Abstract
Photorespiration is initiated by the oxygenase activity of ribulose-1,5-bisphosphate-carboxylase/oxygenase (RUBISCO), the same enzyme that is also responsible for CO2 fixation in almost all photosynthetic organisms. Phosphoglycolate formed by oxygen fixation is recycled to the Calvin cycle intermediate phosphoglycerate in the photorespiratory pathway. This reaction cascade consumes energy and reducing equivalents and part of the afore fixed carbon is again released as CO2. Because of this, photorespiration was often viewed as a wasteful process. Here, we review the current knowledge on the components of the photorespiratory pathway that has been mainly achieved through genetic and biochemical studies in Arabidopsis. Based on this knowledge, the energy costs of photorespiration are calculated, but the numerous positive aspects that challenge the traditional view of photorespiration as a wasteful pathway are also discussed. An outline of possible alternative pathways beside the major pathway is provided. We summarize recent results about photorespiration in photosynthetic organisms expressing a carbon concentrating mechanism and the implications of these results for understanding Arabidopsis photorespiration. Finally, metabolic engineering approaches aiming to improve plant productivity by reducing photorespiratory losses are evaluated.
1. INTRODUCTION
1.1. The Origin and Significance of Photorespiration
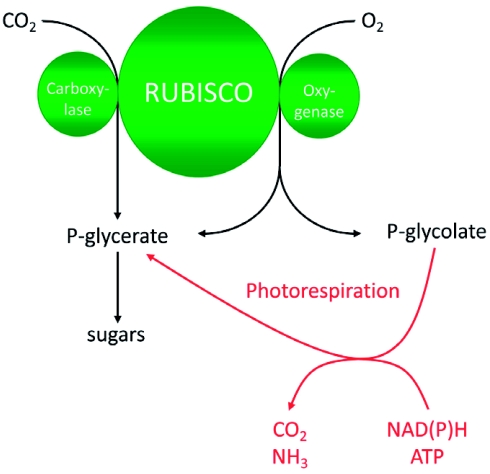
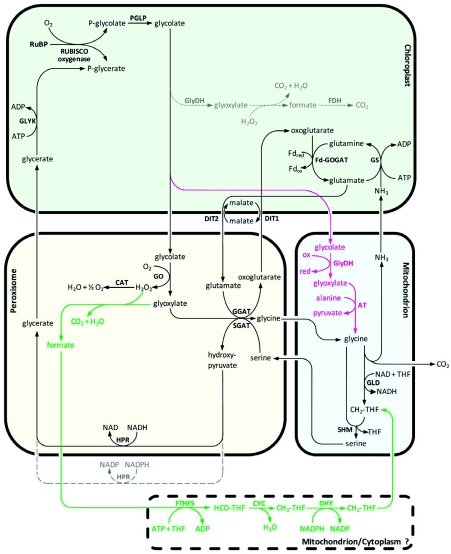
Photorespiration is the process of light-dependent uptake of molecular oxygen (O2) concomitant with release of carbon dioxide (CO2) from organic compounds. The gas exchange resembles respiration and is the reverse of photosynthesis where CO2 is fixed and O2 released. As shown in Figure 1, the entrance reactions to both photosynthesis and photorespiration are catalyzed by the same enzyme: Ribulose-1,5-bisphosphate-carboxylase/oxygenase (RUBISCO, EC 4.1.1.39). Carbon fixation results in the formation of two molecules 3-phosphoglycerate (PGA) that are integrated into the Calvin cycle ultimately to form sugars. During oxygen fixation, one molecule of PGA and one molecule of 2-phosphoglycolate are formed. The latter is converted back to PGA in the photorespiratory cycle. The pathway requires energy (ATP) and reducing (NAD(P)H) equivalents. This is in great part due to the release of CO2 and ammonia (NH3) that have to be refixed (for details see chapter 2).
Figure 1:
Schematic overview of photosynthesis and photorespiration.
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RUBISCO) catalyzes both CO2 and O2 fixation. The product of CO2 fixation is phosphoglycerate (P-glycerate) that enters the Calvin cycle. During oxygenation, equimolar amounts of P-glycerate and phosphoglycolate (P-glycolate) are formed. P-glycolate is recycled to P-glycerate in the photorespiratory pathway. In this reaction cascade, reducing equivalents (NAD(P)H) and energy equivalents are consumed. Ammonia (NH3) and CO2 are released and have to be refixed. Under current atmospheric gas concentrations and moderate environmental conditions, approximately each fourth reaction catalyzed by RUBISCO is an oxygenase reaction.
RUBISCO evolved already early in biotic evolution about 3 billion years ago probably from enzymes involved in sulfur metabolism (Tabita et al., 2007), but is until today the enzyme accounting for the vast amount of net CO2 fixation from the atmosphere.At the time of RUBISCO evolution, the only source of molecular oxygen in the atmosphere was probably photolysis of water by UV light and concentrations were 10-14 below present concentrations (Buick, 2008). At the same time, CO2 concentrations were at least 100-fold higher than today (Kasting and Howard, 2006; Kasting and Ono, 2006). Initial RUBISCO enzymes were probably extremely bad in discriminating CO2 and O2 (Tabita et al., 2007; Badger and Bek, 2008) due to the absence of evolutionary pressure. With the advent of oxygenic photosynthesis in cyanobacteria, huge amounts of CO2 were fixed into biomass that in part sedimented and did not return into the global carbon cycle. Concomitantly, equimolar amounts of O2 were released into the atmosphere, because water was used as the reductant for the photosynthetic electron transport chain (Xiong and Bauer, 2002). Cyanobacteria, the subsequently evolving algae and particularly land plants (Igamberdiev and Lea, 2006) were so successful in doing this that O2 became the second most prominent gas in today's atmosphere and CO2 is extremely scarce. Selection pressure induced some improvement in RUBISCO's specificity for CO2 that might not be further optimized without slowing down catalytic rates (Tcherkez et al., 2006): The more the structure of the bound CO2 molecule resembles a carboxylate group, the better it will be discriminated from O2, but the worse the first intermediate of the condensation can be cleaved into the end products of the reaction. Therefore, O2 uptake by RUBISCO and the subsequent metabolism of the reaction product in the photorespiratory pathway is the second highest metabolite flux in plants behind photosynthesis with flux rates around 25 % of the photosynthetic rate at 25 °C (Sharkey, 1988). Photorespiratory fluxes are even higher under hot and dry growth conditions mainly for three reasons:
The specificity of RUBISCO for CO2 relative to O2 decreases with temperature (Ku and Edwards, 1977a; Jordan and Ogren, 1984; Brooks and Farquhar, 1985).
The solubility of O2 in aqueous solutions such as the cytoplasm and the chloroplast stroma is less affected by higher temperatures than the solubility of CO2 (Ku and Edwards, 1977b).
Plants exposed to limited water supply reduce gas exchange in order to minimize evaporation. CO2 concentrations around RUBISCO rapidly decrease whereas O2 is available in excess (Lawlor and Fock, 1977; Cornic and Briantais, 1991). This favors the oxygenase over the carboxylase reaction.
Under such conditions, the leaf internal CO2 concentration may approach the apparent CO2 compensation point where CO2 uptake by photosynthesis equals CO2 release by photorespiration and respiration. This finally results in an inability of the plant to grow autotrophically (Sage, 2004).
1.2. Photorespiration: An Early Case Study for Arabidopsis Genetics
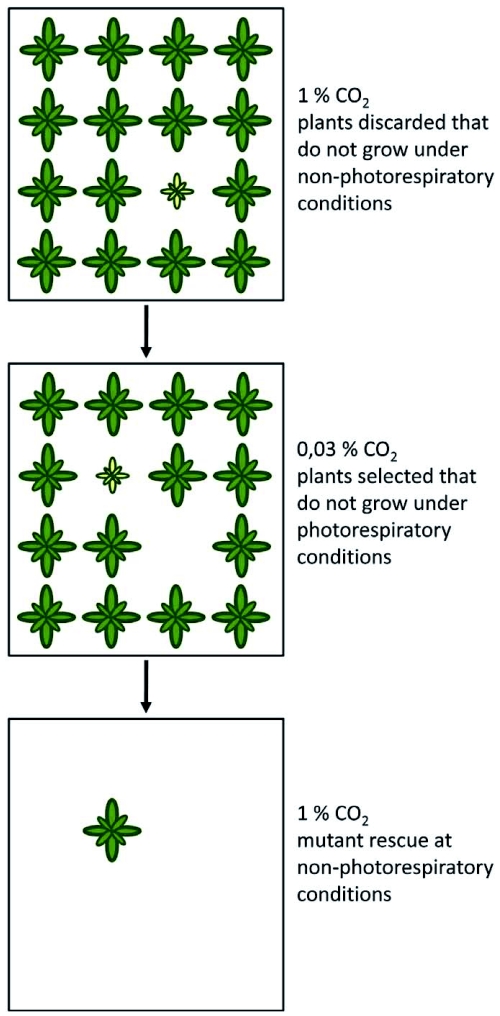
In the early 1970s, Arabidopsis was introduced as a plant model for genetic analyses. Before, the genetics of other plants such as barley or maize had been studied extensively, but a plant with features of a model organism was required: These features included a small genome size, efficient inbreeding, a small space required for growth and a short generation time (Redei, 1975). The necessity of a genetic approach in photorespiration research and the benefits of the Arabidopsis system were early recognized by Chris and Shauna Somerville in William Ogren's laboratory. At that time, there were still ongoing debates whether the substrate for photorespiration was produced by the oxygenase activity of RUBISCO and which biochemical reaction resulted in the release of CO2. The conflicting scientific hypotheses are excellently reviewed in Ogren (2003). The Somervilles' approach was to chemically mutagenize Arabidopsis seeds and to grow the M2 generation derived from this mutagenesis at 1 % CO2 (see Figure 2). The rate of photorespiration was very low under these conditions, thus, mutants in the pathway were expected to grow normally. At seedling stage, all plants showing growth defects or chlorosis were discarded to remove mutants with developmental defects apart from photorespiration. The remaining plants were shifted to normal air with a CO2 concentration of approximately 0.03 %. Plants that developed chlorosis under these conditions were identified as potential mutants in components of the photorespiratory pathway. This was again verified by shifting plants back to 1 % CO2 where photorespiratory mutants should recover from growth inhibition. Using this strategy, they identified “dozens” of plants with an inheritable conditional chlorotic phenotype at ambient CO2 concentrations (Somerville, 2001). Genetic mapping and map-based isolation of the mutated gene (Jander et al., 2002; Peters et al., 2003) was not straightforward in Arabidopsis at these times, thus, enzymatic assays, metabolite profiles and gas exchange measurements were used to characterize the nature of the genetic lesion causing oxygen sensitivity. The first published mutant carried a mutation in phosphoglycolate phosphatase (PGLP, Somerville and Ogren, 1979) which indicated that phosphoglycolate is indeed the source substance for photorespiration. Together with earlier results that molecular oxygen is a competitive inhibitor of CO2 fixation and that RUBISCO can form phosphoglycolate from ribulose-1,5 bisphosphate and O2 (Bowes et al., 1971; Bowes and Ogren, 1972), this result unequivocally linked photorespiration to the oxygenase activity of RUBISCO. Shortly after, analysis of a mutant in mitochondrial serine hydroxymethyltransferase (SHM) defined the major biochemical reaction resulting in photorespiratory CO2 release (Somerville and Ogren, 1981). Mutants deficient in chloroplastic glutamate-oxoglutarate aminotransferase (GLU, Somerville and Ogren, 1980b) and a chloroplast dicarboxylate transporter (DIT, Somerville and Ogren, 1983) confirmed the earlier observations by Keys et al. (1978) that ammonia released during photorespiration is refixed in the chloroplast. These and other mutants are described in more detail in chapter 2. This screen is one of the first examples where the power of Arabidopsis as a tool for plant genetic studies was proven. However, a similar screen performed shortly later in barley identified beside mutants known from Arabidopsis additional mutants in catalase (CAT, Kendall et al., 1983) and glutamine synthetase (GS, Wallsgrove et al., 1987) that have not been recovered in the Arabidopsis screens. Mutants for some of the major photorespiratory genes were never identified. Today, we know that this was in part due to genetic redundancy, e.g. glycolate oxidase (GO) is encoded by multiple genes (Reumann, 2004). Other genes are also required for growth under non-photorespiratory conditions (Engel et al., 2007) or alternative pathways can partially replace the deleted activity (Niessen et al., 2007; Timm et al., 2008). The latter case is discussed in more detail in chapter 4. In summary, the mutant approach clarified many open questions about the major photorespiratory pathway in Arabidopsis. It is now evident that most of the components are shared and that photorespiration is required by all oxygenic photosynthetic organisms (see chapter 5).
Figure 2:
Screening for photorespiratory mutants of Arabidopsis.
A segregating M2 population is grown at a high CO2 concentration that suppresses photorespiration. Stunted or chlorotic plants at this stage probably carry defects in genes unrelated to photorespiration and are discarded. Healthy plants that become chlorotic after the population was shifted to atmospheric CO2 concentrations are candidates for photorespiratory mutants. These mutants are rescued by shifting plants back to a high CO2 concentration.
The work on photorespiration during these years was motivated by the expectation that a reduction of photorespiration could significantly improve plant productivity (Zelitch and Day, 1973; Kelly and Latzko, 1976). However, attempts to identify second-site mutations that could alleviate the deleterious effects of a disrupted photorespiratory pathway were never successful (Somerville and Ogren, 1982b). Metabolic engineering approaches now indicate that deviation of some of the glycolate from photorespiration into novel pathways might indeed enhance biomass production and possibly yield (see chapter 6). Moreover, numerous studies describe photorespiration as a beneficial pathway that supports growth under stress conditions (Wingler et al., 2000) or that provides metabolites for other basal metabolic pathways of the plant (Novitskaya et al., 2002). These positive aspects of photorespiration are exemplified in chapter 3.
2. THE MAJOR PHOTORESPIRATORY PATHWAY IN ARABIDOPSIS
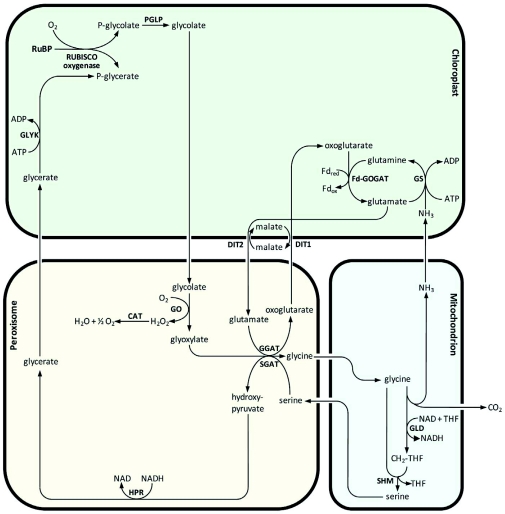
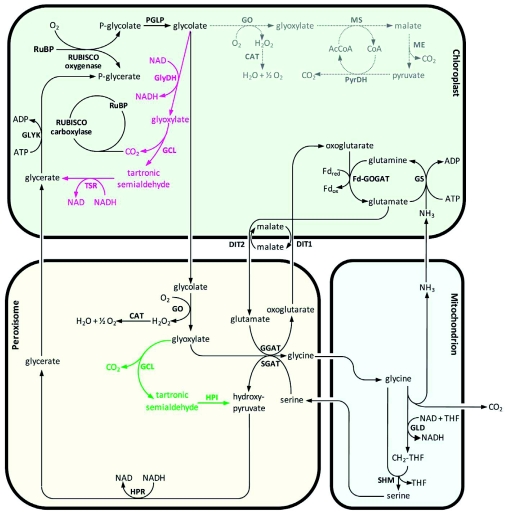
In this chapter, we describe the biochemical pathway that converts the vast amount of 2-phosphoglycolate (PG) to 3-phosphoglycerate (PGA). The pathway components were mainly identified by molecular and biochemical studies in Arabidopsis. Figure 3 provides an overview of the major photorespiratory pathway. Each specific reaction is discussed below.
Figure 3.
The major photorespiratory pathway in Arabidopsis.
Overview of the major photorespiratory pathway in Arabidopsis. For details and chemical structures, see text. RuBP, ribulose-1,5-bisphosphate; RUBISCO, RuBP carboxylase/oxygenase; PGLP, phosphoglycolate phosphatase; GO, glycolate oxidase; CAT, catalase; GGAT, glyoxylate:glutamate aminotransferase; SGAT, serine:glyoxylate aminotransferase; DIT1, dicarboxylate transporter 1; DIT2, dicarboxylate transporter 2; GLD, glycine decarboxylase; SHM, serine hydroxymethyltransferase; HPR, hydroxypyruvate reductase; GLYK, glycerate kinase; GS, glutamine synthetase; Fd-GOGAT, glutamine:oxoglutarate aminotransferase; THF, tetrahydrofolate; CH2-THF, 5, 10-methylene-THF. The stoichiometry of the reactions is not included.
2.1. Enzymatic Reactions in the Major Photorespiratory Pathway
2.1.1. Dephosphorylation of 2-Phosphoglycolate
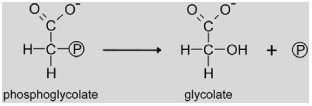 |
2-Phosphoglycolate phosphatase (PGLP, EC 3.1.3.18) catalyzes the entry reaction of photorespiration, dephosphorylation of phosphoglycolate formed by the oxygenase activity of RUBISCO to glycolate. Knock-outs of this gene requiring high CO2 for survival have been identified in both the Arabidopsis and barley mutant screens (see chapter 1) indicating that the functional PGLP is encoded by a single gene. The Arabidopsis mutant CS119 revealed oxygen-sensitive photosynthesis, strong accumulation of phosphoglycolate and absence of PGLP activity (Somerville and Ogren, 1979). The responsible gene has been recently identified by Schwarte and Bauwe (2007) using a candidate gene approach. T-DNA insertion lines for two (At5g36700 encoding AtPGLP1 and At5g47760 encoding AtPGLP2) of a total of 13 annotated potential Pglp genes in Arabidopsis were selected based on homology of the deduced protein to the afore identified Chlamydomonas PGLP. Only one of the two lines showed the typical conditional lethal phenotype. When this gene was sequenced in mutant CS119 lacking PGLP activity, a splice site mutation was identified that resulted in the formation of non-functional transcripts. Thus, At5g36700 encodes the photorespiratory PGLP in Arabidopsis.
There is relatively little known about the regulation of PGLP in Arabidopsis. The gene is expressed in leaves and green parts of flowers. Transcription of the gene is induced by light and repressed by osmotic stress or low nitrate availability (Foyer et al., 2009). Biochemical analyses have been performed with purified enzyme form spinach, tobacco, Chlamydomonas and cyanobacteria. These analyses revealed an usual requirement for both cations such as Mg2+ and anions such as Cl- for enzymatic activity (Husic and Tolbert, 1984; Norman and Colman, 1991). It was suggested that the phosphate moiety of phosphoglycolate is transferred to the protein and that the Cl- ion is required for hydrolysis of the reaction intermediate (Seal and Rose, 1987). Whereas the spinach enzyme was allosterically inhibited by ribose-5-phosphate (Husic and Tolbert, 1984), activation by the same compound was described for the Chlamydomonas enzyme in another study (Mamedov et al., 2001).
2.1.2. Glycolate metabolism in the peroxisome
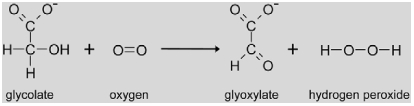 |
Glycolate oxidase (GO, EC 1.1.3.15) belongs to the class of flavindependent 2-hydroxy acid oxidases and catalyzes the oxidation of glycolate to glyoxylate. Molecular oxygen is used as a co-substrate and, thus, H2O2 is produced by the enzyme. The Go gene family in Arabidopsis contains 5 members of which three are potentially localized in the peroxisome (Reumann, 2004; Reumann et al., 2007), Two of these genes (At3g14415 and At3g14420) are transcriptionally co-regulated with other photorespiratory genes with regard to tissue-specific expression as well as positive regulation by light and nitrogen (Reumann and Weber, 2006; Foyer et al., 2009). Probably because of this genetic redundancy, no mutants were identified in the classical Arabidopsis and barley screens (Somerville and Ogren, 1982b; Blackwell et al., 1988). In tobacco and rice, lines with reduced GO activity were generated by co-suppression (Yamaguchi and Nishimura, 2000) and inducible antisense expression (Xu et al., 2009). These studies revealed a positive and linear correlation between GO-activity and the net photosynthetic rate. Once photosynthetic rates fell below 40% of control plants, photoinhibition occurred indicating that GO can exert a strong regulatory effect on photosynthesis, possibly through a reduction in RUBISCO activation state (Xu et al., 2009). Beside metabolic regulation, GO activity is also induced by pathogen attacks. This is probably due to the formation of H2O2 and its importance as a second messenger in plant defense reactions (Taler et al., 2004, see also chapter 3).
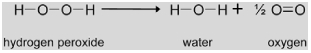 |
H2O2 produced by the glycolate oxidase reaction is disproportionated to H2O and O2 by catalase (CAT, EC 1.11.1.6) in order to prevent accumulation of reactive oxygen species. In Arabidopsis, three catalase genes are known, but only one gene (Cat2, At4g35090) encodes the major enzyme involved in photorespiration. In ambient air, a mutation in Cat2 caused severely decreased rosette biomass, intracellular redox perturbation and activation of oxidative signaling pathways. These effects were absent when mutant plants were grown at high CO2 concentrations (Queval et al., 2007). Cat2 gene expression is light-stimulated and clock-regulated with an induction in the early morning (Zhong et al., 1997; Queval et al., 2007).
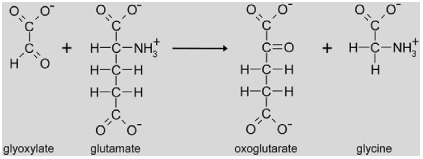 |
The next reaction step in the photorespiratory pathway is the transamination of glyoxylate to glycine by glutamate:glyoxylate aminotransferase (GGAT, EC 2.6.1.4). In Arabidopsis, two loci encode peroxisomal glutamate:glyoxylate aminotransferases (At1g23310 encoding GGAT1 and At1g70580 encoding GGAT2) (Igarashi et al., 2003; Liepman and Olsen, 2003; Igarashi et al., 2006). Ggat1 knock-outs show a weak photorespiratory phenotype and survive in air at moderate light intensities. Residual GGAT activity can be detected in mutant leaf extracts (Igarashi et al., 2003). Furthermore, both genes exhibit a differential diurnal expression pattern. Whereas Ggat2 is induced at the beginning of the photoperiod, Ggat1 is rather repressed. However, GGAT activity in leaves remains constant during the day (Igarashi et al., 2006) suggesting that both loci contribute to the total leaf GGAT content. Ggat1 is not only highly expressed in leaves but also in roots, indicating that its function is not restricted to photorespiration.
2.1.3. Mitochondrial conversion of glycine to serine
The combined reaction of glycine decarboxylase (GLD) and serine hydroxymethyltransferase (SHM) represents the mitochondrial part of the major photorespiratory pathway. Glycine is decarboxylated and deaminated by the multienzyme complex glycine decarboxylase yielding CO2, NH3, NADH and 5, 10-methylenetetrahydrofolate. The latter is used by SHM as a substrate for the transfer of an activated C1 moiety to another molecule of glycine resulting in the formation of serine.
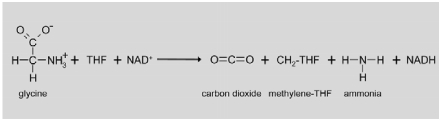 |
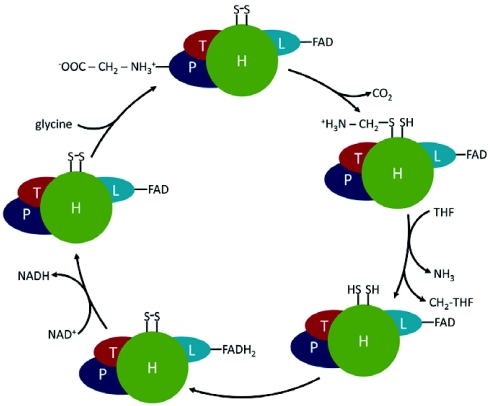
The GLD reaction involves four different proteins (Figure 4). P-protein (EC 1.4.4.2) is a pyridoxal-5-phosphate-containing homodimer, that catalyzes the actual glycine decarboxylation reaction (Bauwe and Kolukisaoglu, 2003). The remaining amino methylene part of glycine is transferred to the distal sulphur atom of the oxidized lipoamide arm of H-protein (Douce et al., 2001). H-protein is not an enzyme component of the complex but a cofactor that interacts with all three enzymes of the complex. The amino methylene group bound to the H-protein is further metabolized by the monomeric aminomethyltransferase T-protein (EC 2.1.2.10) (Douce and Neuburger, 1999). During this reaction, NH3 is released and a methylene group is transferred to tetrahydrofolate resulting in the formation of 5,10-methylene tetrahydrofolate (CH2-THF) (Bauwe and Kolukisaoglu, 2003). Concomitantly, the lipoamide arm of H-protein becomes fully reduced and needs to be re-oxidized for the next reaction cycle. This reaction is catalyzed by L-protein, a homodimeric dihydrolipoamide dehydrogenase (EC 1.8.1.4). Electrons are first transferred to the co-factor FAD that is re-oxidized by NAD+ resulting in the formation of NADH (Bauwe and Kolukisaoglu, 2003). The four proteins are present in unequal molar amounts in the complex. For pea, a subunit stoichiometry of 4P:27H:9T:2L was observed (Oliver et al., 1990), but the functional relevance of this composition of the complex is still elusive. In Arabidopsis, the four proteins are encoded by eight genes, two genes each for P-protein (At4g33010 and At2g26080) and L-protein (At3g17240 and At1g48030), three genes for H-protein (At2g35370, At2g35120 and At1g32470) and one gene for T-protein (At1g11860) (Bauwe and Kolukisaoglu, 2003). Most of these genes are strongly light regulated and some are repressed by low nitrogen concentrations, like most other genes contributing to the photorespiratory pathway (Foyer et al., 2009 and http://www.genevestigator.com).
Figure 4:
The glycine decarboxylase complex in Arabidopsis.
Glycine cleavage by the glycine decarboxylase (GLD) complex. GLD consists of P-, H-, T- and L-protein. THF = tetrahydrofolate; CH2-THF = 5,10-methylene THF. Glycine binds to the pyridoxalphosphate co-factor (not shown) of P-protein, is decarboxylated and transferred to H-protein. T-protein releases NH3 and transfers the methylene group to THF. L-protein catalyzes the re-oxidation of H-protein. The subunit stoichiometry is not shown and the size of the symbols does not imply the size of the proteins. For further details, see main text.
The classical Arabidopsis mutant gld1 showing a photorespiratory phenotype related to loss of function of the GLD complex (Somerville and Ogren, 1982a) was characterized on the molecular level by Ewald et al. (2007). The mutant is deficient in a 3-oxoacyl-(acyl-carrier-protein) synthase (At2g04540, Mt-kas1) that is required for lipoylation of H-protein, Some residual lipoylation by an independent second pathway might be sufficient to rescue the phenotype under non-photorespiratory conditions. In contrast, the combined knock-out of the two genes encoding P-protein is lethal under all growth conditions (Engel et al., 2007) suggesting that the GLD reaction is not only essential for photorespiration, but also for the synthesis of activated C1 moieties required for nucleic acid and amino acid metabolism (Cossins and Chen, 1997)
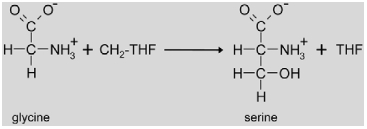 |
The photorespiratory SHM enzyme (EC. 2.1.2.1) is encoded by a single gene (At4g37930) in Arabidopsis. A knock-out of this gene is lethal at ambient CO2 levels, but mutants grow normally at high CO2 (Somerville and Ogren, 1981; Voll et al., 2006). The mRNA abundance is positively regulated by light and the circadian clock, and the gene is not expressed in roots (McClung et al., 2000). Recently, a second class of mutants deficient in mitochondrial SHM activity was identified (Jamai et al., 2009). A mutation in Glu1 (At5g04140) which encodes ferredoxin-dependent glutamine:oxoglutarate aminotransferase (Fd-GOGAT (TAIR abbreviation: GLU); EC 1.4.7.1) reduced mitochondrial SHM activity. It was known before that this protein is involved in ammonia re-assimilation in the chloroplast during photorespiration (Coschigano et al., 1998, see also below), but the new results indicate that Fd-GOGAT is targeted not only to the chloroplast but also to the mitochondria where it physically interacts with SHM. This interaction is necessary for photorespiratory SHM activity (Jamai et al., 2009).
An additional level of regulation of the GLD/SHM complex was recently identified by characterization of plants that were unable to convert 10-formyl-THF, an intermediate of C1 metabolism (Hanson and Roje, 2001), to formate and THF (Collakova et al., 2008). In these plants, the isomer 5-formyl-THF accumulated and inhibited photorespiration resulting in retarded growth and glycine accumulation. Thus, 5-formyl-THF might act as a physiological regulator of GLD/SHM activity in vivo.
2.1.4. Formation of glycerate in the peroxisome
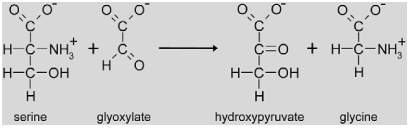 |
Back in the peroxisome, serine is deaminated to hydroxypyruvate by serine:glyoxylate aminotransferase (SGAT (TAIR abbreviation: AGT), EC 2.6.1.45). The amino group is used for the concomitant amination of glyoxylate to glycine. This is important for the stoichiometry of photorespiration, because only one amino donor for the GGAT reaction is recycled during NH3 re-assimilation (see below), but two glycine are required for the mitochondrial formation of serine. In Arabidopsis, SGAT is encoded by a single gene (At2g13360) and its deletion results in plants that are unviable under normal atmospheric conditions (Somerville and Ogren, 1980a; Liepman and Olsen, 2001). A similar phenotype was also described for a barley SGAT mutant (Murray et al., 1987). The Arabidopsis gene is induced by light and repressed by low nitrogen like most of the other photorespiratory genes (Foyer et al., 2009), but transcripts are also detected in flowers and siliques apart from leaves (Liepman and Olsen, 2001).
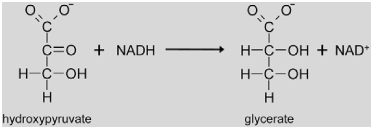 |
Hydroxypyruvate resulting from the SGAT reaction is reduced to glycerate by hydroxypyruvate reductase (HPR, EC 1.1.1.29). In Arabidopsis, the peroxisomal reaction is catalyzed by HPR1 (At1g68010) using NADH as a co-factor (Givan and Kleczkowski, 1992). The Hpr1 gene shows similar regulatory properties like Sgat with positive light regulation and repression under low nitrogen conditions (Foyer et al., 2009). Reducing equivalents for the reaction are probably imported into the peroxisome by a malate/oxaloacetate shuttle and released by a peroxisomal malate dehydrogenase (pMDH, Cousins et al., 2008). However, Arabidopsis mutants with deletions in the genes encoding HPR1 (Timm et al., 2008) and pMDH (Pracharoenwattana et al., 2007) as well as barley HPR mutants (Murray et al., 1989) do not show drastic photorespiratory phenotypes. These data suggest the existence of alternative pathways for the conversion of hydroxypyruvate to glycerate or other metabolites that are further discussed in chapter 4.
2.1.5. Regeneration of phosphoglycerate
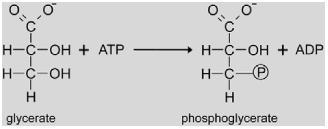 |
The chloroplastic D-glycerate 3-kinase (GLYK, EC 2.7.1.31) catalyzes the phosphorylation of glycerate to 3-phosphoglycerate with ATP as a co-substrate. As no mutants were identified in the classical screens, the gene was instead isolated by purification of the enzymatic activity from Arabidopsis leaf extracts and partial sequencing of the purified protein (Boldt et al., 2005). GLYK is encoded by a single copy gene (At1g80380). Knock-out of this gene results in a conditionally lethal photorespiratory phenotype. The enzyme is the founder of a structurally and phylogenetically novel protein family that is different from known glycerate kinases of non-photosynthetic organisms (Boldt et al., 2005), albeit sequence homologues are also found in yeast (Bartsch et al., 2008). Transcriptional regulation of the Glyk gene is different from most of the other photorespiratory genes. Particularly, Glyk gene expression is not activated by light (Foyer et al., 2009).
2.1.6. Ammonia re-assimilation
During the GLD reaction, one-quarter of the carbon that entered the photorespiratory cycle and an equimolar amount of NH3 are released. Re-assimilation of NH3 takes place in the chloroplast through the sequential action of two enzymes, glutamine synthetase (GS; EC 6.1.3.2) and ferredoxin-dependent glutamine:oxoglutarate aminotransferase (Fd-GOGAT (TAIR abbreviation: GLU); EC 1.4.7.1) (Keys, 2006). NADH-GOGAT (EC 1.4.1.14), that is additionally found in chloroplasts, does not play a significant role in photorespiration (Coschigano et al., 1998).
The net reaction of the GS/Fd-GOGAT cycle is the animation of oxoglutarate to glutamate consuming one ATP and two reduced ferredoxin.
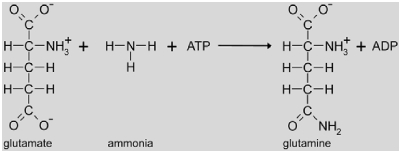 |
First, GS catalyses the amination of glutamate to glutamine which requires one ATP. GS mutants have not been identified in Arabidopsis, but in barley (Wallsgrove et al., 1987). The barley mutant showed the typical requirement for elevated CO2 concentrations emphasizing the major contribution of photorespiration to plant NH3 production and re-assimilation (Keys, 2006). Six Arabidopsis genes encode GS enzymes, but only GS2 (encoded by At5g35630) is targeted to the chloroplast (Peterman and Goodman, 1991). Transcriptional regulation of the gene for GS2 is similar to most photorespiratory genes including light induction and repression by low nitrogen (Peterman and Goodman, 1991; Foyer et al., 2009).
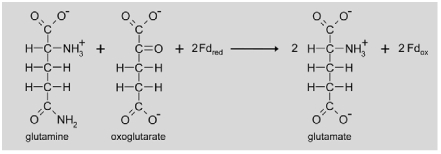 |
Fd-GOGAT uses the glutamine formed by GS as an amino donor for the transamination of oxoglutarate. Two molecules of glutamate are the product of this reaction. Whereas one glutamate is used as a substrate for the next NH3 assimilation reaction by GS, the second is transferred to the peroxisome for transamination of glyoxylate to glycine. Arabidopsis contains two expressed genes for Fd-GOGAT (Glu1 (At5g04140) and Glu2 (At2g41220)), but only mutation of the Glu1 gene resulted in a photorespiratory phenotype (Coschigano et al., 1998). Glu1 mRNA shows highest abundance in leaves and is specifically induced by light and sucrose. In contrast, Glu2 is only slightly expressed in leaves, but the mRNA preferentially accumulates in roots (Coschigano et al., 1998). Although the photorespiratory nitrogen cycle is closed in theory as all the re-fixed NH3 is required for transamination of glyoxylate, some excess NH3 might escape fixation in the GS/Fd-GOGAT cycle. There is physiological evidence that formation of carbamoyl phosphate and ultimately arginine provides an alternative route for NH3 re-assimilation (Potel et al., 2009).
As described above, Fd-GOGAT has been shown to be dual targeted to the chloroplast and the mitochondrion (Jamai et al., 2009). Mitochondrial localization is seemingly not important for NH3 re-assimilation, but rather for regulation of SHM activity. However, dual targeting has also been suggested for GS2 based on the localization of overexpressed fusions of the protein to green fluorescent protein (Taira et al., 2004). The presence of both enzymes would imply that photorespiratory NH3 could also be directly re-assimilated in the mitochondrion. On the other hand, neither Fd-GOGAT nor GS have been identified in proteome analyses of the mitochondrial matrix (Eubel et al., 2007; Jamai et al., 2009). This at least implies that the proteins are scarce in the mitochondrion which would again support a regulatory instead of a catalytic role. Further research is necessary to clarify the role of these dual targeting events in photorespiration.
2.2. Transport of Photorespiratory Intermediates
Due to the complex biochemistry of photorespiration and the involvement of three different compartments, numerous transport steps over membranes are necessary for function of the pathway. However, most transporters have not been identified or even biochemically characterized in detail. The available information on the transporters for photorespiratory intermediates is summarized below. Data on the oxaloacetate/malate shuttles in the membranes of all organelles that are indirectly involved in photorespiration by transferring reducing equivalents during photorespiration (Reumann and Weber, 2006) are not included.
2.2.1. Chloroplast transport
The first transmembrane transport process necessary during the photorespiratory pathway is the transport of glycolate out of the chloroplast into the cytosol. This transporter has not been characterized in Arabidopsis, but was studied before in pea and spinach. Substrate specificity of the chloroplast glycolate transporter was analyzed in isolated pea chloroplasts. The uptake of radiolabeled glycolate into isolated chloroplasts was strongly inhibited by glycerate (Howitz and McCarty, 1983). Based on this, it was concluded that the same transporter is responsible for both glycolate and glycerate transport. This was further supported by reconstitution of transport activity into liposomes. The transporter exchanged glycolate for glycerate (Howitz and McCarty, 1991). The gene encoding this transporter is unknown to date.
During the re-assimilation of ammonia, oxoglutarate enters the chloroplast whereas glutamate leaves the chloroplast. A two-translocator model exists for these transport processes (Weber and Flügge, 2002). The model includes the oxoglutarate/malate translocator (At5g12860 encoding DIT1) and the glutamate/malate translocator (At5g64290 encoding DIT2.1) (Woo et al., 1987). During screening for photorespiratory mutants, Somerville and Ogren (1983) identified a mutant in a potential dicarboxylate translocator that has later been shown to carry a mutation in the gene for DIT2.1 (Renne et al., 2003). In contrast, a knockout of the gene for DIT2.2 (At5g64280) did not result in an apparent phenotype (Renne et al., 2003) and the encoded protein did not show dicarboxylate transport activity (Taniguchi et al., 2002). Hence, its function remains unknown and an involvement in photorespiration is unlikely. Dit1 mutants were not identified in Arabidopsis, but antisense analyses in tobacco verified a role for this protein in photorespiration (Schneidereit et al., 2006). Whereas Dit1 clusters with most other photorespiration genes in transcriptional profile analyses, Dit2.1 shows a broader expression profile especially with regard to tissue specificity and light regulation (Renne et al., 2003; Foyer et al., 2009).
2.2.2. Peroxisomal transport
The high fluxes of photorespiratory intermediates, such as glycolate, glycerate, glutamate, oxoglutarate, glycine, and serine across the membrane of leaf peroxisomes require efficient transport systems (Reumann and Weber, 2006). The presence of an anion-selective porin-like channel was observed in electrophysiological experiments with purified peroxisome membranes in spinach and Ricinus (Reumann et al., 1995; Reumann et al., 1997). Further experiments indicated that the channel is permeable for most photorespiratory intermediates (Reumann et al., 1998). However, such peroxisomal porins have not been unequivocally identified in proteomic studies in Arabidopsis, perhaps because they were misclassified as mitochondrial contaminants, but are in fact dual-targeted to both organelles (Kaur et al., 2009).
2.2.3. Mitochondrial transport
During photorespiration, two molecules of glycine are imported into mitochondria for each serine that is exported. Whereas the transporter has not been identified in Arabidopsis, biochemical data are available from spinach leaf mitochondria (Yu et al., 1983). Based on the biphasic transport kinetics observed, an active transport system is postulated only for low substrate concentrations. Both glycine and serine can inhibit transport of the other compound, respectively, suggesting the presence of a single antiport system for the two amino acids. Passive diffusion might dominate over active transport at higher concentrations of glycine and serine (Yu et al., 1983). This could explain why the transporter has never been identified in screens for photorespiratory mutants.
2.3. Energy Balance Sheet
Photorespiration imposes costs on plant energy metabolism. The oxygenase reaction of RUBISCO does not result in energy or carbon gain for the plant. The ribulose-1,5-bisphosphate molecule for this reaction is therefore “wasted”. Our calculation of the energy costs is based on the assumption that this molecule of ribulose-1,5-bisphosphate has to be completely regenerated to reconstitute the “status quo ante”, i.e. the metabolic state of the Calvin cycle before oxygen fixation. Table 1 summarizes these costs. Possible energetic costs of transport processes are unknown.
Table 1.
The energy balance sheet of the oxygenase reaction of RUBISCO and photorespiration.
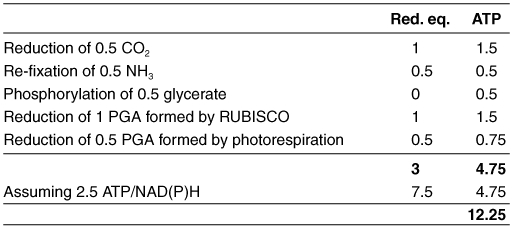
The glycine decarboxylase reaction releases 0.5 CO2 and 0.5 NH3 per phosphoglycolate formed. The carbon atom in CO2 was oxidized from oxidation state +I to +IV during oxygen fixation and glycine decarboxylation and needs to be re-reduced by the carboxylase activity of RUBISCO and the subsequent reactions of the Calvin cycle. This requires 3 ATP and 2 NADPH per CO2 molecule (Benson and Calvin, 1950), thus half of the costs for 0.5 CO2. NH3 has to be re-fixed in the plastidal GS/Fd-GOGAT cycle (see above). For the calculation, 1 reduced ferredoxin is assumed to be equivalent to 0.5 NAD(P)H, because ferredoxin reduction requires one electron, whereas NAD(P)H reduction requires two electrons.
The balance sheet for reducing equivalents is neutral for the subsequent reactions of photorespiration, because one NADH is formed during the re-oxidation of the H protein of GLD and one NADH is used for the oxidation of hydroxypyruvate to glycerate.
Per phosphoglycolate, 0.5 glycerate are formed that are phosphorylated by glycerate kinase for re-integration into the Calvin cycle. This requires 0.5 ATR Both these 0.5 phosphoglycerate (PGA) as well as 1 PGA formed by the oxygenase reaction of RUBISCO are again metabolized in the Calvin cycle for the regeneration of ribulose-1,5 bisphosphate. The Calvin cycle consumes 1 NADPH and 1.5 ATP per PGA. Total costs for undoing oxygen fixation therefore sum up to 4.75 ATP and 3 reducing equivalents.
Reducing equivalents (NAD(P)H) can be converted to energy equivalents (ATP) in the mitochondrial electron transport chain. Exact ratios vary with conditions, but 2.5 ATP are produced from one NADH in average (Ferguson, 1986; Hinkle, 2005). This calculation results in total energy costs of 12.25 ATP.
Based on these calculations, what is the energy burden of the oxygenase reaction on carbon fixation? RUBISCO highly prefers CO2 to O2 as a substrate with a specificity factor of approximately 80 for land plants (Jordan and Ogren, 1981) although this varies between species and is dependent on temperature (Ku and Edwards, 1977a; Jordan and Ogren, 1984). However, atmospheric concentrations of O2 are much higher than CO2 concentrations (250 µM and 8 µM, respectively, at 25 °C) resulting in an oxygen surplus of 31-fold. Therefore, approximately each fourth reaction of RUBISCO is an oxygenase instead of a carboxylase reaction under moderate growth conditions (Ehleringer et al., 1991). Costs for each carbon fixation reaction are 3 ATP and 2 NADPH, or 8 ATP in total if reducing equivalents are converted to energy equivalents as described above. Statistically, the costs for one oxygenation reaction have to be allocated to 3 carboxylations, i.e. 4.1 ATP per carboxylation. This results in extra energy costs of about 50 % for photosynthesis caused by the oxygenase activity of RUBISCO. This calculated number is comparable to the measured about 40–50 % increase of photosynthetic rates in Arabidopsis and other C3 plants in atmospheres containing low O2 concentrations (Hesketh, 1967; Kebeish et al., 2007).
3. THE LIGHT SIDE AND THE DARK SIDE OF PHOTORESPIRATION
As detailed in the last chapter, photorespiration imposes significant extra energy costs on carbon fixation. From this, the view of photorespiration as a wasteful process originated. On the other hand, the pathway rescues ¾ of the carbon in phosphoglycolate that would be otherwise inaccessible for further metabolism. Thus, photorespiration can also be regarded as an important pathway that makes the best of a bad situation caused by RUBISCO's seemingly inevitable oxygenase activity. Beside this, there are several additional arguments for a positive function of photorespiration in plant metabolism:
3.1. Photorespiration Removes Toxic Metabolic Intermediates
Photorespiration is the only pathway in the plant for phosphoglycolate metabolism that would otherwise accumulate. Phosphoglycolate has been shown to strongly inhibit triose phosphate isomerase from pea leaves at concentrations in the low micromolar range (Anderson, 1971). This would interfere with the regeneration of ribulose-1,5-bisphosphate in the Calvin cycle. In addition, phosphoglycolate inhibits the chloroplast phosphofructokinase (Kelly and Latzko, 1976) and, by this, the replenishment of Calvin cycle intermediates from the breakdown of transitory starch. This dual inhibitory function is probably responsible for the low photosynthetic rate of mutants accumulating phosphoglycolate (Somerville and Ogren, 1979).
To our knowledge, no direct inhibitory function has been described for glycolate, the product of phosphoglycolate dephosphorylation, albeit glycolate accumulation has been observed in many photorespiratory mutants. Glycolate is further converted to glyoxylate and this compound is again a strong inhibitor of photosynthesis. Campbell and Ogren (1990) and Chastain and Ogren (1989) reported that micromolar concentrations of glyoxylate inhibited activation of RUBISCO in isolated organelles and in vivo. However, inhibition of the activase enzyme (Campbell and Ogren, 1990) or purified RUBISCO (Cook et al., 1985) required much higher concentrations. Furthermore, glyoxylate was inefficient in rendering the chloroplast envelope permeable to protons as observed for other weak acids before (Flügge et al., 1980). Thus, glyoxylate negatively impacts on the activation state of RUBISCO by a yet unidentified mechanism. The importance of this mechanism is exemplified by the presence of glyoxylate reductase in the cytosol and the chloroplast that might function in protecting the photosynthetic machinery from excess glyoxylate leaking from the peroxisomes (Givan and Kleczkowski, 1992; Allan et al., 2009). Conversely, glyoxylate may be instrumental in reducing RUBISCO activity and, by this, also phosphoglycolate formation under conditions where photorespiration cannot cope with the rates of RUBISCO oxygenase activity. This might be of physiological importance under low nitrogen supply when the transamination reactions converting glyoxylate to glycine are limiting photorespiratory flux. The glyoxylate overflow would simply stop both photosynthesis and photorespiration under such emergency conditions. Alternative mechanisms to metabolize glyoxylate are also discussed explicitly in chapter 4. Taken together, photorespiration is important for the removal of toxic intermediates, but surprisingly little is known about the regulatory function of these intermediates in coordinating photorespiration with other pathways of plant basal metabolism.
3.2. Photorespiration Protects from Photoinhibition
Under stress conditions, such as drought, cold, or high light, NADPH production in the light reactions of photosynthesis often exceeds the demand of the Calvin cycle for reducing power. In that case, energy is dissipated as heat or electrons are transferred from different complexes in the electron transport chain to other acceptor molecules than NADPH. If the acceptor molecule is O2, the reaction results in the formation of reactive oxygen species (ROS, reviewed in Murata et al., 2007). Because of their high reactivity, ROS unspecifically oxidize proteins and lipids. Moreover, ROS also interfere with recycling of specific components of the electron transport chain by inhibiting translation of new proteins (Nishiyama et al., 2004). The chloroplast contains several mechanisms to detoxify ROS. There are compounds that scavenge ROS non-enzymatically like carotenoids and flavonoids, and enzymes, like ascorbate peroxidase, superoxide dismutase and catalase, that detoxify ROS in partly complex pathways (reviewed in Apel and Hirt, 2004). A welcome effect of these pathways is that they ultimately consume reducing equivalents and by this contribute to the provision of fresh NADP+, the terminal electron acceptor of the photosynthetic electron transport chain. Alternatively, excess light energy can be mitigated by the xanthophyll cycle that dissipates the energy of absorbed photons into heat (Yamamoto et al., 1962; Davison et al., 2002). Also photorespiration can act as an electron sink especially under stress conditions (Wingler et al., 2000) by consuming reducing equivalents during the refixation of released ammonia and by exporting reduced components from the chloroplast to the mitochondrion (Igamberdiev and Lea, 2002). This might even result in situations where the mitochondrion becomes over-reduced instead of the chloroplast because of the huge amounts of NADH generated by glycine decarboxylation. To allow for high fluxes through photorespiration under such conditions, mitochondrial electron transport might be uncoupled from ATP synthesis by uncoupling proteins (UCPs) that catalyse proton conductance through the inner mitochondrial membrane and dissipate the energy of the mitochondrial proton gradient as heat (Sweetlove et al., 2006). The importance of photorespiration in protection from photoinhibition was directly tested by Kozaki and Takeba (1996) by measuring the rates of photoinhibition in plants with enhanced or diminished capacities for photorespiration caused by manipulation of the levels of glutamine synthetase (GS). Plants with an improved capacity for photorespiration because of increased GS levels were more tolerant to high light stress. Accordingly, photorespiratory mutants of Arabidopsis showed enhanced photoinhibition (Takahashi et al., 2007). In this study, the authors provide evidence that photoinhibition was rather caused by suppression of the repair of photosystem II (PSII) than by an increase in PSII damage.
Any mechanism that uses photorespiration as a pressure relief valve for reducing power from the chloroplast would ultimately run out of the O2 acceptor molecule ribulose-1,5-bisphosphate because of the net carbon loss associated with photorespiration. Replenishment of O2 acceptor molecules is probably brought about by starch breakdown (Weise et al., 2006). As a consequence, the chloroplast stroma is seemingly less reduced under conditions of high photorespiration than during maximum photosynthesis (Weise et al., 2006). Still, the relative importance of photorespiratory export of reducing equivalents among the multitude of different processes that protect plants from photoinhibition is a matter of ongoing debates and might strongly depend on the conditions (cf. Osmond et al., 1997). Beside its function as an export mechanism for reducing power, photorespiration is also directly involved in the synthesis of sufficient amounts of glycine that is required to cover the strongly enhanced demands for glutathione during stress (Noctor et al., 1998; Noctor et al., 1999).
3.3. Photorespiration Supports Plant Defense Reactions
H2O2 is known to be a signaling molecule in plants involved in both biotic and abiotic stress responses. Because of its participation in multiple pathways, H2O2 is especially suitable for mediating crosstalk between the different resistance mechanisms (reviewed in Neill et al., 2002). During pathogen attack, H2O2 has multiple roles in the so called oxidative burst. On the one hand, it triggers the plant defense response system, including cell wall strengthening and activation of phytoalexin biosynthesis (Wu et al., 1997). On the other hand, H2O2 can damage the pathogen by its reactive potential. Ultimately, H2O2 triggers the hypersensitive response and the attacked cell eventually undergoes programmed cell death (Heath, 2000). As photorespiration is a major source of H2O2 in plants, an involvement in resistance reactions was often discussed (Chamnongpol et al., 1998). This was recently confirmed by the identification of “enzymatic resistance” genes in melon (Taler et al., 2004). Efficient resistance of a wild melon line to the oomycete Pseudoperonospora cubensis was shown to be caused by constitutively enhanced expression of two genes encoding peroxisomal glyoxylate aminotransferases. A mechanism was suggested, in which the elevated activity of these enzymes positively feeds back on glycolate oxidase, thereby enhancing the capacity for H2O2 release.
3.4. Photorespiration is Intimately Integrated Into Primary Metabolism
Photorespiration is obviously tightly integrated into plant primary metabolism. Most photorespiratory intermediates are also part of other metabolic pathways and photorespiration significantly contributes to the synthesis of several amino acids (Novitskaya et al., 2002). Photorespiration connects the metabolic compartments of the cell and is therefore ideally suited to transport information about the energetic state between these compartments (Nunes-Nesi et al., 2008). The conversion of glycine to serine in the mitochondrion is probably essential to supply serine for the cytosolic production of C1-compounds that are in turn required for biosynthetic processes such as methionine synthesis (Mouillon et al., 1999; Engel et al., 2007). Beside this, specific roles have been suggested for photorespiration under certain growth conditions:
Sucrose and starch are the major sinks for photosynthetic products in leaves. Under conditions of high light and carbon availability, synthesis of these sink compounds might not keep pace with production of phosphorylated intermediates in the Calvin cycle. This results in an imbalance of phosphate release during synthesis of sucrose or starch and phosphate use during the synthesis of Calvin cycle intermediates that may ultimately limit photosynthesis (Harley and Sharkey, 1991). Under such conditions, fixation of oxygen by RUBISCO and the subsequent reactions of photorespiration might provide the cell with an additional method for the synthesis of sink products such as glycine and serine instead of sucrose and starch (Harley and Sharkey, 1991). This is consistent with the finding that source leaves start to export glycine and serine instead of sucrose to sink leaves under conditions of high photorespiration (Madore and Grodzinski, 1984).
Another connection of photorespiration and nitrogen metabolism was observed by Rachmilevitch et al. (2004). They found that nitrate assimilation was inhibited in an atmosphere containing 2% O2 where photorespiration is repressed. They proposed a mechanism how photorespiration provides reducing equivalents for cytoplasmic nitrate reductase: The reducing equivalents are exported from the chloroplast into the cytoplasm via the malate/oxaloacetate shuttle to provide reducing power for peroxisomal hydroxypyruvate reduction. Part of these reducing equivalents might then be diverted for nitrate reduction. Reducing equivalents for peroxisomal reactions can alternatively be provided by mitochondria that synthesize the required amounts of NADH during photorespiration (Raghavendra et al., 1998). However, there is evidence that part of the mitochondrial reducing equivalents may rather be used for ATP synthesis (Krömer and Heldt, 1991). The relative importance of these processes requires detailed investigation.
4. METABOLIC COMPLEXITY: ALTERNATIVE PHOTORESPIRATORY PATHWAYS IN ARABIDOPSIS
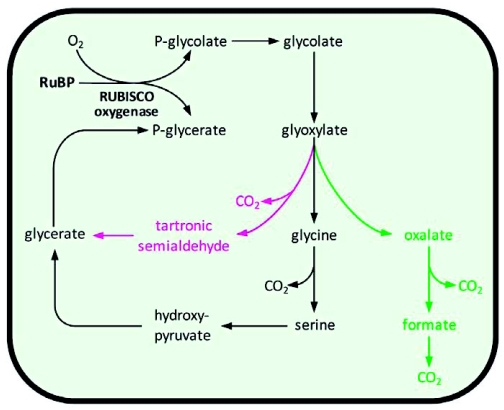
Conflicts about the major routes in phosphoglycolate metabolism were decided by the mutant screens in Arabidopsis and barley (see chapters 1 and 2). Indications for alternative routes were mostly not followed afterwards. However, significant new information about such alternative pathways accumulated during the last few years indicating a metabolic flexibility of photorespiration that is much higher than predicted from the earlier biochemical studies. An overview of possible alternative pathways is given in Figure 5.
Figure 5:
Possible alternative photorespiratory pathways in Arabidopsis.
Overview of the major photorespiratory pathway (black) and four possible alternative photorespiratory pathways (dotted grey, green, red, and dashed grey) in Arabidopsis that are described in the main text. RuBP, ribulose-1,5-bisphosphate; RUBISCO, RuBP carboxylase/oxygenase; PGLP, phosphoglycolate phosphatase; GO, glycolate oxidase; CAT, catalase; GGAT, glyoxylate:glutamate aminotransferase; SGAT, serine:glyoxylate aminotransferase; DIT1, dicarboxylate transporter 1; DIT2, dicarboxylate transporter 2; GLD, glycine decarboxylase; SHM, serine hydroxymethyltransferase; HPR, hydroxypyruvate reductase; GLYK, glycerate kinase; GS, glutamine synthetase; Fd-GOGAT, glutamine:oxoglutarate aminotransferase; GlyDH, glycolate dehydrogenase; AT, aminotransferase; FDH, formate dehydrogenase; FTHFS, 10-formyl-THF-synthetase;CYC, 5,10-methenyl-THF-cyclohydrolase;DHY, 5,10-methenyl-THF-dehydrogenase;THF, tetrahydrofolate; HCO-THF, formyl-THF; CH3-THF, methenyl-THF; CH2-THF, methylene-THF. The stoichiometry of the reactions is not included.
4.1. Possible Alternative Pathways in the Chloroplast
An endogenous pathway for glycolate metabolism in the chloroplast was proposed by Goyal and Tolbert (1996). Using biochemical analyses, they demonstrated the existence of a salicylhydroxamic acid (SHAM)-inhibited light-dependent glycolate-quinone oxidoreductase system that is associated with thylakoid membranes of the chloroplasts of both algae and spinach. By this system, electrons from glycolate oxidation could be transferred to the photosynthetic electron transport chain and used for ATP synthesis. In this scenario, glyoxylate would be reduced back to glycolate by the plastidal glyoxylate reductase (Givan and Kleczkowski, 1992). This would provide an alternative sink for excess electrons under stress conditions (Allan et al., 2009, see also chapter 3). The presence of a glycolate-oxidizing enzyme in chloroplasts of Arabidopsis was recently confirmed by Kebeish et al. (2007). However, the cycle of glycolate oxidation and glyoxylate reduction does not consume glycolate and is therefore not suitable as an alternative pathway for the conversion of the products of RUBISCO's oxygenase activity. Instead, glyoxylate coming from plastidal glycolate oxidation might be further oxidized to CO2 in the chloroplast (Kisaki and Tolbert, 1969; Zelitch, 1972; Oliver, 1981, dotted grey pathway in Figure 5). Slow CO2 release from glyoxylate in chloroplasts from a set of plant species was shown to be stimulated by light and oxygen (Kisaki and Tolbert, 1969; Zelitch, 1972). Although glycolate was not converted by chloroplasts to CO2 in these studies, later analysis in Arabidopsis also revealed coupling of glycolate oxidation and CO2 release (Kebeish et al., 2007). Conversion of glyoxylate to CO2 might be catalyzed by plastidal pyruvate decarboxylase that also accepts glyoxylate as a substrate (Davies and Corbett, 1969). Alternatively, glyoxylate can react non-enzymatically with H2O2 or O2- to formate (Elstner and Heupel, 1973; Halliwell and Butt, 1974, see also chapter 4.2). Such reactive oxygen species are produced in the chloroplast especially under stress conditions (see chapter 3). Formate could be further oxidized to CO2 by a formate dehydrogenase that is dual-targeted to mitochondria and chloroplasts in Arabidopsis (Olson et al., 2000; Herman et al., 2002). Together, this would result in a low-capacity pathway for glycolate oxidation in the chloroplast that is more energy-efficient than the major photorespiratory pathway and that enhances plastidal CO2 concentrations. However, ribulose-1,5-bisphosphate is not recycled in these reactions which may result in a depletion of CO2 acceptor molecules at higher fluxes.
4.2. Possible Alternative Pathways in the Peroxisome
The presence of an alternative pathway for glyoxylate conversion in the peroxisome that does not involve transamination to glycine was suggested by the characterization of Arabidopsis mutants deficient in SHM (Somerville and Ogren, 1981). Although accumulating high amounts of glycine, these mutants still released CO2 from photorespiratory intermediates. The responsible pathway might involve the non-enzymatic decarboxylation of glyoxylate to formate using H2O2 as an oxidizing agent (Oliver, 1981; Igamberdiev and Lea, 2002). Alternatively, catalase can also act as a peroxidase in the oxidation of glyoxylate (Grodzinski, 1978). Peroxisomal glyoxylate decarboxylation could play a physiological role especially under conditions of low nitrogen supply where transamination is inhibited (Somerville and Ogren, 1982b; Singh et al., 1986; Wingler et al., 1999).
Formate resulting from glyoxylate decarboxylation might act a C1 donor in the formation of serine. Based on studies of barley and Amaranthus mutants lacking GLD activity, Wingler et al. (1999) suggested that the C1-tetrahydrofolate (THF) synthase pathway converts photorespiratory formate to 5,10-methylene-THF (green pathway in Figure 5). The enzymes involved in plants are a monofunctional 10-formyl-THF-synthetase (FTHF synthetase, EC 6.3.4.3) and a bifunctional 5,10-methenyl-THF-cyclohydrolase:5,10-methylene-THF dehydrogenase (CYC-DHY, EC 3.5.4.9 and EC 1.5.1.5). FTHF synthetase catalyses the synthesis of 10-formyl-THF, which is converted into 5,10-methenyl-THF by CYC and reduced to 5,10-methylene-THF by DHY. 5,10-methylene-THF is used by SHM to synthesize serine from glycine (Prabhu et al., 1996; Hanson and Roje, 2001). In the barley and Amaranthus GLD mutants, evidence for function of this pathway in photorespiration was provided by the increase in FTHF-synthetase, the accumulation of glyoxylate and formate under photorespiratory conditions, and the ability of the mutants to utilize formate and glycolate for the formation of serine (Wingler et al., 1999). Similar flux analyses were later on also performed with Arabidopsis mutants deficient in GLD activity that confirmed the described results (Li et al., 2003). However, the enzymes catalyzing the conversion of formate to 5,10-methylene-THF have been poorly characterized in Arabidopsis and the subcellular localization is unknown. Enzyme activities have been shown to be mainly associated with the cytosolic fraction in pea leaves (Chen et al., 1997), but isoenzymes are also present in the mitochondrion and the chloroplast (Hanson and Roje, 2001; Christensen and MacKenzie, 2006). In animals and yeast, partially redundant pathways exist in the cytoplasm and the mitochondrion (Piper et al., 2000; Christensen and MacKenzie, 2006). Folate transporters have been identified in the Arabidopsis chloroplast membrane (Bedhomme et al., 2005) and animal mitochondria (McCarthy et al., 2004). The pathway for the conversion of formate resulting from peroxisomal glyoxylate decarboxylation was therefore placed by default in the cytoplasm in Figure 5 (green pathway).
4.3. Possible Alternative Pathways in the Mitochondrion
Metabolic profiling of Go antisense lines in rice (see chapter 2) revealed the expected accumulation of glycolate, but downstream metabolites such as glycine and serine were not reduced in concentration. This confirmed that GO-catalyzed glycolate oxidation is the dominant pathway for glycolate conversion, but also implied the existence of alternative pathways for glyoxylate production (Xu et al., 2009). Moreover, Ggat1 knockout lines (Igarashi et al., 2003, see also chapter 2) showed only weak photorespiratory phenotypes that might not be fully explained by the residual low activity of GGAT2 suggesting the existence of alternative pathways for the conversion of glycolate to glycine.
Leaf-type peroxisomes are absent in chlorophytes, the second big lineage of green algae, that subsumes the classes Chlorophyceae, Ulvophyceae, and Trebouxiophyceae (Frederick et al., 1973). Beside secreting glycolate from the cell, this group uses a mitochondrial glycolate dehydrogenase (GlyDH, EC 1.1.99.14) for the production of glyoxylate that most probably transfers electrons to the respiratory electron transport chain (Paul and Volcani, 1976). Analyses in the trebouxiophycean species Eremosphaera viridis revealed that the subsequent metabolism is similar to photorespiration in higher plants, but that all reactions are located in mitochondria and that the capacity of this pathway for glycolate conversion is lower compared to the major pathway in higher plants (Stabenau and Winkler, 2005). The importance of the mitochondrial GlyDH in chlorophytes has been demonstrated by the characterization of a Chlamydomonas strain carrying a plasmid insertion in the corresponding gene. The mutation is conditionally lethal and the strain is only capable of growing at elevated CO2 concentrations (Nakamura et al., 2005, see also chapter 5).
In charophytes, the nearest relatives to land plants, glycolate oxidation was shifted from the mitochondrion to the peroxisome (Stabenau and Winkler, 2005), most probably because the specific activity of GlyDH is low and allows for low flux rates only. The peroxisomal GO does not show significant homology to GlyDH on the protein level and differs in important enzymatic properties: GlyDH enzymes use D-lactate as an alternative substrate, whereas GO preferentially oxidizes L-lactate. This allows for a precise discrimination of both activities.
Studies in Arabidopsis questioned the clear differentiation of glycolate metabolism in higher plants and chlorophytes (Bari et al., 2004). An enzyme with homology to bacterial and algal GlyDH enzymes (At5g06580) was shown to be targeted to the mitochondrion. The recombinant enzyme complemented Escherichia coli mutants deficient in GlyDH activity and oxidized glycolate to glyoxylate in vitro. Knock-out of At5g06580 drastically reduced the capacity of isolated Arabidopsis mitochondria to convert glycolate to CO2 and reduced metabolite flux through photorespiration (Niessen et al., 2007). Inhibitor studies suggested that a mitochondrial alanine:glyoxylate aminotransferase might further convert glyoxylate resulting from the GlyDH reaction to glycine (Niessen et al., 2007, red pathway in Figure 5). Alternatively, this reaction might also be catalyzed by mitochondrial γ-aminobutyrate transaminase that has been shown to accept glyoxylate as a substrate. The accumulation ofγ-aminobutyrate during stress is a well documented phenomenon and would provide enough amino donor for the transamination of glyoxylate (Clark et al., 2009). By any of the aminotransferase reactions discussed, mitochondrial glycolate oxidation can be linked to the GLD/SHM complex of the major pathway.
The main advantage of the mitochondrial pathway is the direct coupling of glycolate oxidation to the electron transport chain that allows to recover energy equivalents (Paul and Volcani, 1976; Stabenau and Winkler, 2005). The combination with the high capacity major pathway in a single cell provides the plant with an optimized adaptability to changes in environmental conditions. However, neither knock-out of GlyDH nor γ-aminobutyrate transaminase resulted in an obvious phenotype under normal growth conditions (Niessen et al., 2007; Clark et al., 2009). In addition, the view that Arabidopsis GlyDH is involved in photorespiration was recently challenged by characterization of the enzymatic properties (Engqvist et al., 2009). In vitro data revealed a 2000-fold higher preference for D-lactate compared to glycolate due to a very low catalytic rate with the latter substrate. The enzyme was necessary for growth of Arabidopsis on media containing high concentrations of D-lactate, but not of glycolate. However, glycolate might have been detoxified by the peroxisomal glycolate oxidase in such a setup and this would mask a possible supplementary function of GlyDH activity. Further research is necessary to explain the conflicting data sets. As At5g06580 is expressed in all plant organs, the function of the enzyme might also differ between roots and leaves.
4.4. Possible Alternative Pathway in the Cytosol
As specified in chapter 2, Hpr1 encodes the peroxisomal hydroxypyruvate reductase involved in the major photorespiratory pathway. However, knock-out in Arabidopsis (Timm et al., 2008) or barley (Murray et al., 1989) did not result in severe photorespiratory phenotypes. It was therefore speculated that cytosolic HPR2 (Kleczkowski et al., 1991) could provide a bypass for the peroxisomal reaction (dashed grey pathway in Figure 5). A double mutant with T-DNA insertions in the genes encoding both HPR1 and HPR2 was recently characterized by Timm et al. (2008). The double knock-out showed the typical air-sensitivity and drastically reduced photosynthetic performance of photorespiratory mutants. Thus, the cytosolic pathway seemed to have the capacity to fully replace the peroxisomal pathway at least under moderate growth conditions. However, metabolic profiling and gas exchange revealed clearly stronger perturbances of photosynthesis and basal metabolism in the HPR1 compared to the HPR2 mutant (Timm et al., 2008). This suggests that peroxisomal HPR1 plays a dominant role under moderate growth conditions whereas HPR2 provides an overflow mechanism for the utilization of excess hydroxypyruvate leaking from peroxisomes under conditions of very high photorespiratory flow. Such a function in protection and/or improvement of the energy balance under non-standard growth conditions might be a joint feature of all alternative pathways and explain why these pathways have been conserved in evolution.
5. METABOLIC COMPLEXITY: PHOTORESPIRATION IS NECESSARY FOR ALL ORGANISMS PERFORMING OXYGENIC PHOTOSYNTHESIS
This chapter is not about photorespiration in Arabidopsis. However, recent results on the significance of photorespiration in other photosynthetic organisms shed new light on the evolution and the major function of photorespiration and are therefore discussed here.
The use of RUBISCO for CO2 fixation is common to all organisms with oxygenic photosynthesis: cyanobacteria, algae, lower and higher plants. Despite billion years of evolution, all RUBISCO enzymes also share oxygenase activity and, therefore, produce phosphoglycolate when exposed to oxygen as a substrate (Anderson, 2008, see also chapter 1). However, several lineages independently evolved mechanisms to reduce the probability of oxygen fixation:
Cyanobacteria and many algae concentrate CO2 in the ultimate vicinity of RUBISCO (Giordano et al., 2005; Price et al., 2007; Spalding, 2007). Both mechanisms are based on a tight association of RUBISCO with carbonic anhydrase and a shell made from proteins or starch, respectively, around these complexes. CO2 that enters the cell is converted to bicarbonate, diffuses into the vicinity of RUBISCO and is reconverted to CO2 immediately before fixation by RUBISCO. It has been argued that the carbon concentrating mechanism (CCM) mainly serves the saturation of RUBISCO at low availability of gaseous CO2 in an aqueous environment (Giordano et al., 2005; Raven et al., 2008). Nevertheless, suppression of oxygen fixation is at least a welcome side effect.
In higher plants, evolutionary pressure resulted in an enhancement of the specificity factor of RUBISCO. This parameter indicating the relative preference of RUBISCO for CO2 over O2 increased in evolution of the green lineage from cyanobacteria over green algae and is highest for RUBISCOs from higherplants (Jordan and Ogren, 1981), although some even more specific RUBISCO enzymes evolved in non-green algae perhaps as an adaptation to specific ecological niches (Badger and Bek, 2008). However, there is a negative correlation between specificity and the catalytic rate of RUBISCO (Zhu et al., 2004): Highly specific enzymes are mostly slow catalysts (see also chapter 1) although there is some variation around this general rule also in higher plants (Galmes et al., 2005). This might have limited the further evolution of RUBISCOs with even higher specificities.
Some land plants that were exposed to conditions of high photorespiratory pressure, evolved biochemical pumps that concentrate CO2 at the site of fixation. These mechanisms are all based on a primary fixation of bicarbonate into organic acids by phosphoenolpyruvate carboxylase (PEPC). In a second step, decarboxylation of these organic acids enhances the CO2 concentration around RUBISCO resulting in a strong decrease in the probability of oxygen fixation. Primary and secondary CO2 fixation can be either spatially (C4 metabolism) or temporally (crassulacean acid metabolism (CAM)) separated. For informative reviews on these mechanisms, see Sage (2004) and Dodd et al. (2002), respectively.
Despite of the presence of mechanisms that reduce oxygen fixation, there is evidence for functional photorespiratory pathways in all these organisms. Mutant analyses analogous to Somerville's studies on Arabidopsis in the 1970s (see chapter 1) imply that these pathways are not only an evolutionary relict, but required for survival:
Despite knowledge about the presence and activity of photorespiratory enzymes (Frederick et al., 1973) and the description of oxygen-dependent CO2 release in green algae (Birmingham et al., 1982), metabolic flux through the photorespiratory pathway has only been shown recently (Stabenau and Winkler, 2005). In screens for Chlamydomonas mutants that require high CO2 for growth, mostly mutations in components of the CCM were identified (Van et al., 2001). However, Suzuki et al. (1990) described a mutant that accumulated high concentrations of phosphoglycolate and showed a high CO2-requiring phenotype although the CCM was fully functional. A second mutant with a transposon insertion in the gene encoding glycolate dehydrogenase (GlyDH) showed a similar phenotype and excreted large amounts of glycolate under conditions that favor phosphoglycolate formation (Nakamura et al., 2005). Thus, at least the species Chlamydomonas requires photorespiration for growth at normal CO2 concentrations.
In cyanobacteria, knowledge about photorespiration was very limited until recently. Some photorespiratory enzymes were shown to be present by biochemical methods (Grodzinski and Colman, 1976; Norman and Colman, 1991, 1992) and glycolate excretion was observed under photorespiratory conditions (Renström and Bergman, 1989). A survey of the Synechocystis genome sequence revealed that genes for enzymes of both the higher plant photorespiratory pathway and a bacterial pathway for conversion of glycolate to glycerate were present (Eisenhut et al., 2006, Figure 6). The higher plant pathway plays the major role in phosphoglycolate metabolism, but can be partially replaced by the bacterial pathway. Nevertheless, even double mutants, where both pathways were disrupted, were able to grow under ambient CO2 concentrations, which was attributed to the activity of the CCM (Eisenhut et al., 2006). Instead, the same group identified a third pathway for glycolate metabolism that includes a series of decarboxylations: Glyoxylate is first converted to oxalate, then to formate and finally to CO2 (Eisenhut et al., 2008, Figure 6). This pathway actually resembles possible alternative routes for glycolate metabolism in Arabidopsis chloroplasts and/or peroxisomes (see chapter 4). Simultaneous knock-out of all the three routes for glycolate conversion in a triple mutant finally caused a high CO2-requiring phenotype. Beside indicating the importance of photorespiration in cyanobacteria, these data also imply that not only photosynthesis, but also photorespiration was transferred from cyanobacteria to plants during endosymbiosis (Eisenhut et al., 2008).
Figure 6.
The three pathways for glyoxylate conversion in Synechocystis.
Overview of the metabolism of photorespiratory glyoxylate in Synechocystis. The pathways are homologous to the major pathway in Arabidopsis (black), the bacterial glycerate pathway (red), or involve the complete oxidation of glyoxylate to CO2 (green), respectively. RuBP, ribulose-1,5-bisphosphate; RUBISCO, RuBP carboxylase/oxygenase. The stoichiometry of the reactions is not included.
C4 metabolism evolved on top of C3 photosynthesis. Relocation of photorespiratory CO2 release to the bundle sheath was probably one major step during the evolution of C4 plants (Ohnishi et al., 1985; Sage, 2004). Photorespiratory enzymes were detected in C4 plants (e.g. Popov et al., 2003; Majeran et al., 2005) and inhibition of C4 photosynthesis by O2 could be observed under certain conditions (Chollet and Ogren, 1975; Dai et al., 1993; Martinelli et al., 2007). In isolated bundle sheath strands of both maize and Panicum, operation of the photorespiratory cycle could be shown by metabolic flux labeling (Farineau et al., 1984). All this argued for presence and function of photorespiration in C4 plants, however, flux rates were mostly believed to be very low to insignificant. Again, identification of a mutant confirmed that photorespiration is also essential for C4 plants (Zelitch et al., 2008). A maize line with a transposon insertion in the Go1 gene, that encodes the major bundle sheath glycolate oxidase, required elevated CO2 for growth and accumulated glycolate to high concentrations when shifted from permissive CO2 concentrations to ambient air. This was accompanied by a clear reduction in photosynthetic rates.
Based on these results, it can now be concluded that photorespiration is essential for growth of most if not all organisms that perform oxygenic photosynthesis. As phosphoglycolate formation is low in species with efficient CO2 concentrating mechanisms, carbon loss in the absence of a recycling mechanism is probably not problematic. It rather seems that the accumulation of intermediates that are inhibiting photosynthesis (see chapter 3) is causal for the severe phenotype of photorespiratory mutants in these species. These findings are also important for understanding the role of photorespiration in C3 plants such as Arabidopsis.
METABOLIC ENGINEERING: MANIPULATING PHOTORESPIRATION
Early during the identification of the photorespiratory cycle, researchers realized that a reduction in photorespiration would show considerable promise to enhance plant carbon fixation, growth and yield. This was one major motivation to invest significant research efforts into the elucidation of the photorespiratory pathway (Ogren, 2003). Indeed, the biochemical inhibition of glycolate synthesis (and consequently photorespiration) by glycidate in tobacco leaf disks resulted in a strong enhancement of photosynthesis (Zelitch, 1974) although some of these results were heavily discussed and could not be reproduced by others in later experiments (Chollet, 1977). In a complementary setup, experiments with tobacco varieties suggested that lines with lower rates of photorespiration had enhanced photosynthesis (Zelitch and Day, 1973). However, as described in chapter 1, mutants with reduced photorespiration and enhanced photosynthesis were never recovered from extensive mutant screens in Arabidopsis. Chris Somerville concluded that changing the specificity of RUBISCO for oxygen relative to carbon dioxide would be the only way to reduce photorespiratory losses (Somerville and Ogren, 1982b). Since then, many scientific efforts have been pursued in this direction and major progress has been obtained in understanding the dual function of RUBISCO and in manipulating the enzyme (Zhu et al., 2004; Mueller-Cajar and Whitney, 2008). However, the final breakthrough of engineering a plant with a better RUBISCO and higher photosynthesis has still not been accomplished.
In parallel, several groups tried to establish components of the C4 CO2 concentrating mechanism in Arabidopsis and other C3 plants in order to reduce photorespiration (Haeusler et al., 2002). As described in chapter 5, C4 plants invest energy to concentrate CO2 around RUBISCO. By this, they reduce oxygen fixation and the subsequent photorespiratory losses. The trade-off between energy investment and the reduction of photorespiratory losses is positive when photorespiratory rates are high, i.e. under hot and dry growth conditions. Enzymes involved in C4 metabolism were well-characterized and, thus, researchers started to overexpress the encoding genes in C3 plants (reviewed in Matsuoka et al., 2001; Leegood, 2002). This type of approach based on C4-like pathways that were not separated into two different tissues for primary and secondary CO2 fixation showed very limited success in reducing photorespiration so far. Most scientists now agree that it will be essential to understand the genetic control of the establishment of C4 leaf anatomy if an efficient CO2 pump should be set up in C3 plants. Efforts in this direction have recently been reinitiated with the long-term aim to convert rice into a C4 plant and, by this, to cover the ever growing demands for food in developing countries (Hibberd et al., 2008).
Beside the improvement of RUBISCO's specificity and the transfer of a C4-like mechanism, a third different approach has been tested recently. The idea is based on redirecting photorespiration instead of reducing it. The novel pathways should provide alternatives for the metabolism of photorespiratory intermediates that result in an improved energy balance. Chapter 4 described a considerable biochemical plasticity of photorespiration that received little attention after the identification of the major pathway components. The presence of alternative photorespiratory pathways indicates that a deviation of metabolites from the major pathway might be acceptable or even beneficial for plants under certain environmental conditions. Templates for new metabolic pathways are available from non-photosynthetic microorganisms that are capable of growing on glycolate as a sole carbon source by converting it to glycerate (Lord, 1972) or malate (Kornberg and Sadler, 1961), respectively. As shown in Figure 7 (red pathway), installation of the glycerate pathway from Escherichia coli in the plant chloroplast establishes a novel pathway that starts and ends with intermediates of photorespiration. By this, a shortcircuit is created that re-directs photorespiratory metabolism. The pathway is made up from three different enzymes: The first reaction resembles the peroxisomal glycolate oxidase reaction in producing glyoxylate from glycolate, but the bacterial glycolate dehydrogenase (GlyDH) uses organic compounds as co-factors instead of molecular oxygen. Unfortunately, the GlyDH enzyme from E. coli is made up from several subunits from which the three subunits D, E, and F are necessary for function (Pellicer et al., 1996). Thus, installation of this enzymatic activity requires the simultaneous over-expression of three genes. In step 2 of the pathway, two molecules of glyoxylate are ligated by glyoxylate carboligase (GCL) forming tartronic semialdehyde. This reaction produces CO2 in the chloroplast similar to the mitochondrial CO2 release in the major photorespiratory pathway. Tartronic semialdehyde is reduced to glycerate by tartronic semialdehyde reductase (TSR) and the glycerate kinase reaction of the endogenous pathway is used to recycle phosphoglycerate. Installation of this shortcircuit in Arabidopsis by repeated transformation and genetic crossing resulted in an improvement of biomass production of about 30 % under short-day growth conditions (Kebeish et al., 2007). As shown in Figure 8, the effect was even stronger at longer growth periods and additional nutrient supply. Three possible explanations are available why the shortcircuit is superior to the endogenous pathway:
The endogenous pathway combusts reducing power when forming H2O2 instead of organic reducing equivalents during the conversion of glycolate to glyoxylate. Dependent on the availability of reducing equivalents, this may limit growth.
The shortcircuit does not include NH3 loss and refixation, again saving energy both in the form of ATP and reducing equivalents. In sum, 1. and 2. have the potential of reducing photorespiratory energy requirements from 4.75 ATP and 3 reducing equivalents to 4.25 ATP and only 2 reducing equivalents. Thus, the photorespiratory “burden” on CO2 fixation would be reduced from 50 % to 37 % (cf. chapter 2) assuming the all photorespiratory glycolate would be redirected into the shortcircuit.
CO2 release is shifted from the mitochondrion to the chloroplast. Based on physiological measurements, this enhances the CO2 concentration in the chloroplast, improves the probability of refixation of the released CO2 molecule, and reduces the probability of the next oxygenase reaction (Kebeish et al., 2007).
Figure 7.
Transgenic pathways for the reduction of photorespiratory losses in Arabidopsis.
Overview of the major photorespiratory pathway (black) and different transgenic approaches for the reduction of photorespiratory losses (red, dotted grey, and green). The red pathway shows the glycerate pathway from E. coli integrated into the chloroplast, the dotted grey pathway shows the alternative complete oxidation of glycolate inside the chloroplast, and the green pathway shows a shortcircuit inside the peroxisome. RuBP, ribulose-1,5-bisphosphate; RUBISCO, RuBP carboxylase/oxygenase; PGLP, phosphoglycolate phosphatase; GO, glycolate oxidase; CAT, catalase; GGAT, glyoxylate:glutamate aminotransferase; SGAT, serine:glyoxylate aminotransferase; DIT1, dicarboxylate transporter 1; DIT2, dicarboxylate transporter 2; GLD, glycine decarboxylase; SHM, serine hydroxymethyltransferase; HPR, hydroxypyruvate reductase; GLYK, glycerate kinase; GS, glutamine synthetase; Fd-GOGAT, glutamine:oxoglutarate aminotransferase; GlyDH, glycolate dehydrogenase; GCL, glyoxylate carboligase; TSR, tartronic semialdehyde reductase; MS, malate synthase; ME, malic enzyme; PyrDH, pyruvate dehydrogenase; AcCoA, acetylated Coenzyme A; CoA, Coenzyme A; HYI, hydroxypyruvate isomerase; THF, tetrahydrofolate; CH2-THF, methylene-THF. The stoichiometry of the reactions is not included.
Figure 8.
Photorespiratory bypass increases biomass production in Arabidopsis.
Arabidopsis plants were grown at 100 µE light and a 8h light/16 h dark light regime for 12 weeks. After 8 weeks, plants were repotted from 8 cm pots to 15 cm pots. The upper right and lower left plants express the photorespiratory bypass (red pathway in Figure 7), the upper left and lower right plants are wildtype controls.
Interestingly, overexpression of GlyDH alone was already sufficient to induce the growth phenotype in large part (Kebeish et al., 2007). This provides independent evidence that chloroplasts are capable of further metabolizing glyoxylate as already suggested in chapter 4.
The alternative conversion of glycolate to malate instead of glycerate (dotted grey pathway in Figure 7) has also been tested (Maurino and Flügge, 2009). In this approach, glycolate is oxidized by glycolate oxidase and the resulting H2O2 is detoxified by catalase. Malate synthase forms malate from glyoxylate and Acetyl-CoA present in the chloroplast. Through two additional endogenous reactions, Acetyl-CoA is recycled and glyoxylate fully converted to CO2. Also installation of this pathway in Arabidopsis resulted in an enhanced growth phenotype (Maurino and Flügge, 2009).
A third possible synthetic pathway to improve the energy balance of photorespiration was suggested by Parry et al. (2007) (green pathway in Figure 7). The authors aimed at bypassing photorespiratory NH3 release by installation of a shortcircuit in the peroxisome. This pathway contained two enzymes, glyoxylate carboligase and hydroxypyruvate isomerase. The former enzyme ligates two molecules of glyoxylate to form tartronic semialdehyde as described above and the latter converts tartronic semialdehyde to hydroxypyruvate that is again integrated into the endogenous pathway. Overexpression of this pathway in tobacco resulted in plants with chlorotic lesions for unknown reasons.
Results from these studies suggest that manipulation of photorespiration can in fact improve plant photosynthesis as hoped during the early days of photorespiration research. It is fascinating to see that two of the recently developed engineering approaches resemble those pathways that nature established in Synechocystis as alternatives to the major pathway. This knowledge was not available when the synthetic pathways were designed, but the coincidence hopefully further motivates scientists to use natural variation as a template for genetic engineering. However, the results from the third study also sound a note of caution that any synthetic interference with basal metabolism might result in unexpected phenotypes as long as the metabolic integration of photorespiration is not fully understood.
7. CONCLUSIONS
In this review, we summarized the current knowledge about photorespiration in Arabidopsis. Although the initial identification of the process dates to the pre-Arabidopsis era of plant research, work in Arabidopsis allowed for the detailed characterization of most of the pathway components. Future work in this direction will hopefully lead to the identification of the multiple metabolite transporters necessary for photorespiration. More ecophysiological work is also necessary to understand regulation and metabolic integration of the pathway and the significance of alternative detours under certain growth conditions.
The ease of Arabidopsis transformation allowed for testing synthetic pathways that deviate some of the phosphoglycolate from photorespiration. Promising initial results currently motivate researchers to test the applicability of these and related approaches in crop improvement (Hibberd et al., 2008; Peterhansel et al., 2008; Sage and Sage, 2009). But what is the significance of photorespiration in a future earth atmosphere with high CO2 concentrations? Current projections expect CO2 concentrations to rise by more than 40 % compared to today until 2050. An upper limit of perhaps double the current CO2 concentration may be reached until 2100 (Intergovernmental panel on climate change, 2007). At first sight, doubling the atmosphere's CO2 content would reduce photorespiration to one half (Sharkey, 1988) which would, in turn, diminish the importance of photorespiration in the control of plant performance. However, the situation is much more complex. An expected increase in average atmospheric temperatures by 3 °C (Intergovernmental panel on climate change, 2007) alone will reduce the specificity of RUBISCO for CO2 by approximately 10 % (Jordan and Ogren, 1984). Actual leaf temperatures might increase much stronger because higher CO2 concentrations tend to induce stomatal closure and by this reduce transpirational leafcooling (Ainsworth and Rogers, 2007; Bernacchi et al., 2007). Furthermore, the frequency and intensity of droughts is predicted to increase (Li et al., 2009), growth conditions at which photorespiration is high. Independent of climate change, the ever growing world population will demand food production on less favourable grounds, in particular in hot and dry areas (Bruinsma, 2009; Eckardt et al., 2009). Thus, beside the fascination provided by the intricate complexity of plant carbon metabolism, there are very good reasons for plant researchers to continue studying photorespiration.
ACKNOWLEDGEMENTS
Work in the authors' laboratory on photorespiration is funded by the Deutsche Forschungsgemeinschaft as part of the German Photorespiration Network Promics (FOR 1186).
Footnotes
Citation: Peterhansel C., Horst I., Niessen M., Blume C., Kebeish R., Kürkcüoglu S., and Kreuzaler F. (2010) Photorespiration. The Arabidopsis Book 8:e0130. doi:10.1199/tab.0130
elocation-id: e0130
First published on March 23, 2010
REFERENCES
- Ainsworth E.A., Rogers A. The response of photosynthesis and stomatal conductance to rising CO2: mechanisms and environmental interactions. Plant Cell Environ. 2007;308(1):258–270. doi: 10.1111/j.1365-3040.2007.01641.x. [DOI] [PubMed] [Google Scholar]
- Allan W.L., Clark S.M., Hoover G.J., Shelp B.J. Role of plant glyoxylate reductases during stress: a hypothesis. Biochem. J. 2009;4238(1):15–22. doi: 10.1042/BJ20090826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.E. Chloroplast and cytoplasmic enzymes II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta. 1971;2358(1):237–244. doi: 10.1016/0005-2744(71)90051-9. [DOI] [PubMed] [Google Scholar]
- Andersson I. Catalysis and regulation in Rubisco. J. Exp. Bot. 2008;598(1):1555–1568. doi: 10.1093/jxb/ern091. [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;558(1):373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Badger M.R., Bek E.J. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008;598(1):1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- Bari R., Kebeish R., Kalamajka R., Rademacher T., Peterhänsel C. A glycolate dehydrogenase in the mitochondria of Arabidopsis thaliana. J. Exp. Bot. 2004;558(1):623–630. doi: 10.1093/jxb/erh079. [DOI] [PubMed] [Google Scholar]
- Bartsch O., Hagemann M., Bauwe H. Only plant-type (GLYK) glycerate kinases produce D-glycerate 3-phosphate. FEBS Lett. 2008;5828(1):3025–3028. doi: 10.1016/j.febslet.2008.07.038. [DOI] [PubMed] [Google Scholar]
- Bauwe H., Kolukisaoglu U. Genetic manipulation of glycine decarboxylation. J. Exp. Bot. 2003;548(1):1523–1535. doi: 10.1093/jxb/erg171. [DOI] [PubMed] [Google Scholar]
- Bedhomme M., Hoffmann M., McCarthy E.A., Gambonnet B., Moran R.G., RebeIlle F., Ravanel S. Folate metabolism in plants: An Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 2005;2608(1):34823–34831. doi: 10.1074/jbc.M506045200. [DOI] [PubMed] [Google Scholar]
- Benson A.A., Calvin M. Carbon dioxide fixation by green plants. Annu. Rev. Plant Physiol. 1950;18(1):25–42. [Google Scholar]
- Bernacchi C.J., Kimball B.A., Queries D.R., Long S.P., Ort D.R. Decreases in stomatal conductance of soybean under openair elevation of CO2 are closely coupled with decreases in ecosystem evapotranspiration. Plant Physiol. 2007;1438(1):134–144. doi: 10.1104/pp.106.089557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham B.C., Coleman J.R., Colman B. Measurement of photorespiration in algae. Plant Physiol. 1982;698(1):259–262. doi: 10.1104/pp.69.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell R., Murray A., Lea P., Kendall A., Hall N., Turner J., Wallsgrove R. The value of mutants unable to carry out photorespiration. Photosynth. Res. 1988;168(1):155–176. doi: 10.1007/BF00039491. [DOI] [PubMed] [Google Scholar]
- Boldt R., Edner C., Kolukisaoglu U., Hagemann M., Weckwerth W., Wlenkoop S., Morgenthal K., Bauwe H. D-glycerate 3-kinase, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell. 2005;178(1):2413–2420. doi: 10.1105/tpc.105.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes G., Ogren W.L. Oxygen inhibition and other properties of soybean ribulose 1,5-diphosphate carboxylase. J. Biol. Chem. 1972;2478(1):2171–2176. [PubMed] [Google Scholar]
- Bowes G., Ogren W.L., Hageman R.H. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 1971;458(1):716–722. doi: 10.1016/0006-291x(71)90475-x. [DOI] [PubMed] [Google Scholar]
- Brooks A., Farquhar G.D. Effect of the temperature on the CO2/O2 specifity of ribulose-1,5-bisphosphate carboxylase/oxygenase and the rate of respiration in the light. Planta. 1985;1658(1):397–406. doi: 10.1007/BF00392238. [DOI] [PubMed] [Google Scholar]
- Bruinsma J. The resource outlook to 2050. Expert meeting on how to feed the world in. 2009;8(1):2050. Food and Agriculture Organization of the United Nations, ftp://ftp.fao.org/docrep/fao/012/ak971e/ak971e000.pdf. [Google Scholar]
- Buick R. When did oxygenic photosynthesis evolve? Phil. Trans. R. Soc. Lond. Biol. Sci. 2008;3638(1):2731–2743. doi: 10.1098/rstb.2008.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W.J., Ogren W.L. Glyoxylate inhibition of ribulosebisphosphate carboxylase-oxygenase: Activation in intact, lysed and reconstituted chloroplasts. Photosynth. Res. 1990;238(1):257–268. doi: 10.1007/BF00034856. [DOI] [PubMed] [Google Scholar]
- Chamnongpol S., Willekens H., Moeder W., Langebartels C., Sandermann H., Jr, Van Montagu M., Inze D., Van Camp W. Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. USA. 1998;958(1):5818–5823. doi: 10.1073/pnas.95.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain C.J., Ogren W.L. Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation state in vivo. Plant Cell Physiol. 1989;308(1):937–944. [Google Scholar]
- Chen L., Chan S.Y., Cossins E.A. Distribution of folate derivatives and enzymes for synthesis of 10-formyltetrahydrofolate in cytosolic and mitochondrial fractions of pea leaves. Plant Physiol. 1997;1158(1):299–309. doi: 10.1104/pp.115.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R. The biochemistry of photorespiration. Trends Biochem. Sci. 1977;28(1):155–159. [Google Scholar]
- Chollet R., Ogren W.L. Regulation of photorespiration in C3 and C4 species. Bot. Rev. 1975;418(1):137–179. [Google Scholar]
- Christensen K.E., MacKenzie R.E. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. Bioessays. 2006;288(1):595–605. doi: 10.1002/bies.20420. [DOI] [PubMed] [Google Scholar]
- Clark S.M., Di Leo R., Dhanoa P.K., Van Cauwenberghe O.R., Mullen R.T., Shelp B.J. Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis g-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J. Exp. Bot. 2009;608(1):1743–1757. doi: 10.1093/jxb/erp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova E., Goyer A., Naponelli V., Krassovskaya I., Gregory J.F., Hanson A.D., III, Shachar-Hill Y. Arabidopsis 10-formyl tetrahydrofolate deformylases are essential for photorespiration. Plant Cell. 2008;208(1):1818–1832. doi: 10.1105/tpc.108.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C.M., Mulligan R.M., Tolbert N.E. Inhibition and stimulation of ribulose-1,5-bisphosphate carboxylase/oxygenase by glyoxylate. Arch. Biochem. Biophys. 1985;2408(1):392–401. doi: 10.1016/0003-9861(85)90044-x. [DOI] [PubMed] [Google Scholar]
- Cornic G., Briantais J.-M. Partitioning of photosynthetic electron flow between CO2 and O2 reduction in a C3 leaf (Phaseolus vulgaris L.) at different CO2 concentrations and during drought stress. Planta. 1991;1838(1):178–184. doi: 10.1007/BF00197786. [DOI] [PubMed] [Google Scholar]
- Coschigano K.T., Melo-Oliveira R., Lim J., Coruzzi G.M. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell. 1998;108(1):741–752. doi: 10.1105/tpc.10.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins E.A., Chen L. Folates and one-carbon metabolism in plants and fungi. Phytochem. 1997;458(1):437–452. doi: 10.1016/s0031-9422(96)00833-3. [DOI] [PubMed] [Google Scholar]
- Cousins A.B., Pracharoenwattana I., Zhou W., Smith S.M., Badger M.R. Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol. 2008;1488(1):786–795. doi: 10.1104/pp.108.122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Ku M., Edwards G.E. C4 photosynthesis (The CO2-concentrating mechanism and photorespiration). Plant Physiol. 1993;1038(1):83–90. doi: 10.1104/pp.103.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D.D., Corbett R.J. Glyoxylate decarboxylase activity in higher plants. Phytochem. 1969;88(1):529–542. [Google Scholar]
- Davison P.A., Hunter C.N., Horton P. Overexpression of beta-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature. 2002;4188(1):203–206. doi: 10.1038/nature00861. [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Borland A.M., Haslam R.P., Griffiths H., Maxwell K. Crassulacean acid metabolism: plastic, fantastic. J. Exp. Bot. 2002;538(1):569–580. doi: 10.1093/jexbot/53.369.569. [DOI] [PubMed] [Google Scholar]
- Douce R., Neuburger M. Biochemical dissection of photorespiration. Curr. Opin. Plant Biol. 1999;28(1):214–222. doi: 10.1016/S1369-5266(99)80038-7. [DOI] [PubMed] [Google Scholar]
- Douce R., Bourguignon J., Neuburger M., Rebeille F. The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 2001;68(1):167. doi: 10.1016/s1360-1385(01)01892-1. [DOI] [PubMed] [Google Scholar]
- Eckardt N.A., Cominelli E., Galbiati M., Tonelli C. The future of science: Food and water for life. Plant Cell. 2009;218(1):368–372. doi: 10.1105/tpc.109.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehleringer J.R., Sage R.F., Flanagan L.B., Pearcy R.W. Climate change and the evolution of C4 photosynthesis. Trends Ecol. Evol. 1991;68(1):95–99. doi: 10.1016/0169-5347(91)90183-X. [DOI] [PubMed] [Google Scholar]
- Eisenhut M., Ruth W., Haimovich M., Bauwe H., Kaplan A., Hagemann M. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. USA. 2008;1058(1):17199–17204. doi: 10.1073/pnas.0807043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M., Kahlon S., Hasse D., Ewald R., Lieman-Hurwitz J., Ogawa T., Ruth W., Bauwe H., Kaplan A., Hagemann M. The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 2006;1428(1):333–342. doi: 10.1104/pp.106.082982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstner E.F., Heupel A. On the decarboxylation of a-keto acids by isolated chloroplasts. Biochim. Biophys. Acta. 1973;3258(1):182–188. doi: 10.1016/0005-2728(73)90164-3. [DOI] [PubMed] [Google Scholar]
- Engel N., van den Daele K., Kolukisaoglu U., Morgenthal K., Weckwerth W., Parnik T., Keerberg O., Bauwe H. Deletion of glycine decarboxylase in Arabidopsis is lethal under non-photorespiratory conditions. Plant Physiol. 2007;1448(1):1328–1335. doi: 10.1104/pp.107.099317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M., Drincovich M.F., Flugge U.-I., Maurino V.G. Two D-2-hydroxyacid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and b-oxidation pathways. J. Biol. Chem. 2009;2648(1):25026–25037. doi: 10.1074/jbc.M109.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H., Lee C,P., Kuo J., Meyer E.H., Taylor N.L., Millar A.H. Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J. 2007;528(1):583–594. doi: 10.1111/j.1365-313X.2007.03253.x. [DOI] [PubMed] [Google Scholar]
- Ewald R., Kolukisaoglu U., Bauwe U., Mikkat S., Bauwe H. Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol. 2007;1458(1):41–48. doi: 10.1104/pp.107.104000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farineau J., Lelandias M., Morot-Gaudry J.F. Operation of the glycolate pathway in isolated bundle sheath strands of maize (Zea mays) cultivar Inra-258 and Panicum maximum. Physiol. Plant. 1984;608(1):208–214. [Google Scholar]
- Ferguson S.J. The ups and downs of P/O ratios. Trends Biochem. Sci. 1986;118(1):351–353. [Google Scholar]
- Flügge U.I., Freisl M., Heldt H.W. The mechanism of the control of carbon fixation by the pH in the chloroplast stroma. Planta. 1980;1498(1):48–51. doi: 10.1007/BF00386226. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009;608(1):455–484. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- Frederick S.E., Gruber P.J., Tolbert N.E. The occurence of glycolate dehydrogenase and glycolate oxidase in green plants: An evolutionary survey. Plant Physiol. 1973;528(1):318–323. doi: 10.1104/pp.52.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmes J., Flexas J., Keys A.J., Cifre J., Mitchell R.A.C., Madgwick P.J., Haslam R.P., Medrano H., Parry M.A.J. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ. 2005;288(1):571–579. [Google Scholar]
- Giordano M., Beardall J., Raven J.A. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 2005;568(1):99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- Givan C.V., Kleczkowski L.A. The enzymic reduction of glyoxylate and hydroxypyruvate in leaves of higher plants. Plant Physiol. 1992;1008(1):552–556. doi: 10.1104/pp.100.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A., Tolbert N.E. Association of glycolate oxidation with photosynthetic electron transport in plant and algal chloroplasts. Proc. Natl. Acad. Sci. USA. 1996;938(1):3319–3324. doi: 10.1073/pnas.93.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzinski B. Glyoxylate decarboxylation during photorespiration. Planta. 1978;1448(1):31. doi: 10.1007/BF00385004. [DOI] [PubMed] [Google Scholar]
- Grodzinski B., Colman B. Intracellular localization of glycolate dehydrogenase in a blue-green alga. Plant Physiol. 1976;588(1):199–202. doi: 10.1104/pp.58.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler R.E., Hirsch H.J., Kreuzaler F., Peterhansel C. Overexpression of C4-cycle enzymes in transgenic C3 plants: a biotechnological approach to improve C3 photosynthesis. J. Exp. Bot. 2002;538(1):591–607. doi: 10.1093/jexbot/53.369.591. [DOI] [PubMed] [Google Scholar]
- Halliwell B., Butt V.S. Oxidative decarboxylation of glycollate and glyoxylate by leaf peroxisomes. Biochem. J. 1974;1388(1):217–224. doi: 10.1042/bj1380217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson A.D., Roje S. One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;528(1):119–137. doi: 10.1146/annurev.arplant.52.1.119. [DOI] [PubMed] [Google Scholar]
- Harley P.C., Sharkey T.D. An improved model of C3 photosynthesis at high CO2: Reversed O2 sensitivity explained by lack of glycerate reentry into the chloroplast. Photosynth. Res. 1991;278(1):169–178. doi: 10.1007/BF00035838. [DOI] [PubMed] [Google Scholar]
- Heath M.C. Hypersensitive response-related death. Plant Mol. Biol. 2000;448(1):321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- Herman P.L., Ramberg H., Baack R.D., Markwell J., Osterman J.C. Formate dehydrogenase in Arabidopsis thaliana: overexpression and subcellular localization in leaves. Plant Sci. 2002;1638(1):1137–1145. [Google Scholar]
- Hesketh J. Enhancement of photosynthetic CO2 assimilation in the absence of oxygen, as dependent upon species and temperature. Planta. 1967;768(1):371–374. doi: 10.1007/BF00387543. [DOI] [PubMed] [Google Scholar]
- Hibberd J.M., Sheehy J.E., Langdale J.A. Using C4 photosynthesis to increase the yield of rice - rationale and feasibility. Curr. Opin. Plant Biol. 2008;118(1):228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Hinkle P.C. P/O ratios of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta. 2005;17068(1):1–11. doi: 10.1016/j.bbabio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Howitz K.T., McCarty R. Evidence for a glycolate transporter in the envelope of pea chloroplasts. FEBS Lett. 1983;1548(1):339–342. [Google Scholar]
- Howitz K.T., McCarty R.E. Solubilization, partial purification, and reconstitution of the glycolate/glycerate transporter from chloroplast inner envelope membranes. Plant Physiol. 1991;968(1):1060–1069. doi: 10.1104/pp.96.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husic H.D., Tolbert N.E. Anion and divalent cation activation of phosphoglycolate phosphatase from leaves. Arch. Biochem. Biophys. 1984;2298(1):64–72. doi: 10.1016/0003-9861(84)90130-9. [DOI] [PubMed] [Google Scholar]
- Igamberdiev A.U., Lea P.J. The role of peroxisomes in the integration of metabolism and evolutionary diversity of photosynthetic organisms. Phytochem. 2002;608(1):651–674. doi: 10.1016/s0031-9422(02)00179-6. [DOI] [PubMed] [Google Scholar]
- Igamberdiev A.U., Lea P.J. Land plants equilibrate O2 and CO2 concentrations in the atmosphere. Photosynth. Res. 2006;878(1):177–194. doi: 10.1007/s11120-005-8388-2. [DOI] [PubMed] [Google Scholar]
- Igarashi D., Tsuchida H., Miyao M., Ohsumi C. Glutamate:glyoxylate aminotransferase modulates amino acid content during photorespiration. Plant Physiol. 2006;1428(1):901–910. doi: 10.1104/pp.106.085514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D., Miwa T., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Ohsumi C. Identification of photorespiratory glutamate:glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J. 2003;338(1):975–987. doi: 10.1046/j.1365-313x.2003.01688.x. [DOI] [PubMed] [Google Scholar]
- Intergovernmental panel on climate change. 2007. Climate change 2007: Synthesis report, http://www.ipcc.ch/publications_and_data/publications_ipcc_fourth_assessment_report_synthesis_report.htm.
- Jamai A., Salome P.A., Schilling S.H., Weber A.P., McClung C.R. Arabidopsis photorespiratory serine hydroxymethyltransferase activity requires the mitochondrial accumulation of ferredoxindependent glutamate synthase. Plant Cell. 2009;218(1):595–606. doi: 10.1105/tpc.108.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;1298(1):440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan D.B., Ogren W.L. Species variation in the specificity of ribulose biphosphate carboxylase/oxygenase. Nature. 1981;2918(1):513–515. [Google Scholar]
- Jordan D.B., Ogren W.L. The CO2/O2 specificity of ribulose 1,5-bisphosphate carboxylase/oxygenase. Planta. 1984;1618(1):308–313. doi: 10.1007/BF00398720. [DOI] [PubMed] [Google Scholar]
- Kasting J.F., Howard M.T. Atmospheric composition and climate on the early earth. Phil. Trans. R. Soc. Lond. Biol. Sci. 2006;3618(1):1733–1742. doi: 10.1098/rstb.2006.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasting J.F., Ono S. Palaeoclimates: the first two billion years. Phil. Trans. R. Soc. Lond. Biol. Sci. 2006;3618(1):917–929. doi: 10.1098/rstb.2006.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Reumann S., Hu J. Peroxisome biogenesis and function. The Arabidopsis Book; Rockville, MD: 2009. The American Society of Plant Biologists, doi:10.1199/tab.0123, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebeish R., Niessen M., Thiruveedhi K., Bari R., Hirsch H.-J., Rosenkranz R., Stäbler N., Schönfeld B., Kreuzaler F., Peterhansel C. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat. Biotech. 2007;258(1):593–599. doi: 10.1038/nbt1299. [DOI] [PubMed] [Google Scholar]
- Kelly G.J., Latzko E. Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett. 1976;688(1):55–58. doi: 10.1016/0014-5793(76)80403-6. [DOI] [PubMed] [Google Scholar]
- Kendall A.C., Keys A.J., Turner J.C., Lea P.J., Miflin B.J. The isolation and characterisation of a catalase-deficient mutant of barley (Hordeum vulgare L.). Planta. 1983;1598(1):505–511. doi: 10.1007/BF00409139. [DOI] [PubMed] [Google Scholar]
- Keys A.J. The re-assimilation of ammonia produced by photorespiration and the nitrogen economy of C3 higher plants. Photosynth. Res. 2006;878(1):165–175. doi: 10.1007/s11120-005-9024-x. [DOI] [PubMed] [Google Scholar]
- Keys A.J., Bird I.F., Cornelius M.J., Lea P.J., Wallsgrove R.M., Miflin B.J. Photorespiratory nitrogen cycle. Nature. 1978;2758(1):741–743. [Google Scholar]
- Kisaki T., Tolbert N.E. Glycolate and glyoxylate metabolism by isolated peroxisomes or chloroplasts. Plant Physiol. 1969;448(1):242–250. doi: 10.1104/pp.44.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski L.A., Randall D.D., Edwards G.E. Oxalate as a potent and selective inhibitor of spinach (Spinacia oleracea) leaf NADPH-dependent hydroxypyruvate reductase. Biochem. J. 1991;276 (Pt 18(1):125–127. doi: 10.1042/bj2760125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H.L., Sadler J.R. The metabolism of C2-compounds in microorganisms. VIII. A dicarboxylic acid cycle as a route for the oxidation of glycollate by Escherichia coli. Biochem. J. 1961;818(1):503–513. doi: 10.1042/bj0810503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A., Takeba G. Photorespiration protects C3 plants from photooxidation. Nature. 1996;3848(1):557–560. [Google Scholar]
- Krömer S., Heldt H.W. On the role of mitochondrial oxidative posphorylation in photosynthesis metabolism as studied by the effect of oligomycin on photosynthesis in protoplasts and leaves of barley (Hordeum vulgare). Plant Physiol. 1991;958(1):1270–1276. doi: 10.1104/pp.95.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S.B., Edwards G.E. Oxygen inhibition of photosynthesis: II. kinetic characteristics as affected by temperature. Plant Physiol. 1977a;598(1):991–999. doi: 10.1104/pp.59.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S.B., Edwards G.E. Oxygen inhibition of photosynthesis. I. temperature dependence and relation to O2/CO2 solubility ratio. Plant Physiol. 1977b;598(1):986–990. doi: 10.1104/pp.59.5.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor D.W., Fock H. Water stress induced changes in the amounts of some photosynthetic assimilation products and respiratory metabolites of sunflower leaves. J. Exp. Bot. 1977;288(1):329–337. [Google Scholar]
- Leegood R.C. C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. J. Exp. Bot. 2002;538(1):581–590. doi: 10.1093/jexbot/53.369.581. [DOI] [PubMed] [Google Scholar]
- Li R., Moore M., King J. Investigating the regulation of one-carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2003;448(1):233–241. doi: 10.1093/pcp/pcg029. [DOI] [PubMed] [Google Scholar]
- Li Y., Ye W., Wang M., Yan X. Climate change and drought: a risk assessment of crop-yield impacts. Climate Res. 2009;398(1):31–46. [Google Scholar]
- Liepman A.H., Olsen L.J. Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 2001;258(1):487–498. doi: 10.1046/j.1365-313x.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- Liepman A.H., Olsen L.J. Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 2003;1318(1):215–227. doi: 10.1104/pp.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J.M. Glycolate oxidoreductase in Escherichia coli. Biochim. Biophys. Acta. 1972;2678(1):227–237. doi: 10.1016/0005-2728(72)90111-9. [DOI] [PubMed] [Google Scholar]
- Madore M., Grodzinski B. Effect of oxygen concentration on C-photoassimilate transport from leaves of Salvia splendens L. Plant Physiol. 1984;768(1):782–786. doi: 10.1104/pp.76.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Cai Y., Sun Q., van Wijk K.J. Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell. 2005;178(1):3111–3140. doi: 10.1105/tpc.105.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamedov T.G., Suzuki K., Miura K., Kucho K.-I., Fukuzawa H. Characteristics and sequence of phosphoglycolate phosphatase from a eukaryotic green alga Chlamydomonas reinhardtii. J. Biol. Chem. 2001;2768(1):45573–45579. doi: 10.1074/jbc.M103882200. [DOI] [PubMed] [Google Scholar]
- Martinelli T., Whittaker A., Masclaux-Daubresse C., Farrant J.M., Brilli F., Loreto F., Vazzana C. Evidence for the presence of photorespiration in desiccation-sensitive leaves of the C4 ‘resurrection’ plant Sporobolus stapfianus during dehydration stress. J. Exp. Bot. 2007;588(1):3929–3939. doi: 10.1093/jxb/erm247. [DOI] [PubMed] [Google Scholar]
- Matsuoka M., Furbank R.T., Fukayama H., Miyao M. Molecular engineering of C4 photosynthesis. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 2001;528(1):297–314. doi: 10.1146/annurev.arplant.52.1.297. [DOI] [PubMed] [Google Scholar]
- Maurino V.G., Flügge U.-I. Means for improving agrobiological traits in a plant by providing a plant cell comprising in its chloroplasts enzymatic activities for converting glycolate into malate. Patent application WO 2009/103762 A1. 2009.
- McClung C.R., Hsu M., Painter J.E., Gagne J.M., Karlsberg S.D., Salome P.A. Integrated temporal regulation of the photorespiratory pathway. Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol. 2000;1238(1):381–392. doi: 10.1104/pp.123.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy E.A., Titus S.A., Taylor S.M., Jackson-Cook C., Moran R.G. A mutation inactivating the mitochondrial inner membrane folate transporter creates a glycine requirement for survival of Chinese hamster cells. J. Biol. Chem. 2004;2798(1):33829–33836. doi: 10.1074/jbc.M403677200. [DOI] [PubMed] [Google Scholar]
- Mouillon J.-M., Aubert S., Bourguignon J., Gout E., Douce R., Rébeillé F. Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J. 1999;208(1):197–205. doi: 10.1046/j.1365-313x.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O., Whitney S. Directing the evolution of Rubisco and Rubisco activase: first impressions of a new tool for photosynthesis research. Photosynth. Res. 2008;988(1):667–675. doi: 10.1007/s11120-008-9324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta. 2007;17678(1):414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Murray A.J.S., Blackwell R.D., Lea P.J. Metabolism of hydroxypyruvate in a mutant of barley lacking NADH-dependent hydroxypyruvate reductase, an important photorespiratory enzyme activity. Plant Physiol. 1989;918(1):395–400. doi: 10.1104/pp.91.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.J.S., Blackwell R.D., Joy K.W., Lea P.J. Photorespiratory N donors, aminotransferase specificity and photosynthesis in a mutant of barley deficient in serine:glyoxylate aminotransferase activity. Planta. 1987;1728(1):106–113. doi: 10.1007/BF00403035. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kanakagirl S., Van K., He W., Spalding M. Disruption of the glycolate dehydrogenase gene in the high-CO2-requiring mutant HCR89 of Chlamydomonas reinhardtii. Can. J. Bot. 2005;638(1):820–833. [Google Scholar]
- Neill S., Desikan R., Hancock J. Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 2002;58(1):388–395. doi: 10.1016/s1369-5266(02)00282-0. [DOI] [PubMed] [Google Scholar]
- Niessen M., Thiruveedhi K., Rosenkranz R., Kebeish R., Hirsch H.-J., Kreuzaler F., Peterhansel C. Mitochondrial glycolate oxidation contributes to photorespiration in higher plants. J. Exp. Bot. 2007;588(1):2709–2715. doi: 10.1093/jxb/erm131. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Allakhverdiev S.I., Yamamoto H., Hayashi H., Murata N. Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry. 2004;438(1):11321–11330. doi: 10.1021/bi036178q. [DOI] [PubMed] [Google Scholar]
- Noctor G., Arisi A.-C.M., Jouanin L., Foyer C.H. Photorespiratory glycine enhances glutathione accumulation in both the chloroplastic and cytosolic compartments. J. Exp. Bot. 1999;508(1):1157–1167. [Google Scholar]
- Noctor G., Arisi A., Jouanin L., Kunert K., Rennenberg H., Foyer C. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J. Exp. Bot. 1998;498(1):623–647. [Google Scholar]
- Norman E.G., Colman B. Purification and characterization of phosphoglycolate phosphatase from the cyanobacterium Coccochloris peniocystis. Plant Physiol. 1991;958(1):693–698. doi: 10.1104/pp.95.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman E.G., Colman B. Formation and metabolism of glycolate in the cyanobacterium Coccochloris peniocystis. Arch. Microbiol. 1992;1578(1):375–380. [Google Scholar]
- Novitskaya L., Trevanlon S.J., Driscoll S., Foyer C.H., Noctor G. How does photorespiration modulate leaf amino acid contents? A dual approach through modelling and metabolite analysis. Plant Cell Environ. 2002;258(1):821–835. [Google Scholar]
- Nunes-Nesi A., Sulpice R., Gibon Y., Fernie A.R. The enigmatic contribution of mitochondrial function in photosynthesis. J. Exp. Bot. 2008;598(1):1675–1684. doi: 10.1093/jxb/ern002. [DOI] [PubMed] [Google Scholar]
- Ogren W.L. Affixing the O to Rubisco: discovering the source of photorespiratory glycolate and its regulation. Photosynth. Res. 2003;768(1):53–63. doi: 10.1023/A:1024913925002. [DOI] [PubMed] [Google Scholar]
- Ohnishi J.I., Yamazaki M., Kanal R. Differentiation of photorespiratory activity between mesophyll and bundle sheath cells of 4-carbon pathway photosynthesis plants. II. Peroxisomes of Panicum miliaceum. Plant Cell Physiol. 1985;268(1):797–804. [Google Scholar]
- Oliver D.J. Role of glycine and glyoxylate decarboxylation in photorespiratory CO2 release. Plant Physiol. 1981;688(1):1031–1034. doi: 10.1104/pp.68.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D.J., Neuburger M., Bourguignon J., Douce R. Interaction between the component enzymes of the glycine decarboxylase multienzyme complex. Plant Physiol. 1990;948(1):833–839. doi: 10.1104/pp.94.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson B.J.S.C, Skavdahl M., Ramberg H., Osterman J.C., Markwell J. Formate dehydrogenase in Arabidopsis thaliana: characterization and possible targeting to the chloroplast. Plant Sci. 2000;1598(1):205. doi: 10.1016/s0168-9452(00)00337-x. [DOI] [PubMed] [Google Scholar]
- Osmond B., Badger M., Maxwell K., Björkman O., Leegood R.C. Too many photons: photorespiration, photoinhibition and photooxidation. Trends Plant Sci. 1997;28(1):119–121. [Google Scholar]
- Parry M.A.J., Madgwick P.J., Carvalho J.F.C., Andralojc P.J. Prospects for increasing photosynthesis by overcoming the limitations of Rubisco. J. Agr. Sci. 2007;1458(1):31–43. [Google Scholar]
- Paul J.S., Volcani B.E. A mitochondrial glycolate:cytochrome C reductase in Chlamydomonas reinhardii. Planta. 1976;1298(1):59–61. doi: 10.1007/BF00390914. [DOI] [PubMed] [Google Scholar]
- Pellicer M.T., Badia J., Aguilar J., Baldoma L. glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol. 1996;1788(1):2051–2059. doi: 10.1128/jb.178.7.2051-2059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhansel C., Niessen M., Kebeish R.M. Metabolic engineering towards the enhancement of photosynthesis. Photochem. Photobiol. 2008;848(1):1317–1323. doi: 10.1111/j.1751-1097.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- Peterman T.K., Goodman H.M. The glutamine synthetase gene family of Arabidopsis thaliana light-regulation and differential expression in leaves, roots and seeds. Mol. Gen. Genet. 1991;2308(1):145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Peters J.L., Cnudde F., Gerats T. Forward genetics and map-based cloning approaches. Trends Plant Sci. 2003;88(1):484–491. doi: 10.1016/j.tplants.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Piper M.D., Hong S.-P., Ball G.E., Dawes I.W. Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2000;2758(1):30987–30995. doi: 10.1074/jbc.M004248200. [DOI] [PubMed] [Google Scholar]
- Popov V.N., Dmitrieva E.A., Eprintsev A.T., Igamberdiev A.U. Glycolate oxidase isoforms are distributed between the bundle sheath and mesophyll tissues of maize leaves. J. Plant Physiol. 2003;1608(1):851–857. doi: 10.1078/0176-1617-01105. [DOI] [PubMed] [Google Scholar]
- Potel F., Valadier M.-H., Ferrario-Méry S., Grandjean O., Morin H., Gaufichon L., Boutet-Mercey S., Lothier J., Rothstein S.J., Hirose N., Suzuki A. Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana: roles of glutamate synthases and carbamoylphosphate synthetase in leaves. FEBS J. 2009;2768(1):4061–4076. doi: 10.1111/j.1742-4658.2009.07114.x. [DOI] [PubMed] [Google Scholar]
- Prabhu V., Chatson K.B., Abrams G.D., King J. 13C nuclear magnetic resonance detection of interactions of serine hydroxymethyltransferase with C1-tetrahydrofolate synthase and glycine decarboxylase complex activities in Arabidopsis. Plant Physiol. 1996;1128(1):207–216. doi: 10.1104/pp.112.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J.E., Smith S.M. Arabidopsis peroxisomal malate dehydrogenase functions in b-oxidation but not in the glyoxylate cycle. Plant J. 2007;508(1):381–390. doi: 10.1111/j.1365-313X.2007.03055.x. [DOI] [PubMed] [Google Scholar]
- Price G.D., Badger M.R., Woodger F.J., Long B.M. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 2007;598(1):1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gaklère B., Vanacker H., Miginiac-Maslow M., Breusegem F.V., Noctor G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007;528(1):640–657. doi: 10.1111/j.1365-313X.2007.03263.x. [DOI] [PubMed] [Google Scholar]
- Rachmilevitch S., Cousins A.B., Bloom A.J. Nitrate assimilation in plant shoots depends on photorespiration. Proc. Natl. Acad. Sci. USA. 2004;1018(1):11506–11510. doi: 10.1073/pnas.0404388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A.S., Reumann S., Heldt H.W. Participation of mitochondrial metabolism in photorespiration. Reconstituted system of peroxisomes and mitochondria from spinach leaves. Plant Physiol. 1998;1168(1):1333–1337. doi: 10.1104/pp.116.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven J.A., Cockell C.S., De La Rocha C.L. The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Phil. Trans. R. Soc. Lond. Biol. Sci. 2008;3638(1):2641–2650. doi: 10.1098/rstb.2008.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redel G.P. Arabidopsis as a genetic tool. Annu. Rev. Genet. 1975;98(1):111–127. doi: 10.1146/annurev.ge.09.120175.000551. [DOI] [PubMed] [Google Scholar]
- Renne P., Dressen U., Hebbeker U., Hille D., Flugge U.I., Westhoff P., Weber A.P. The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 2003;358(1):316–331. doi: 10.1046/j.1365-313x.2003.01806.x. [DOI] [PubMed] [Google Scholar]
- Renström E., Bergman B. Glycolate metabolism in cyanobacteria. Physiol. Plant. 1989;758(1):137–143. [Google Scholar]
- Reumann S. Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 2004;1358(1):783–800. doi: 10.1104/pp.103.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Weber A.P.M. Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled -others remain. Biochim. Biophys. Acta. 2006;17638(1):1496. doi: 10.1016/j.bbamcr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Reumann S., Maier E., Benz R., Heldt H.W. The membrane of leaf peroxisomes contains a porin-like channel. J. Biol. Chem. 1995;2708(1):17559–17565. doi: 10.1074/jbc.270.29.17559. [DOI] [PubMed] [Google Scholar]
- Reumann S., Bettermann M., Benz R., Heldt H.W. Evidence for the presence of a porin in the membrane of glyoxysomes of castor bean. Plant Physiol. 1997;1158(1):891–899. doi: 10.1104/pp.115.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Maier E., Heldt H.W., Benz R. Permeability properties of the porin of spinach leaf peroxisomes. Eur. J. Biochem. 1998;2518(1):359–366. doi: 10.1046/j.1432-1327.1998.2510359.x. [DOI] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Silemsen T., Antonicelli G.E., Rasche N., Luder F., Weckwerth W., Jahn O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell. 2007;198(1):3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R.F. The evolution of C4 photosynthesis. New Phytol. 2004;1618(1):341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- Sage T.L., Sage R.F. The functional anatomy of rice leaves: Implications for refixation of photorespiratory CO2 and efforts to engineer C4 photosynthesis into rice. Plant Cell Physiol. 2009;508(1):756–772. doi: 10.1093/pcp/pcp033. [DOI] [PubMed] [Google Scholar]
- Schneidereit J., Hausler R.E., Fiene G., Kaiser W.M., Weber A.P. Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J. 2006;458(1):206–224. doi: 10.1111/j.1365-313X.2005.02594.x. [DOI] [PubMed] [Google Scholar]
- Schwarte S., Bauwe H. Identification of the photorespiratory 2-phosphoglyoolate phosphatase, PGLP1, in Arabidopsis. Plant Physiol. 2007;1448(1):1580–1586. doi: 10.1104/pp.107.099192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S.N., Rose Z.B. Characterization of a phosphoenzyme intermediate in the reaction of phosphoglycolate phosphatase. J. Biol. Chem. 1987;2628(1):13496–13500. [PubMed] [Google Scholar]
- Sharkey T.D. Estimating the rate of photorespiration in leaves. Physiol. Plant. 1988;738(1):147–152. [Google Scholar]
- Singh P., Kumar P.A., Abrol Y.P., Naik M.S. Photorespiratory nitrogen cycle - A critical evaluation. Physiol. Plant. 1986;668(1):169–176. [Google Scholar]
- Somerville C.R. An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol. 2001;1258(1):20–24. doi: 10.1104/pp.125.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C.R., Ogren W.L. A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature. 1979;2608(1):833–836. [Google Scholar]
- Somerville C.R., Ogren W.L. Photorespiration mutants of Arabidopsis thaliana deficient in serine-glyoxylate aminotransferase activity. Proc. Natl. Acad. Sci. USA. 1980a;778(1):2684–2687. doi: 10.1073/pnas.77.5.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C.R., Ogren W.L. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature. 1980b;2868(1):257–259. [Google Scholar]
- Somerville C.R., Ogren W.L. Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 1981;678(1):666–671. doi: 10.1104/pp.67.4.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C.R., Ogren W.L. Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem. J. 1982a;2028(1):373–380. doi: 10.1042/bj2020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C.R., Ogren W.L. Genetic modification of photorespiration. Trends Biochem. Sci. 1982b;78(1):171–174. [Google Scholar]
- Somerville S.C., Ogren W.L. An Arabidopsis thaliana mutant defective in chloroplast dicarboxylate transport. Proc. Natl. Acad. Sci. USA. 1983;808(1):1290–1294. doi: 10.1073/pnas.80.5.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M.H. Microalgal carbon-dioxide-concentrating mechanisms: Chlamydomonas inorganic carbon transporters. J. Exp. Bot. 2007;598(1):1463–1473. doi: 10.1093/jxb/erm128. [DOI] [PubMed] [Google Scholar]
- Stabenau H., Winkler U. Glycolate metabolism in green algae. Physiol. Plant. 2005;1238(1):235–245. [Google Scholar]
- Suzuki K., Marek L.F., Spalding M.H. A photorespiratory mutant of Chlamydomonas reinhardtii. Plant Physiol. 1990;938(1):231–237. doi: 10.1104/pp.93.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Lytovchenko A., Morgan M., Nunes-Nesi A., Taylor N.L., Baxter C.J., Elckmeier I., Fernie A.R. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA. 2006;1038(1):19587–19592. doi: 10.1073/pnas.0607751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita F.R., Hanson T.E., Li H., Satagopan S., Singh J., Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 2007;718(1):576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira M., Valtersson U., Burkhardt B., Ludwig R. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts Plant Cell. 2004;168(1):2048–2058. doi: 10.1105/tpc.104.022046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Bauwe H., Badger M. Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 2007;1448(1):487–494. doi: 10.1104/pp.107.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taler D., Galperin M., Benjamin I., Cohen Y., Kenigsbuch D. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell. 2004;168(1):172–184. doi: 10.1105/tpc.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Taniguchi Y., Kawasaki M., Takeda S., Kato T., Sato S., Tabata S., Miyake H., Sugiyama T. Identifying and characterizing plastidic 2-oxoglutarate/malate and dicarboxylate transporters in Arabidopsis thaliana. Plant Cell Physiol. 2002;438(1):706–717. doi: 10.1093/pcp/pcf109. [DOI] [PubMed] [Google Scholar]
- Tcherkez G.G., Farquhar G.D., Andrews T.J. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. USA. 2006;1038(1):7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Nunes-Nesl A., Parnik T., Morgenthal K., Wienkoop S., Keerberg O., Weckwerth W., Kleczkowski L.A., Fernie A.R., Bauwe H. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell. 2008;208(1):2848–2859. doi: 10.1105/tpc.108.062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K., Wang Y., Nakamura Y., Spalding M.H. Insertional mutants of Chlamydomonas reinhardtii that require elevated CO2 for survival. Plant Physiol. 2001;1278(1):607–614. [PMC free article] [PubMed] [Google Scholar]
- Voll L.M., Jamai A., Renne P., Voll H., McClung C.R., Weber A.P.M. The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 2006;1408(1):59–66. doi: 10.1104/pp.105.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallsgrove R.M., Turner J.C., Hall N.P., Kendall A.C., Bright S.W. Barley mutants lacking chloroplast glutamine synthetasebiochemical and genetic analysis. Plant Physiol. 1987;838(1):155–158. doi: 10.1104/pp.83.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A., Flügge U.-I. Interaction of cytosolic and plastidic nitrogen metabolism in plants. J. Exp. Bot. 2002;538(1):865–874. doi: 10.1093/jexbot/53.370.865. [DOI] [PubMed] [Google Scholar]
- Weise S.E., Schrader S.M., Kleinbeck K.R., Sharkey T.D. Carbon balance and circadian regulation of hydrolytic and phosphorolytic breakdown of transitory starch. Plant Physiol. 2006;1418(1):879–886. doi: 10.1104/pp.106.081174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A., Lea P.J., Leegood R.C. Photorespiratory metabolism of glyoxylate and formate in glycine-accumulating mutants of barley and Amaranthus edulis. Planta. 1999;2078(1):518–526. [Google Scholar]
- Wingler A., Lea P.J., Quick W.P., Leegood R.C. Photorespiration: metabolic pathways and their role in stress protection. Phil. Trans. R. Soc. Lond. Biol. Sci. 2000;3558(1):1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K.C., Flugge U.I., Heldt H.W. A two-translocator model for the transport of 2-oxoglutarate and glutamate in chloroplasts during ammonia assimilation in the light. Plant Physiol. 1987;848(1):624–632. doi: 10.1104/pp.84.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Shortt B.J., Lawrence E.B., Leon J., Fitzsimmons K.C., Levine E.B., Raskin I., Shah D.M. Activation of host defense mechanisms by elevated production of H2O2 in transgenic plants. Plant Physiol. 1997;1158(1):427–435. doi: 10.1104/pp.115.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Bauer C.E. Complex evolution of photosynthesis. Annu. Rev. Plant Biol. 2002;538(1):503–521. doi: 10.1146/annurev.arplant.53.100301.135212. [DOI] [PubMed] [Google Scholar]
- Xu H., Zhang J., Zeng J., Jiang L., Liu E., Peng C., He Z., Peng X. Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. J. Exp. Bot. 2009;608(1):1799–1809. doi: 10.1093/jxb/erp056. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Nishimura M. Reduction to below threshold levels of glycolate oxidase activities in transgenic tobacco enhances photoinhibition during irradiation. Plant Cell Physiol. 2000;418(1):1397–1406. doi: 10.1093/pcp/pcd074. [DOI] [PubMed] [Google Scholar]
- Yamamoto H.Y., Nakayama T.O., Chichester C.O. Studies on the light and dark interconversions of leaf xanthophylls. Arch. Biochem. Biophys. 1962;978(1):168–173. doi: 10.1016/0003-9861(62)90060-7. [DOI] [PubMed] [Google Scholar]
- Yu C., Claybrook D.L., Huang A.H.C. Transport of glycine, serine, and proline into spinach leaf mitochondria. Arch. Biochem. Biophys. 1983;2278(1):180–187. doi: 10.1016/0003-9861(83)90361-2. [DOI] [PubMed] [Google Scholar]
- Zelitch I. The photooxidation of glyoxylate by envelope-free spinach chloroplasts and its relation to photorespiration. Arch. Biochem. Biophys. 1972;1508(1):698–707. doi: 10.1016/0003-9861(72)90088-4. [DOI] [PubMed] [Google Scholar]
- Zelitch I. The effect of glycidate, an inhibitor of glycolate synthesis, on photorespiration and net photosynthesis. Arch. Biochem. Biophys. 1974;1638(1):367–377. doi: 10.1016/0003-9861(74)90488-3. [DOI] [PubMed] [Google Scholar]
- Zelitch I., Day P.R. The effect on net photosynthesis of pedigree selection for low and high rates of photorespiration in tobacco. Plant Physiol. 1973;528(1):33–37. doi: 10.1104/pp.52.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelitch I., Schultes N.P., Peterson R.B., Brown P., Brutnell T.P. High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 2008;1498(1):195–204. doi: 10.1104/pp.108.128439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H.H., Resnick A.S., Straume M., McClung C.R. Effects of synergistic signaling by phytochrome A and oryptoohromel on circadian clock-regulated catalase expression. Plant Cell. 1997;98(1):947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.-G., Portis A.R., Long S.P. Would transformation of C3 crop plants with foreign Rubisco increase productivity? A computational analysis extrapolating from kinetic properties to canopy photosynthesis. Plant Cell Environ. 2004;278(1):155–165. [Google Scholar]