1146/annurev.genet.42.110807.091541. [DOI] [PubMed] [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5:e251. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayón Y, Trinidad AG, de la Puerta ML, Del Carmen Rodríguez M, Bogetz J, Rojas A, De Pereda JM, Rahmouni S, Williams S, Matsuzawa S-I, et al. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008;275:3900–3910. doi: 10.1111/j.1742-4658.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbely AA. A two process model of sleep regulation. Human neurobiology. 1982;1:195–204. [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic Mutants Have Reduced Sleep and Impaired Memory. Journal of Neuroscience. 2007;27:5384–539. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Hughes KA, Tononi G, Cirelli C. Sleep, aging, and lifespan in Drosophila. BMC Neurosci. 2010;11:56. doi: 10.1186/1471-2202-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neuroscience and biobehavioral reviews. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Chung B, Kilman V, Keath J, Pitman J, Allada R. The GABA(A) Receptor RDL Acts in Peptidergic PDF Neurons to Promote Sleep in Drosophila. Curr Biol. 2009 doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. Genetic analysis of sleep. Genes Dev. 2010;24:1220–1235. doi: 10.1101/gad.1913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smaele E, Di Marcotullio L, Moretti M, Pelloni M, Occhione MA, Infante P, Cucchi D, Greco A, Pietrosanti L, Todorovic J, et al. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia (New York, NY. 2011;13:374–385. doi: 10.1593/neo.101630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dement WC. History of sleep physiology and medicine. In: Kryger R, MH T, Dement WC, editors. Principles and Practice of Sleep Medicine. Elsevier Saunders; Philadelphia: 2005. pp. 1–12. [Google Scholar]
- Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, Grigorieff N, Goldstein SAN. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009;387:175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC. Invited review: Sleeping flies don't lie: the use of Drosophila melanogaster to study sleep and circadian rhythms. J Appl Physiol. 2003;94:1660–1672. doi: 10.1152/japplphysiol.00904.2002. discussion 1673. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Ho KS, Sehgal A. Drosophila melanogaster: an insect model for fundamental studies of sleep. Meth Enzymol. 2005;393:772–793. doi: 10.1016/S0076-6879(05)93041-3. [DOI] [PubMed] [Google Scholar]
- Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kaiser W. Busy bees need rest, too: Behavioral and electromyographical sleep signs in honeybees. Journal of Comparative physiology A. 1988;163:565–584. [Google Scholar]
- Kaiser W, Steiner-Kaiser J. Neuronal correlates of sleep, wakefulness and arousal in a diurnal insect. Nature. 1983;301:707–709. doi: 10.1038/301707a0. [DOI] [PubMed] [Google Scholar]
- Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jan YN, Jan LY. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Mistry H, Wilson BA, Roberts IJH, O'kane CJ, Skeath JB. Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech Dev. 2004;121:1495–1507. doi: 10.1016/j.mod.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002;12:1934–1940. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- Ou C-Y, Lin Y-F, Chen Y-J, Chien C-T. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJL, Kang K, Liu X, Garrity PA, Rosbash M, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson J-LF, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- Pitman JL, Mcgill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Ramón F, Hernández-Falcón J, Nguyen B, Bullock TH. Slow wave sleep in crayfish. Proc Natl Acad Sci USA. 2004;101:11857–11861. doi: 10.1073/pnas.0402015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–184. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- Sauer S, Herrmann E, Kaiser W. Sleep deprivation in honey bees. J Sleep Res. 2004;13:145–152. doi: 10.1111/j.1365-2869.2004.00393.x. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- Shen NV, Chen X, Boyer MM, Pfaffinger PJ. Deletion analysis of K+ channel assembly. Neuron. 1993;11:67–76. doi: 10.1016/0896-6273(93)90271-r. [DOI] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJS, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I. Effect of forced locomotion on the rest-activity cycle of the cockroach. Behav Brain Res. 1983;8:351–360. doi: 10.1016/0166-4328(83)90180-8. [DOI] [PubMed] [Google Scholar]
- Tobler I, Neuner-Jehle M. 24-h variation of vigilance in the cockroach Blaberus giganteus. J Sleep Res. 1992;1:231–239. doi: 10.1111/j.1365-2869.1992.tb00044.x. [DOI] [PubMed] [Google Scholar]
- Tobler I, Stalder J. Rest in the scorpion—a sleep-like state? Journal of Comparative Physiology A: Neuroethology. 1988;163:227–235. [Google Scholar]
- Vanswinderen B, Nitz D, Greenspan R. Uncoupling of Brain Activity from Movement Defines Arousal States in Drosophila. Current Biology. 2004;14:81–87. [PubMed] [Google Scholar]
- Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin T-H, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- Zhu S, Perez R, Pan M, Lee T. Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J Neurosci. 2005;25:4189–4197. doi: 10.1523/JNEUROSCI.0149-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.