Abstract
In Alzheimer’s disease (AD) and other neurodegenerative disorders, proteins accumulate into ordered aggregates, called amyloids. Recent evidence suggests that these structures include both large, insoluble fibrils and smaller, prefibrillar structures, such as dimers, oligomers, and protofibrils. Recently, focus has shifted to the prefibrillar aggregates because they are highly neurotoxic and their levels appear to correlate with cognitive impairment. Thus, there is interest in finding methods for specifically quantifying these structures. One of the classic ways of detecting amyloid formation is through the fluorescence of the benzothiazole dye, thioflavin T (ThT). This reagent has been a “workhorse” of the amyloid field because it is robust and inexpensive. However, one of its limitations is that it does not distinguish between prefibrillar and fibrillar aggregates. We screened a library of 37 indoles for those that selectively change fluorescence in the presence of prefibrillar amyloid-β(Aβ). From this process, we selected the most promising example, tryptophanol (TROL), to use in a quantitative “thioflavin-like” assay. Using this probe in combination with electron microscopy, we found that prefibrils are largely depleted during Aβ aggregation in vitro but that they remain present after the apparent saturation of the ThT signal. These results suggest that a combination of TROL and ThT provides greater insight into the process of amyloid formation by Aβ. In addition, we found that TROL also recognizes other amyloid-prone proteins, including ataxin-3, amylin, and CsgA. Thus, this assay might be an inexpensive spectroscopic method for quantifying amyloid prefibrils in vitro.
Keywords: Alzheimer’s disease, amyloid-β, oligomers, prefibrils, thioflavin T
Introduction
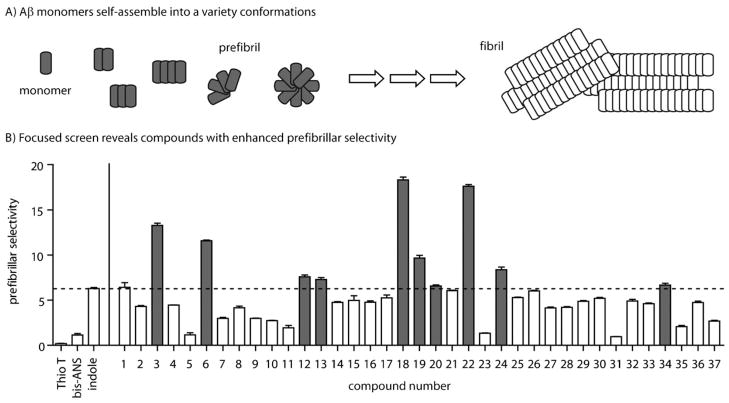
Many neurodegenerative disorders, including Alzheimer’s and Parkinson’s diseases, are characterized by the age-dependent accumulation of aggregated protein, known as amyloid, in the brain.[1, 2] In each disease, a different protein is implicated; for example, in AD, the amyloid-β(Aβ) peptide self-associates to form insoluble fibrils that deposit in the classic neuritic plaque (NP) pathology. These visually striking fibrils were initially thought to be linked to neurotoxicity but, more recently, it has been found that Aβ forms a variety of other conformations, including dimers, trimers, and globular oligomers (Figure 1A). Collectively, these prefibrillar structures appear to be of special importance in disease. For instance, AD pathology is specifically correlated with the presence of prefibrils but not fibrils.[3–5] Also, recent studies show that Aβ dimers and trimers are more neurotoxic than fibrils to cultured neurons.[6–10] Together, these observations and others have generated interest in finding reagents that specifically detect prefibrillar amyloids.[3, 11, 12]
Figure 1.
Assessment of prefibrillar selectivity in a focused indole collection. A) Schematic of the Aβ-aggregation pathway highlights some prefibril structures (e.g., monomers, dimers, trimers, tetramers, protofibrils, and globular oligomers) that are believed to eventually form fibrils. This schematic is not meant to imply any particular order to the aggregation event, only to show that multiple types of structures are observed and that these can be broadly classified as either prefibrillar or fibrillar. B) A library of 37 indole-containing compounds was screened for changes in fluorescence in the presence of either Aβ prefibrils or fibrils. Prefibrillar selectivity is defined as the ratio (prefibril/fibril) of the fluorescence change. The results from ten compounds (gray bars) that showed greater prefibrillar selectivity relative to the initial “hit” indole (6.3; dashed line), are highlighted. All compounds were tested in triplicate and the error bars represent the standard deviation.
Small organic dyes, such as thioflavin T (ThT) and Congo Red, have been commonly used to quantify the total amount of aggregated Aβ in vitro.[13–22] Briefly, ThT becomes fluorescent in the presence of aggregated, but not monomeric, amyloid.[14] Thus, its fluorescence can be used to robustly and inexpensively monitor the aggregation process. However, although ThT can report on aggregation, it does not distinguish between prefibrillar and fibrillar Aβ. Given the current focus on understanding the biology of prefibrils, we sought to develop a robust, spectroscopic, “ThT-like” assay to selectively detect pre-fibrils in complex Aβ mixtures.
Toward this goal, we recently screened a small library of chemically diverse Aβ ligands and identified five indole-containing compounds whose fluorescence was quenched in the presence of prefibrillar Aβ, but not Aβ fibrils.[23] Here, we expand on this initial observation by screening a focused collection of 37 indoles. Using the most promising probe (tryptophanol, TROL), we developed an assay that quantifies prefibrillar Aβ, even in complex mixtures. We also found that TROL is sensitive to the prefibrillar forms of other amyloids, including ataxin-3, amylin, and CsgA. Thus, this probe appears to provide an inexpensive and robust way to quantify prefibrillar material in vitro.
Results
A focused screen identifies TROL as a reagent with 18-fold selectivity for prefibrillar Aβ
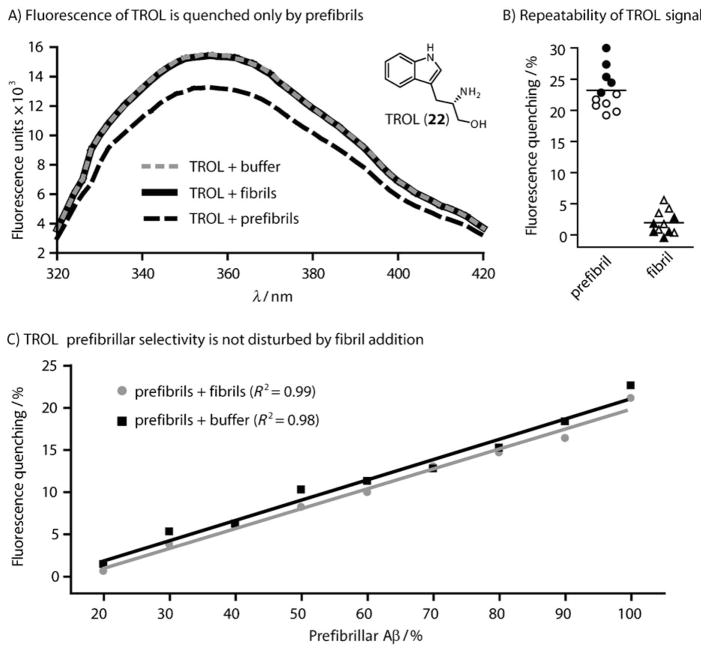
Based on the results of a pilot screen,[24] our objective was to empirically identify indole derivatives with improved signal intensity and selectivity. We collected 37 substituted indoles (see Table S1 in the Supporting Information) and incubated 100 μM of each of them with solutions enriched for either Aβ prefibrils or Aβ fibrils. As previously reported, the prefibrillar solution contained a mixture of spherical oligomers and smaller aggregates.[23, 24] The fibril sample contained exclusively elongated fibrils from aged Aβ preparation. Following a 60-minute incubation, the fluorescence of each indole-treated sample was recorded and compared to that of the indole alone. We first determined the percent change in the presence of either prefibrils or fibrils. Then we calculated the ratio of the fluorescence change between these samples (Δfluorescence prefibril sample/Δfluorescence fibril sample) and termed this value “prefibrillar selectivity”. This analysis confirmed[23, 25] that ThT and another common probe, bis-ANS, had poor prefibrillar selectivity (values of 0.2 and 1.1, respectively; Figure 1B). However, we identified ten compounds (3, 6, 12, 13, 18, 19, 20, 22, 24, and 34) with better selectivity than the initial indole (Figure 1B and Table S1). We decided to focus on compound 22 (tryptophanol; TROL) because of its combination of high fluorescence intensity, selectivity (ca. 18-fold), and relatively large quenching (ca. 25 %; Figure 2A).
Figure 2.
TROL selectively detects prefibrils. A) The fluorescence spectra of TROL (100 μM, λex = 280 nm) in the presence of 25 μM Aβ prefibrils (black dashed), fibrils (black), or PBS (gray dashed) shows the quenching effect of prefibrils. Inset: chemical structure of tryptophanol. B) The TROL quenching effect is robust and repeatable. Aliquots of either Aβ(1–40) (closed symbols) or (1–42) (open symbols) were tested. These data include 11 samples from two different vendors and stock solutions. The average quench is shown by the black bar. C) Increasing amounts of Aβ fibrils (gray) or PBS (black) were titrated into a solution of prefibrils. The change in TROL fluorescence shows that fibrils do not disrupt the signal. Each data point was performed in triplicate and the error bars represent the standard deviation. Note that error bars are often smaller than the data symbols.
Tryptophanol (TROL) assay optimization and repeatability
To scrutinize TROL as a potential fluorescent probe, we first evaluated the influence of a range of potential assay parameters. We focused on the parameters that have previously been shown to be important in the ThT protocol, including 1) the concentration of probe, 2) the buffer conditions, 3) the incubation time, and 4) incubation temperature. We varied these factors and selected the conditions under which the greatest signal was achieved (Figure S1). This search resulted in the optimized protocol shown in Table 1. Briefly, we found that the fluorescence equilibrates within 15 min following a brief, five-minute incubation at 37 °C, as long as the TROL concentration was 10 μM, the volume was 100 μL, and the TROL stock solution was used within 7 h of its preparation (Figure S1). We also noticed that gently mixing the solutions with a pipette accelerated the equilibration of the signal. After incorporating these optimized procedures, we were pleased to find that the general procedures for the TROL and ThT assays were roughly parallel (Table 1); this suggested that they could be conveniently employed side-by-side.
Table 1.
Comparison of the TROL and ThT protocols.
| Tryptophanol (TROL) assay | Thioflavin T (ThT) assay |
|---|---|
| 1) add 10 μL of amyloid solution to 96-well plate[a] | 1) add 10 μL of amyloid solution to 96-well plate[a] |
| 2) add 100 μL TROL (10 μM) per well (50 mM glycine, pH 8.2, 0.01 % DMSO) | 2) add 200 μL filtered (0.22 μm) ThT (5 μM) per well (50 mM glycine, pH 8.2) |
| 3) mix by pipetting, incubate 5 min (37 °C) and then cool 15 min (RT) | 3) mix by pipetting and incubate 15 min (RT) |
| 4) read fluorescence (λex = 280/λem = 355) | 4) read fluorescence (λex = 446/λem = 490) |
96-well black, opaque, flat-bottomed microplate.
Because Aβ aggregation reactions can be heterogeneous, variability often arises when comparing samples from different stock solutions or vendors. Therefore, we wanted to explicitly study the repeatability of the TROL signal. Using several batches of both Aβ(1–40) and Aβ(1–42) obtained from two vendors, we performed reproducibility profiling experiments. These studies revealed that prefibrils consistently quenched TROL fluorescence by over 20 %, while fibrils routinely gave little signal (Figure 2B). Further, the TROL signal was similar regardless of whether Aβ(1–40) or (1–42) was used.
TROL detects prefibrillar Aβ in the presence of preformed fibrils
For the TROL assay to be applicable in monitoring aggregation, the probe must be able to reliably detect prefibrils in mixtures of different amyloid conformations. To address this possibility, we added increasing amounts of prefibrillar Aβ(1–40) to solutions of fibrils. These well-defined mixtures were then immediately incubated with TROL. Using this approach, we found that the TROL signal could reliably detect prefibrils even in mixtures (Figure 2C). We also estimated that the lower limit for the detection of prefibrillar Aβ is approximately 7.5 μM (or 30 % of the mixture), after which the TROL signal does not change significantly. It is important to note that, because it is difficult to determine the concentration of prefibrils accurately, this method is likely not accurate at determining the absolute concentration of these structures de novo. Based upon these findings, we predicted that TROL might be used to quantify prefibrillar Aβ during amyloid formation, analogous to how ThT is used to track total aggregation.
Monitoring the depletion of prefibrillar Aβ during aggregation by using TROL
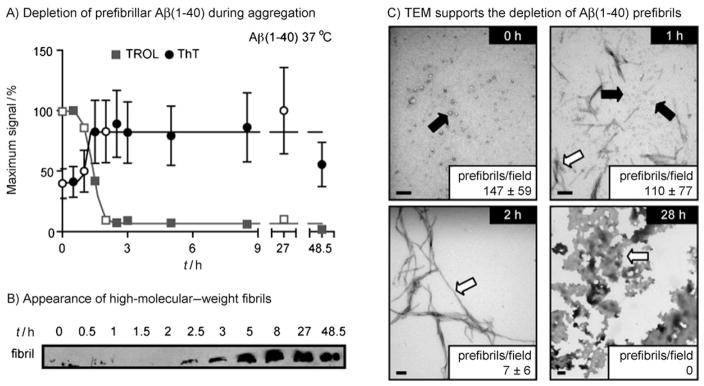
One use of ThT is to monitor the kinetics of aggregate formation.[26–30] Similarly, we hypothesized that the TROL signal might dissipate during depletion of its target, revealing when prefibrils progress to fibrils during the aggregation pathway. To test this idea, Aβ(1–40) was incubated at 37 °C and, at the indicated times, 10 μL aliquots were removed and plated in two sets of triplicates. The TROL and ThT assays were then performed concurrently. From these experiments, we observed a roughly inverse relationship between the signals (Figure 3A). As expected, at early time points (0, 0.5, and 1 h), both signals were relatively stable, with the TROL assay showing maximum prefibrillar content and ThT indicating low amounts of total aggregated Aβ. Between 1 and 1.5 h, the TROL signal decreased dramatically, while the ThT signal increased by 30 %; this suggests that more advanced Aβ aggregates begin to accumulate during this time. Interestingly, the TROL signal remained clearly present after the ThT signal reached equilibrium (e.g., 1.5 h). After TROL signal equilibration (~2.0 to 2.5 h), the signal remains slightly above zero until 48 h. We hypothesize that prefibrils remain during this time, and could possibly exist within a dynamic equilibrium with the mature fibrils. These findings suggest that the fibrillization process (and, specifically, depletion of prefibrillar content) remains active after the ThT signal has equilibrated. Similar trends were observed for the longer peptide, Aβ(1–42) (Figure S2).
Figure 3.
Monitoring the decrease of prefibrillar Aβ over time by using TROL. A) 25 μM Aβ(1–40) was suspended in PBS and incubated at 37 °C with shaking. At the indicated time points, 10 μL was removed and tested for either TROL or ThT reactivity in triplicate. Open symbols correspond to the samples used in the TEM experiments. Error bars represent standard deviation. In some cases the error is smaller than the data symbol. B) A general anti-Aβ antibody (6E10) was used to monitor high-molecular-weight fibrils. At the indicated time points, samples were separated by electrophoresis, and the interface between the stacking and resolving gels was blotted. C) TEM was used to evaluate the content of Aβ mixtures. The levels of prefibrils were roughly quantified at each time point (average prefibrils per 13.2 μm2 field). Prefibrils were defined as globular oligomers and short protofibrils. Scale bar = 100 nm. Arrows indicate prefibrils (black arrow) and fibrils (open arrow).
Neither the TROL nor the ThT signal is sufficient to draw any definitive conclusions about prefibrillar or fibrillar content. Rather, complementary techniques, such as gel electrophoresis and TEM, are commonly employed to supplement studies of Aβ aggregation.[24, 31, 32] For example, one feature of large fibrils is that they are retained at the stacking gel during electrophoresis. We used this method to probe when these structures form in relation to the TROL and ThT signals. Using the general anti-Aβ antibody (6E10), we observed a marked increase in a high-molecular-weight band during aggregation (Figure 3B). Specifically, this band emerged after 2.5 h, and its relative intensity increased until approximately 8 h. It then remained throughout the rest of the time course. In comparison, the TROL response equilibrated at around 2.0 h; this suggests that this probe’s signal is exclusive of the formation of high-molecular-mass Aβ fibrils. Interestingly, the ThT fluorescence was saturated approximately 1 hour prior to the appearance of the large fibrils; it had reached ~90 % of its maximum signal at 1.5 h. Thus, these results suggest that the disappearance of the TROL signal better correlated with the appearance of the fibrils, likely because ThT was unable to distinguish between prefibrillar and fibrillar conformations.
Next, we used TEM to analyze the prefibrillar and fibrillar content at various times during Aβ(1–40) aggregation. We specifically focused on the 1 and 2 h times, because changes in the sensitivity to TROL and ThT seemed to occur in this period. To allow quantitative comparisons, the average number of prefibrillar, spherical oligomers per field (13.2 μM2) was determined. As expected, many prefibrillar structures were observed a few minutes after the initiation of aggregation (147 ± 59 prefibrils per field; Figure 3C). By 1 h, this number had decreased to 110 ± 77 prefibrils per field, corresponding with the appearance of some short fibrils. At 2 h, the total level of prefibrils was depleted dramatically (~7 per field), and the sample consisted largely of mature fibrils (average length >500 nm) (Figure 3C). By 27 h, large amorphous deposits predominated and the smaller structures had been entirely depleted. Together, these results support a model in which TROL recognizes prefibrillar Aβ.
TROL signal diminishes during aggregation of ataxin-3, amylin, and CsgA, but not α-synuclein
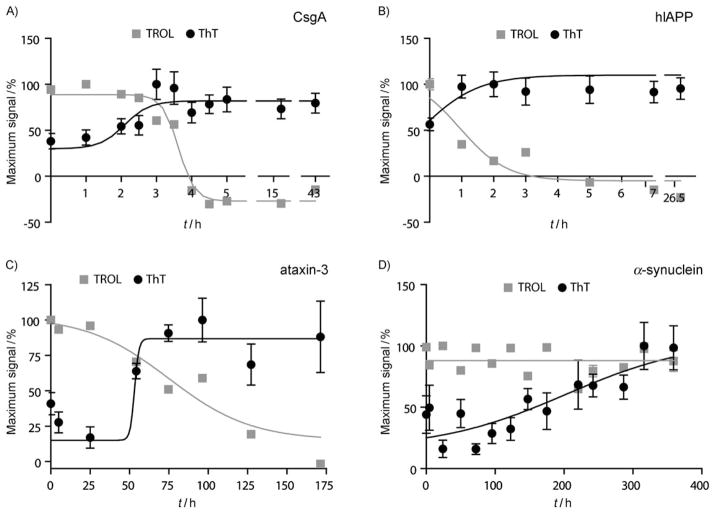
Many proteins involved in aggregation disorders, such as human islet amyloid polypeptide (hIAPP or amylin) and α-synuclein, assemble into amyloids whose morphologies are nearly indistinguishable from those formed by Aβ.[3, 33–35] In addition, a growing number of functional amyloids are being described, including those assembled from the bacterial protein CsgA.[36] Despite the fact that none of these amyloid-forming proteins share obvious sequence homology, they all form β-sheet-rich fibrils and they interact with ThT.[37] To test whether TROL can also recognize multiple amyloid-forming proteins, we performed assays on α-synuclein, amylin (hIAPP), ataxin-3, and CsgA, using known procedures (see the Experimental Section). Interestingly, we found that the TROL signal diminished during the aggregation of CsgA, amylin, and ataxin-3 (Figure 4A–C), but not during α-synuclein self-assembly (Figure 4D). For each target, especially those that contain tryptophan residues, we conducted side-by-side controls in the absence of the TROL reagent, but found that the signal from the TROL dominated the fluorescence signature. For the sensitive amyloids, TROL fluorescence decreased concurrently with the increase in ThT fluorescence, qualitatively similar to what we observed with Aβ(1–40). For α-synuclein, we did not observe any change in fluorescence; this suggests that either these fibrils quench TROL fluorescence or that prefibrils remain during the entire time course. Regardless, these results demonstrate that the TROL response is not restricted to Aβ, but that it likely responds to a common prefibrillar characteristic of multiple amyloid systems.
Figure 4.
TROL is sensitive to prefibrillar forms of several amyloidogenic proteins The TROL assay was conducted with four amyloid-forming proteins. For each experiment, ThT and TROL fluorescence was recorded side-by-side. TROL reacts with all four amyloids and the signal dramatically decreases in the presence of A) CsgA (25 μM), B) hIAPP/amylin (20 μM), and C) ataxin-3 (10 μM). D) TROL fluorescence is not responsive to the presence of α-synuclein throughout the entire time-course. Each data point is the average of three experiments. Error bars represent the standard deviation and are often smaller than the data symbols. For each data set, results were fit to arbitrary sigmoidal or linear curves only to facilitate visual interpretation.
Discussion
Existing chemical dyes for the rapid in vitro quantification of amyloids, such as ThT and bis-ANS, have been invaluable for studying the aggregation process, but they do not distinguish between prefibrillar and fibrillar Aβ. Here, we report the development of the TROL assay: an analogous, fluorescence-based method for the specific detection of prefibrillar Aβ in vitro. We found that prefibrils quenched TROL fluorescence and that this effect was not disrupted by the presence of fibrils. When TROL was used together with ThT, we found evidence that Aβ prefibrils continue to be present after equilibration of the ThT signal, at least under the conditions we employed (Figure 3A). Thus, the Aβ-aggregation process is likely ongoing, producing mature fibrils from pools of prefibrillar structures, a conclusion that is supported by the electrophoresis (Figure 3B) and electron microscopy studies (Figure 3C) as well as previous literature reports.[38–41] We further showed that the TROL assay is applicable to several amyloid systems (Figure 4). However, it is important to note that some amyloid-forming proteins, such as ataxin-3, contain intrinsic tryptophan residues and therefore require additional controls. When envisioning other applications of this reagent, it is important to note that TROL is likely not suitable for use in vivo. Several groups have noted that indole fluorescence is altered by nonamyloidogenic proteins, such as BSA, thus limiting its use to in vitro applications.[42, 43] Despite this limitation, there are likely multiple potential applications of the TROL protocol, such as studying the contributions of individual residues to prefibril formation or rapidly quantifying prefibrillar content prior to measuring relative toxicity.
Prefibrillar Aβ levels are thought to correlate with AD pathology better than the amount of fibrils.[5, 8] Despite this emerging consensus, there is still considerable debate as to what exactly constitutes the key neurotoxic structure(s). Because there is good evidence for a number of these structures, including dimers, spherical oligomers of various sizes, and other intermediate assemblies,[1, 8, 44] it seems plausible that multiple Aβ conformations contribute to disease. These challenges highlight the need to develop multiple classes of informative amyloid reporters, including those that selectively recognize individual conformations (e.g., oligomers) and those that more broadly encompass the known neurotoxic structures (e.g., dimers, oligomers, etc.). In response to the first of these needs, a series of useful antibodies has been developed against specific antigens.[3, 11] Towards the second aim, assays such as TROL might provide a survey of total prefibrillar content.
What feature of an amyloid prefibril is recognized by TROL ? This probe appears to react with multiple structures, likely encompassing multiple types of oligomeric and monomeric Aβ(Figure S3), but its main feature is that it fails to recognize fibrillar Aβ. We hypothesize that TROL might either recognize a unique structural feature of prefibrillar amyloids or, more likely, that its normal binding site is precluded or otherwise altered in fibrils. Further work is needed to differentiate between these possibilities. Additional details of the structure and binding site(s) of this reagent might lead to rational design of probes that discriminate even better between amyloid morphologies.
Experimental Section
Materials
Compounds were purchased from Sigma, TCI America (Portland, OR), or Anaspec (San Jose, CA). Tryptophanol (TROL) from Sigma and TCI America yielded identical results. Aβ(1–40) was purchased from Anaspec and Aβ(1–42) was purchased from Anaspec and EZBiolab (Westfield, IN). Fluorescence readings were taken on a SpectraMax M5 multi-mode plate reader (Molecular Devices). All fluorescence assays were performed in black, opaque, flat-bottom 96-well plates (Corning). Sealing films were purchased from Nalge Nunc International (Rochester, NY).
Amyloid-β preparation
Aβ(1–40) or Aβ(1–42) was initially suspended in hexafluoroisopropanol (HFIP) and aliquoted, then the HFIP was removed under a stream of nitrogen. The aliquots were stored at −30 °C. We followed established protocols to obtain predominantly prefibrils or fibrils.[23, 24] Briefly, to obtain prefibrils, Aβ was suspended in 1 % DMSO followed by DMEM-F12 cell culture medium (Gibco, Carlsbad, CA) to a final concentration of 25 μM, vortexed, and sonicated for 90 s. The sample was then incubated for 2 days at 4 °C without agitation. To obtain fibrils, Aβ was suspended in 1 % DMSO followed by PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.2) to a final concentration of 25 μM, vortexed, sonicated for 90 s, and agitated for 7–10 days at 37 °C.
Screen of the indole library
Prior to screening, the excitation and emission maxima of each indole derivative (100 μM) in 0.01 % DMSO and glycine (50 mM, pH 8.2) were determined (Table S1). Then, compound (150 μL, 100 μM) was added to black 96-well plates with either prefibrils or fibrils (10 μL, 25 μM). As a control, the fluorescence of each compound was also determined by adding either DMEM-F12 or PBS (10 μL) in place of prefibrils or fibrils, respectively. Plates were sealed and incubated in the dark for 60 min at room temperature, after which the fluorescence was recorded at the appropriate excitation and emission maximum.
TROL assay protocol
TROL (100 μL, 10 μM) in 0.01 % DMSO and glycine (50 mM, pH 8.2) was added to a sample of amyloid (e.g., Aβ(1–40) 10 μL, 25 μM in PBS) or corresponding buffer in black 96-well plates. These samples were mixed three times by gentle pipetting. The plate was then sealed, incubated at 37 °C for 5 min, and cooled for 15 min at room temperature in the dark. Following gentle tapping of the plate, the fluorescence was recorded (λex = 280 nm, λem = 355 nm).
Thioflavin T (ThT) assay
The ThT assay was performed by using previously established methods.[15, 23, 45, 46] Briefly, protein or the corresponding buffer (10 μL) was plated in triplicate in a 96-well black plate. ThT (5 μM) was prepared in glycine (50 mM, pH 8.2) and filtered (0.22 μm). ThT (200 μL) was then added, the solution was mixed by pipetting, and then the plates were incubated at room temperature for 15 min. Following gentle tapping, the fluorescence was recorded (λex = 446 nm, λem = 490 nm).
Monitoring amyloid aggregation over time
Aβ(1–40) or Aβ(1–42) was suspended in PBS containing 1 % DMSO to a final concentration of 25 μM. Aβ(1–40) was incubated at 37 °C with shaking, and Aβ(1–42) was incubated at room temperature without shaking. The ThT signal and kinetics of its appearance correlate well with previous reports under identical conditions.[47] Recombinant ataxin-3 (Q80) was expressed and purified as previously reported.[48] A 75 μM stock solution was diluted to 10 μM in PBS and incubated at 37 °C with shaking for aggregation.[49] hIAPP/amylin peptide (Anaspec, San Jose, CA) was suspended in HFIP, aliquoted, and lyophilized. The lyophilized powder was then brought up in PBS containing 1 % DMSO to a final concentration of 20 μM and incubated at 37 °C with shaking.[50] Recombinant CsgA was expressed and purified as reported.[51] A 68 μM stock solution was diluted to 25 μM in PBS and incubated at room temperature without shaking for aggregation.[52] Recombinant α-synuclein was expressed and purified as reported.[53] This protein was suspended in PBS to yield a 20 μM solution and agitated at 37 °C.[54] For each of these amyloid-forming proteins, an aliquot (10 μL) was plated in triplicate at each time point. Prior to plating, each sample was inverted twice, except for CsgA, which was also vortexed. The TROL and ThT assays were then performed according to the optimized protocols described (Table 1).
6E10 Western blot
Aβ(1–40) samples from the indicated times were flash frozen and stored at −80 °C. Once thawed, an aliquot (15 μL) of each sample was separated on a 10–20 % tris-tricine gradient gel (Invitrogen) by using nondenaturing, nonreducing loading buffer (300 mM tris-HCl, 8 % glycerol, 0.01 % bromophenol blue). The gel was transferred to nitrocellulose (1 h, 4 °C, 175 mA) and blocked with 3 % BSA in TBS-T (140 mM sodium chloride, 25 mM tris, 0.1 % Tween-20) for 90 min at room temperature. The membrane was washed with TBS-T (3 × 5 min) and incubated overnight in 1:2000 6E10 (Covance, Dedham, MA) containing 1.5 % BSA in TBS-T at 4 °C. The membrane was then washed again (3 × 5 min), incubated for 1 h with HRP-conjugated goat anti-mouse IgG (Abcam, Cambridge, MA) in TBS-T, washed (3 × 5 min), and developed by using the Western Lightning Plus-ECL kit according to the manufacturer’s protocol (PerkinElmer).
Transmission electron microscopy
Each sample (3 μL) was applied to Formvar/carbon 300-mesh copper grids (Electron Microscopy Sciences, Hatfield, PA) and incubated for 1 min. Excess sample was wicked away with filter paper, and the grid was washed twice with doubly deionized H2O. 1 % uranylacetate in methanol (3 μL) was then added to the grid for 1 min. Excess solution was wicked away with filter paper, and the grids were dried for 15 min at room temperature. Samples were visualized on a Phillips CM-100 transmission electron microscope at an accelerating voltage of 80 kV with magnification settings ranging from 10 500–92 000 ×. The levels of prefibrils per field were quantified by tallying the number of spherical oligomers and short protofibrils in each micrograph. The average number of prefibrils was then calculated from between 5 and 16 micrographs.
Supplementary Material
Acknowledgments
The authors would like to thank Ari Gafni for supplying amylin, Elizabeth Rhoades for providing α-synuclein, Matthew Chapman and Yizhou Zhou for the CsgA, and Henry Paulson and Matthew Scaglione for the ataxin. The authors also thank Paul Smecker and Kathryn McMenimen for helpful comments on the manuscript. A.A.R. was supported by a fellowship from the Biogerontology NIA Training Grant (AG000114). This work was also supported by a grant from the Alzheimer’s Association (NIRG-08-89471).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201000358.
References
- 1.Caughey B, Lansbury PT. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 2.Haass C, Selkoe DJ. Nat Rev Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 3.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 4.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 5.Walsh DM, Klyubin I, Fadeeva JV, Rowan MJ, Selkoe DJ. Biochem Soc Trans. 2002;30:552–557. doi: 10.1042/bst0300552. [DOI] [PubMed] [Google Scholar]
- 6.Hung LW, Ciccotosto GD, Giannakis E, Tew DJ, Perez K, Masters CL, Cappai R, Wade JD, Barnham KJ. J Neurosci. 2008;28:11950–11958. doi: 10.1523/JNEUROSCI.3916-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klyubin I, Betts V, Welzel AT, Blennow K, Zetterberg H, Wallin A, Lemere CA, Cullen WK, Peng Y, Wisniewski T, Selkoe DJ, Anwyl R, Walsh DM, Rowan MJ. J Neurosci. 2008;28:4231–4237. doi: 10.1523/JNEUROSCI.5161-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono K, Condron MM, Teplow DB. Proc Natl Acad Sci USA. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Townsend M, Shankar GM, Mehta T, Walsh DM, Selkoe DJ. J Physiol. 2006;572:477–492. doi: 10.1113/jphysiol.2005.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glabe CG. Trends Biochem Sci. 2004;29:542–547. doi: 10.1016/j.tibs.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Mamikonyan G, Necula M, Mkrtichyan M, Ghochikyan A, Petrushina I, Movsesyan N, Mina E, Kiyatkin A, Glabe CG, Cribbs DH, Agadjanyan MG. J Biol Chem. 2007;282:22376–22386. doi: 10.1074/jbc.M700088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klunk WE, Pettegrew JW, Abraham DJ. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 14.LeVine H., III Protein Sci. 2008;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeVine H., III Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 16.Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, Cole GM. J Biol Chem. 2005;280:5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 17.Frid P, Anisimov SV, Popovic N. Brain Res Rev. 2007;53:135–160. doi: 10.1016/j.brainresrev.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Naiki H, Higuchi K, Hosokawa M, Takeda T. Anal Biochem. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 19.Biancalana M, Koide S. Biochim Biophys Acta, Proteins Proteomics. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Findeis MA. Biochim Biophys Acta Mol Basis Dis. 2000;1502:76–84. doi: 10.1016/s0925-4439(00)00034-x. [DOI] [PubMed] [Google Scholar]
- 21.Ono K, Hasegawa K, Naiki H, Yamada M. J Neurosci Res. 2004;75:742–750. doi: 10.1002/jnr.20025. [DOI] [PubMed] [Google Scholar]
- 22.Walsh DM, Hartley DM, Kusumoto Y, Fezoui Y, Condron MM, Lomakin A, Benedek GB, Selkoe DJ, Teplow DB. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 23.Reinke AA, Seh HY, Gestwicki JE. Bioorg Med Chem Lett. 2009;19:4952–4957. doi: 10.1016/j.bmcl.2009.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 25.Lindgren M, Sörgjerd K, Hammarström P. Biophys J. 2005;88:4200–4212. doi: 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colby DW, Zhang Q, Wang S, Groth D, Legname G, Riesner D, Prusiner SB. Proc Natl Acad Sci USA. 2007;104:20914–20919. doi: 10.1073/pnas.0710152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasegawa K, Yamaguchi I, Omata S, Gejyo F, Naiki H. Biochemistry. 1999;38:15514–15521. doi: 10.1021/bi991161m. [DOI] [PubMed] [Google Scholar]
- 28.Jan A, Gokce O, Luthi-Carter R, Lashuel HA. J Biol Chem. 2008;283:28176–28189. doi: 10.1074/jbc.M803159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maji SK, Ogorzalek Loo RR, Inayathullah M, Spring SM, Vollers SS, Condron MM, Bitan G, Loo JA, Teplow DB. J Biol Chem. 2009;284:23580–23591. doi: 10.1074/jbc.M109.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams AD, Shivaprasad S, Wetzel R. J Mol Biol. 2006;357:1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 31.Bowerman CJ, Ryan DM, Nissan DA, Nilsson BL. Mol BioSyst. 2009;5:1058–1069. doi: 10.1039/b904439f. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson MR. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahimi F, Shanmugam A, Bitan G. Curr Alzheimer Res. 2008;5:319–341. doi: 10.2174/156720508784533358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi Y, Okamoto Y, Popiel HA, Fujikake N, Toda T, Kinjo M, Nagai Y. J Biol Chem. 2007;282:24039–24048. doi: 10.1074/jbc.M704789200. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Chapman MR. Prion. 2008;2:57–60. doi: 10.4161/pri.2.2.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. J Mol Biol. 2009;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 38.Ban T, Yamaguchi K, Goto Y. Acc Chem Res. 2006;39:663–670. doi: 10.1021/ar050074l. [DOI] [PubMed] [Google Scholar]
- 39.Shahi P, Sharma R, Sanger S, Kumar I, Jolly RS. Biochemistry. 2007;46:7365–7373. doi: 10.1021/bi7001136. [DOI] [PubMed] [Google Scholar]
- 40.Yagi H, Ban T, Morigaki K, Naiki H, Goto Y. Biochemistry. 2007;46:15009–15017. doi: 10.1021/bi701842n. [DOI] [PubMed] [Google Scholar]
- 41.Zhu M, Han S, Zhou F, Carter SA, Fink AL. J Biol Chem. 2004;279:24452–24459. doi: 10.1074/jbc.M400004200. [DOI] [PubMed] [Google Scholar]
- 42.Bogdan M, Pirnau A, Floare C, Bugeac C. J Pharm Biomed Anal. 2008;47:981–984. doi: 10.1016/j.jpba.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Singh AK, Asefa A. Luminescence. 2009;24:123–130. doi: 10.1002/bio.1085. [DOI] [PubMed] [Google Scholar]
- 44.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrêne YF, Narayanaswami V, Goormaghtigh E, Ruysschaert JM, Raussens V. Biochem J. 2009;421:415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 45.Evans CG, Wisen S, Gestwicki JE. J Biol Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 46.Reinke AA, Gestwicki JE. Chem Biol Drug Des. 2007;70:206–215. doi: 10.1111/j.1747-0285.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 47.Qi W, Zhang A, Patel D, Lee S, Harrington JL, Zhao L, Schaefer D, Good TA, Fernandez EJ. Biotechnol Bioeng. 2008;100:1214–1227. doi: 10.1002/bit.21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Todi SV, Laco MN, Winborn BJ, Travis SM, Wen HM, Paulson HL. J Biol Chem. 2007;282:29348–29358. doi: 10.1074/jbc.M704126200. [DOI] [PubMed] [Google Scholar]
- 49.Ellisdon AM, Thomas B, Bottomley SP. J Biol Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 50.Koo BW, Miranker AD. Protein Sci. 2005;14:231–239. doi: 10.1110/ps.041051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Zhou Y, Ren JJ, Hammer ND, Chapman MR. Proc Natl Acad Sci USA. 2010;107:163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Smith DR, Jones JW, Chapman MR. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trexler AJ, Rhoades E. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waxman EA, Giasson BI. J Neurochem. 2010;113:374–388. doi: 10.1111/j.1471-4159.2010.06592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.