Abstract
Fundamental questions remain to be answered on how lineages split and new species form. The Arabidopsis genus, with several increasingly well characterized species closely related to the model system A. thaliana, provides a rare opportunity to address key questions in speciation research. Arabidopsis species, and in some cases populations within a species, vary considerably in their habitat preferences, adaptations to local environments, mating system, life history strategy, genome structure and chromosome number. These differences provide numerous open doors for understanding the role these factors play in population divergence and how they may cause barriers to arise among nascent species. Molecular tools available in A. thaliana are widely applicable to its relatives, and together with modern comparative genomic approaches they will provide new and increasingly mechanistic insights into the processes underpinning lineage divergence and speciation. We will discuss recent progress in understanding the molecular basis of local adaptation, reproductive isolation and genetic incompatibility, focusing on work utilizing the Arabidopsis genus, and will highlight several areas in which additional research will provide meaningful insights into adaptation and speciation processes in this genus.
INTRODUCTION
Explaining how the diversity of organisms populating Earth arises remains a fascinating challenge. Extensive progress has been made in recent years, and numerous informative reviews have been written on this topic (e.g. Coyne and Orr, 2004; Lowry et al. 2008a; Rieseberg and Willis, 2007; Rieseberg and Blackman, 2010; Schluter, 2009; Schluter and Conte, 2009; Sobel et al. 2009), and a number of conclusions can be drawn from work on speciation in recent decades. For example, it is increasingly clear that pre-zygotic barriers (factors that prevent mating), such as adaptation to divergent habitats or distinct pollinator preferences, likely play starring roles in diversification and speciation (Lowry et al., 2008a; Rieseberg and Blackman, 2010; Rieseberg and Willis, 2007; Schluter, 2009; Schluter and Conte, 2009; Sobel et al. 2009). Though they may contribute less to overall isolation, it is also clear that postzygotic barriers (e.g. genetic incompatibilities) also play an important role in restricting gene flow among species (Lowry et al., 2008a), and recent years have seen the identification of numerous causal genes, particularly in plants (Rieseberg and Blackman, 2010). It has been pointed out also that the majority of “speciation genes” identified to date are post-zygotic, and comparatively little is known about the molecular basis of pre-zygotic barriers (with the exception of pollinator preferences; Rieseberg and Blackman, 2010).
Implicit in contemporary views of speciation is the notion that ecological and genetic factors play important, and often inextricably intertwined roles in population divergence and isolation (e.g. Lowry et al., 2008a; Rieseberg and Willis, 2007; Schluter, 2009; Schluter and Conte, 2009). Relevant factors in speciation thus include causes of population fragmentation and divergence (such as geographic barriers, temporal isolation or adaptations to different habitats), genes that mediate environmental adaptation, and the genetic barriers that accumulate between diverging populations, whether they result from selection or drift. This underscores the importance of gaining a detailed understanding of multiple aspects of species biology, including ecology, population genetic patterns, demographic history and the genetics of extrinsic or intrinsic reproductive isolating barriers. Perhaps most important (and in some ways most difficult) is to clearly assess and quantify current, and where possible historic impediments to gene flow. Such studies are still rare, but those that have been done provide insights into the relative importance of different barriers to gene flow in natural populations (Lowry et al., 2008a).
Molecular tools available in model systems can be extended with increasing ease to relatives, providing us with “model genera” or even “model families” (Bomblies and Weigel, 2007a, 2010; Crosby et al. 2007) that can help get around some of the biases inherent in studying just a single model species, while the relatedness to traditional models keeps open the door to a wealth of molecular tools. The Arabidopsis genus, with a manageable number of closely related, genetically tractable and sometimes inter-fertile species, provides excellent opportunities to study processes relevant to speciation in molecular detail (Bomblies and Weigel, 2007a, 2010). In this review, we will do two things: First, we will discuss work relevant to speciation and population divergence that has utilized the Arabidopsis genus either as a way of gaining molecular insight into barriers known from other species, or to understand barriers active in this genus. Second, we will point to ongoing and future opportunities for more fully capitalizing on the power of this genus for studies in speciation. Since previous reviews have covered additional aspects of speciation research in Arabidopsis, we will not cover all areas here (see Bomblies and Weigel 2007a, 2010).
Species and Speciation History in the Arabidopsis Genus
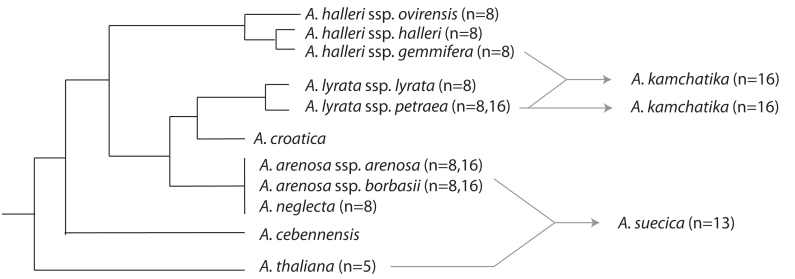
The taxonomy, evolution and ecology of the Arabidopsis genus have been extensively reviewed (see e.g. Al-Shehbaz and O'Kane, 2002; Clauss and Koch, 2006; Hoffmann, 2005; Koch et al., 2008; Koch and Matschinger, 2007; Mitchell-Olds, 2001; Shimizu, 2002) and we will thus include only a brief summary here to provide context relevant for the remainder of the paper. Aside from several rare or locally endemic species, the Arabidopsis genus consists of four major lineages: A. thaliana, A. lyrata, A. arenosa, and A. halleri with the latter three each divided into two or more sub-species (Figure 1). Species in these Arabidopsis lineages generally have eight chromosomes, or sixteen in tetraploids, with the exception of A. thaliana which has 5 chromosomes, and A. suecica, which has 13 chromosomes (Al-Shehbaz and O'Kane, 2002). Most species are self-incompatible, though A. thaliana, A. suecica and A. kamchatika are self-fertile.
Figure 1:
Phylogeny of Arabidopsis genus based on Al-Shehbaz and O'Kane, 2002 and Hoffman, 2005. Chromosome numbers given are from Al-Shehbaz and O'Kane, 2002. Hybrid species are shown with grey arrows indicating parental species. Note that A. kamchatika originated both as a hybrid of A. halleri x A. lyrata, and as a polyploidy derivate of A. lyrata.
The earliest diverging lineage is A. thaliana, which split from the rest of the genus about 3.8–5 million years ago (MYA). The remaining species radiated approximately 2 MYA (Clauss and Koch, 2006; Hoffmann, 2005; Koch and Matschinger, 2007). Arabidopsis thaliana is primarily self-fertilizing and inhabits a variety of low competition habitats, primarily human-associated cultivated or waste areas, but also sparse meadows, rubble slopes and riverbanks (Al-Shehbaz and O'Kane, 2002). Being the model system of choice for many molecular biologists means that a genome sequence, a wealth of molecular information and extensive resources are available. Large-scale genome sequencing and characterization efforts are adding to the store of knowledge about variation in genome sequences and organization, and opening the door to understanding how this correlates with phenotypic variation, poising A. thaliana to become an increasingly powerful tool for understanding the molecular basis of evolutionary processes (e.g. Clark et al. 2007; Weigel and Mott, 2009; Zhang et al. 2006).
Arabidopsis lyrata is perennial and is made up of two recognized geographically isolated subspecies: A. lyrata ssp. petraea occurs in Eurasia, while A. lyrata ssp. lyrata occurs in North America (Al-Shehbaz and O'Kane, 2002; Clauss and Koch, 2006). There are populations in temperate regions of Europe and North America, but A. lyrata is primarily an arctic species (Al-Shehbaz and O'Kane, 2002; Hoffmann, 2005). It is diploid (n=8) throughout most of its range, but tetraploid (n=16) populations are known from Austria (Dart et al., 2004). The A. lyrata genome has recently been sequenced, greatly facilitating molecular studies and opening numerous doors for detailed investigation of mechanisms relevant to speciation and adaptation (http://genome.jgi-psf.org/Araly1/Araly1.info.html).
Arabidopsis arenosa is currently divided into two recognized subspecies that both occur in Europe: ssp. borbasii tends to be perennial and inhabits mountainous or rocky habitats, while ssp. arenosa is annual or biennial and more often inhabits ruderal sites (Al-Shehbaz and O'Kane, 2002; Scholz, 1962). Arabidopsis arenosa is tetraploid (n=16) through much of its range, but diploid populations (n=8) are known from the Balkans and Carpathian mountains (MA Koch et al. personal communication; Měsíček, 1967; Schmickl and Koch in preparation). The genome of a diploid A. arenosa is also being sequenced, which will greatly expand the opportunities for comparative molecular approaches with multiple members of the Arabidopsis genus (Comai and Rokshar et al.; http://www.jgi.doe.gov/sequencing/statusreporter/psr.php?projectid=402430).
Arabidopsis halleri is currently divided into five recognized sub-species (Koch et al., 2008; Kolník and Marhold, 2006). It is diploid (n = 8) and populations are patchily distributed throughout Eurasia (Hoffmann, 2005; Kolník and Marhold, 2006). Unlike its congeners, A. halleri is tolerant of shading and competition and occurs primarily in mesic meadow habitats (Clauss and Koch, 2006; Pauwels et al., 2005). This species is also tolerant of high levels of heavy metals in soil, which has allowed it to invade many habitats polluted by human mining activities (Clauss and Koch, 2006).
Hybrid zones can be very important models for understanding processes of introgression, adaptation, speciation, and gene flow barriers (e.g. Barton and Hewitt, 1989; Rieseberg et al., 1999). In Eastern Austria tetraploid (n = 16) populations of A. arenosa and A. lyrata co-occur and a hybrid zone has recently been found (Schmickl and Koch in preparation; MA Koch et al. personal communication), promising exciting opportunities to study the molecular and phenotypic consequences of gene flow and the mechanisms of maintenance of species barriers. There is molecular evidence of hybridization in several other Arabidopsis species as well, though no current hybrid zones have been identified. For example, there is evidence of historical gene flow between A. halleri and A. lyrata (Ramos-Onsins et al., 2004), though while crosses in the lab between these species are possible, hybrid fitness is lower than that of the parents. This suggests that intrinsic incompatibilities have arisen since their divergence, which may explain why there are no modern hybrid zones known (Ramos-Onsins et al., 2004). Gene flow also seems to have occurred historically between A. arenosa and A. croatica (Koch and Matschinger, 2007).
Hybridization can also give rise to new species. This mechanism is thought to be a very important factor in plant speciation (Buerkle et al. 2000; Hegarty and Hiscock, 2005). The Arabidopsis genus contains two hybrid species that provide interesting models (Figure 1): A single hybridization event between A. thaliana and A. arenosa spawned a natural allopolyploid hybrid species, A. suecica, with 13 chromosomes, eight of which come from A. arenosa, and five from A. thaliana (Hylander, 1957; Jakobsson et al., 2006; Měsíček, 1967; Säll et al., 2003). Arabidopsis kamchatika is a more recently recognized hybrid Arabidopsis species that seems to have arisen via several independent hybridization events between A. halleri ssp. gemmifera and A. lyrata in Eastern Asia, though some lineages recognized as A. kamchatika appear to be autopolyploid derivatives of A. lyrata (Clauss and Koch, 2006; Koch and Matschinger, 2007; Schmickl et al., 2010; Shimizu-Inatsugi et al., 2009; Watanabe et al., 2005).
Overall, the species in the Arabidopsis genus provide many opportunities for studying processes relevant to adaptation, divergence and speciation. In this review we will discuss how they have already contributed to our understanding of these processes, as well as where more research is needed. Table 1 provides a list of gene flow barriers relevant to plants, primarily following previously utilized classification schemes (e.g. Coyne and Orr, 2004; Rieseberg and Blackman, 2010), and provides a brief summary of what is known in the Arabidopsis genus, which will be discussed in more detail below, as well as pointing out areas where more research is needed.
Table 1.
Barriers relevant to plant speciation
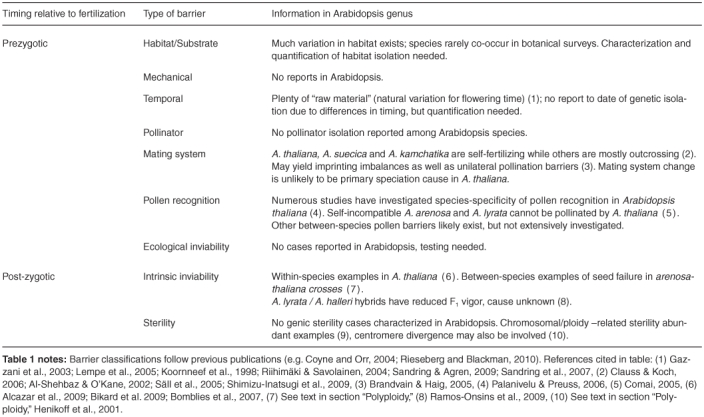
PREZYGOTIC FACTORS
Edaphic adaptation: Adaptations to habitat or soil type differences (edaphic adaptation) may be very important in divergence and ecological speciation in plants (Schluter, 2009; Schluter and Conte, 2009; Rajakaruna, 2004; Rieseberg and Willis, 2007). While several genes important for adaptation are known, it has yet to be demonstrated whether these adaptations lead to reproductive isolation; thus genes that might truly be considered “edaphic isolation genes” remain as yet unknown (Rieseberg and Blackmann, 2010). The Arabidopsis genus provides excellent opportunities in this area. Despite extensive range overlap and broad similarities in habitat preferences, botanical surveys rarely name multiple Arabidopsis species co-occurring in the same plant communities. This suggests that at least some habitat isolation exists among these species. It will be crucial for future research in speciation in this genus to investigate which local adaptations lead to gene flow barriers among species or populations in nature.
Arabidopsis species have colonized a wide range of primarily low-competition habitats. Arabidopsis lyrata, for example, occurs in sand dunes, tundra, stream banks, lakeshores, and rocky slopes, and can be found on calcareous/alkaline substrates (e.g. limestone, dolomite or gypsum), silicaceous/acidic substrates (e.g. sandstone and granite), and even serpentine (Al-Shehbaz and O'Kane, 2002; Černý et al., 2006; Clauss and Koch, 2006; Stelfox, 1970; Turner et al., 2008). It occurs from temperate regions into the high arctic (Hoffmann, 2005). These habitats likely vary in numerous important factors, such as nutrient content and availability, rhizosphere communities, temperature, light incidence, growing season length, competition, and pathogen and herbivore prevalence. The closely related A. arenosa is also found in a diverse array of habitats (Boublík et al., 2007; Clauss and Koch, 2006; Husová, 1967; Justin, 1993; Polatschek, 1965), but has also adapted to more extreme substrates than those reported for A. lyrata, such as strongly acidic soils (Lawesson and Mark, 2000) and heavy metal contaminated sites (Kucharski et al., 2005; Przedpełska and Wierzbicka, 2007). Arabidopsis halleri is unusual in that it is tolerant of shading and can fare well in high competition habitats such as mesic meadow sites (Clauss and Koch, 2006; Pauwels et al., 2005). There are many adaptations that are no doubt relevant to gene flow in and among Arabidopsis species - for example, adaptation from ancestral alpine habitats to lower altitude habitats was likely important in the diversification of A. arenosa and A. halleri subspecies (Al-Shehbaz and O'Kane, 2002).
Little is known in this genus about the role of edaphic factors in isolating gene pools. However, Arabidopsis species have already provided important insights into the molecular mechanisms of adaptation, which provide valuable information for filling out future studies of edaphic isolation. Adaptation to challenging substrates has been particularly well studied. As an illustrative example, we will discuss heavy metal tolerance and accumulation in A. halleri, which is an adaptation that is especially well understood at the molecular level (Bert et al., 2002; Macnair, 2002; Macnair et al., 1999). In A. halleri, adaptation to metal-contaminated sites has occurred multiple times, indicating a predisposition to achieving this ecological transition (e.g. Bert et al., 2002; Pauwels et al., 2005; Punz and Mucina, 1997). Several genes important in adaptation to heavy metal-containing soils have been identified. For example, HMA4 is a metal transporter that contributes to Zn and Cd tolerance differences between A. lyrata and A. halleri (Courbot et al., 2007; Hanikenne et al., 2008; Willems et al., 2007); its ortholog is also implicated in heavy metal tolerance and hyperaccumulation in T. caerulescens (Bernard et al., 2004; Papoyan and Kochian, 2004). Non-tolerant species and A. halleri differ in HMA4 expression, with the higher level in A. halleri attributable to a combination of gene copy number increase and cis-regulatory changes (Hanikenne et al., 2008). Another zinc transporter, MTP1, and at least two additional genes implicated in hypertolerance also show evidence of copy number and expression level increases (Dräger et al., 2004; Talke et al., 2006). Adaptation in A. lyrata to another challenging substrate, serpentine soil, also showed evidence of copy number expansion in several substrate-associated genomic regions, though causal genes remain to be characterized (Turner et al., 2008; Turner et al., 2010). Salt tolerance in A. thaliana accessions was similarly traced to a natural variant with altered expression in root tissues of the sodium transporter HKT1 (Rus et al., 2006). These studies suggest that expression changes of causal genes already found in ancestral populations will be a common theme at least in substrate adaptation. To what degree these confer gene flow barriers, and whether the effects are direct or indirect, provides interesting questions for follow-up studies.
In the future, traditional QTL mapping (where crosses are possible), and high throughput genomic approaches will likely provide additional insights into adaptation to the diverse habitats in which Arabidopsis species are found. TILING-array analysis and high-throughput sequencing of genotypes inhabiting alternate habitats can be used to identify genomic regions differentiated by habitat, providing a list of candidate loci that can be tested for involvement in local adaptation and habitat isolation (e.g. Turner et al., 2008; Turner et al., 2010). One important example likely to contribute significantly to our understanding of substrate adaptation in particular is “ionomics” - a strategy designed specifically to accelerate the identification of the genetic basis of nutrient acquisition (Baxter, 2009; Salt et al., 2008). While these approaches have been currently applied primarily to soil adaptations, there is no reason that modified strategies could not be employed for studying adaptations to other environmental factors, such as shading, temperature, altitude, moisture availability, and pathogen or herbivore repertoires. Large-scale approaches hold promise for speeding up the identification of the molecular basis of habitat adaptations. When combined with studies of how these adaptations in turn affect gene flow, such studies will greatly expand our understanding of how edaphic adaptation and gene flow barriers may inter connect.
Plasticity and trade-offs in temporal isolation: Gene flow barriers due to temporal non-overlap of reproduction may be very important in speciation and maintenance of species boundaries in both animals and plants (Coyne and Orr, 2004). There is abundant evidence in the plant literature of differences in flowering time among populations or related species leading to non-overlapping flowering times. In numerous examples flowering time differences are associated with edaphic adaptations (e.g. examples in Hall and Willis, 2006; Levin, 2009; Lowry et al., 2008b; Savolainen et al., 2006; Silvertown et al., 2005; Snaydon and Davies, 1982). For example, recent speciation of palms on Lord Howe island seems to have involved both adaptation to distinct substrates, and associated flowering time differences (Savolainen et al., 2006), and a long-term study on Anthoxanthum odoratum populations growing under distinct artificially-applied edaphic regimes showed significant local adaptation within 150 years (Snaydon and Davies, 1982) and sometimes also associated differences in flowering time (Silvertown et al., 2005). The two Lord Howe Island palm species are especially intriguing. They have strong reproductive isolation with almost no overlap in flowering times, but reproductive isolation in this case arises from a plastic response to the environment — if the now-diverged palms happen to co-occur on the same substrate, the flowering time difference disappears (Savolainen et al., 2006). Plastic responses to the environment can promote divergence if growth on different substrates is inextricably (e.g. physiologically) linked to divergence in flowering time. This may be a very important factor in plant speciation (Levin, 2009). A mathematical model built on the Howe Island palm scenario suggested that speciation indeed becomes particularly likely when there is a direct environmental effect on flowering time and a small number of loci involved in adaptation (Gavrilets and Vose, 2007).
The Arabidopsis genus provides tests for the predictions of these ideas. While we are not aware of direct evidence in the Arabidopsis genus of temporal isolation restricting gene flow among Arabidopsis species or populations, the raw material — genetic variation for flowering time — is widely available, at least in A. thaliana (Gazzani et al., 2003; Koornneef et al., 1998; Lempe et al., 2005) and A. lyrata (Riihimäki and Savolainen, 2004; Sandring and Agren, 2009; Sandring et al., 2007). In A. thaliana the genetic basis of flowering time regulation has been intensely investigated with mutants as well as natural variants (see for reviews e.g. Koornneef et al., 1998; Simpson and Dean, 2002; Yant et al., 2009), and it is clear that a small number of mutations can have a large impact on flowering time, as is predicted to facilitate speciation in the Gavrilets and Vose models (2007).
Given that temporal isolation is also greatly aided if there is developmental plasticity of flowering time in response to environmental cues that differ among habitats, it is intriguing that a number of different abiotic factors (e.g. soil type, nutrient availability, pH, moisture, and temperature) can alter maturation and flowering times in a wide range of species (see Levin, 2009; Wielgolaski, 2001). In A. thaliana environment can also directly affect flowering time, and provides the rare opportunity to understand the molecular basis of this plasticity (e.g. Juenger et al., 2005; Lempe et al., 2005; Pigliucci and Schlichting, 2002; Stinchcombe and Schmitt, 2004; Wilczek et al., 2009). Several studies have highlighted the importance of hormones in linking environment and flowering time. For example, salicylic acid (SA), which is induced by environmental or pathogen stress, can accelerate flowering (Korves and Bergelson, 2003; Martínez et al., 2004). Elevated SA also increases tolerance of some abiotic stresses (e.g. high heavy metal content in soil or drought), but can decrease tolerance of other stresses (e.g. temperature and salinity; Chini et al., 2004; Freeman et al., 2005). Natural accessions differ in their level of response to SA (van Leeuwen et al., 2007), suggesting that the utility of SA signaling for adaptation, and thus the magnitude of any effects of edaphic factors on flowering time will likely vary among strains. In contrast to SA, Nitric oxide (NO), whose production is also induced by various biotic and abiotic stresses, delays flowering (He et al., 2004). This knowledge gives a good framework for understanding and interpreting patterns observed in the wild in other species.
Alternative strategies for adaptation to similar environments could also conceivably lead to reproductive isolation during population divergence (“non-ecological speciation”; Schluter, 2009). In A. thaliana, experimental evidence exists supporting the plausibility of this in flowering time differentiation: A trade-off between drought escape via early flowering and drought resistance via water use efficiency is caused by pleiotropic effects of flowering-time genes (McKay et al., 2003). This means that two viable alternate adaptation strategies (escape by early flowering versus tolerance due to increased water use efficiency) to the same environmental challenge (drought) could result in temporal isolation as a pleiotropic effect of environmental adaptation. Whether these differences isolate populations in nature remains to be tested.
Given how much is known about the mechanisms of flowering time regulation and plasticity in Arabidopsis, it will be interesting to investigate whether temporal isolation occurs in this genus in nature.
Mating system: Mating system differences may affect gene flow among diverging populations and could thus isolate nascent lineages (Grant, 1971; Jain, 1976; Rieseberg and Willis, 2007). In the related genus Capsella, speciation of C. rubella is attributable to isolation due to self-fertilization arising in a single individual (Foxe et al. 2009; Guo et al. 2009). In the Arabidopsis genus, there are three self-fertilizing species: A. thaliana, A. suecica and A. kamchatika. In the case of A. thaliana, the transition to self-fertilizing is estimated to have occurred between 400,000 and 1 million years ago (Bechsgaard et al., 2006; Tang et al., 2007), and multiple alleles of the self-incompatibility locus still exist, speaking against a selective sweep as observed in Capsella rubella (Boggs et al., 2009; Sherman-Broyles et al., 2007; Tang et al., 2007). These findings argue that the loss of self-incompatibility is unlikely to have been the primary cause of speciation of A. thaliana, though it may have contributed to strengthening isolation at later stages. For A. kamchatika and A. suecica the timing of the origin of selfing relative to speciation is unknown, but it is possible that, as has been previously observed for hybrids in Arabidopsis and Capsella, selfing may have arisen in these species as a direct consequence of polyploidization and/or hybridization via epigenetic silencing of the self-incompatibility locus (Nasrallah et al., 2007). In these instances, the transition to self-fertilization may indeed contribute directly to speciation. Transitions from self-incompatibility to self-fertility are commonly observed in association with polyploid speciation (Comai, 2005).
Species-specificity of pollination: Fertilization is often highly species-specific and rapid evolution of species-specificity in gamete recognition and fertilization is observed in virtually all taxa that have been examined (Clark et al., 2006; Swanson et al., 2004; Swanson and Vacquier, 2002). Several hypotheses have been put forth to explain the extremely rapid evolution of proteins involved in reproduction, including gamete competition, sexual selection, and sexual conflict (Brandvain and Haig, 2005; Clark et al., 2006; Swanson et al., 2004; Swanson and Vacquier, 2002). Under these models, reproductive isolation arises as a by-product.
In plants, species-specificity of gamete recognition is mediated at numerous stages during pollination, including adhesion, germination, pollen tube growth, and guidance to the ovules (Swanson et al., 2004). While any of these barriers individually may not provide complete isolation, acting together they can confer strong species-specificity. In the absence of strong barriers, there is still commonly an advantage of conspecific pollen over heterospecific pollen in mixed matings (conspecific pollen precedence), which may also be a very important factor in isolation and speciation (Howard, 1999).
Arabidopsis species, especially A. thaliana, have been used to investigate the molecular mechanisms resulting in species-specificity of pollen recognition. Pollen of other Arabidopsis species can germinate successfully on A. thaliana stigmas (Palanivelu and Preuss, 2006), suggesting that primary recognition events are unlikely to have contributed to speciation. Pollen tube growth and guidance to the ovules, however, do decline with phylogenetic distance (Palanivelu and Preuss, 2006). That immature pistils show much less ability to discriminate pollen than mature pistils, suggests that there is an active production during pistil maturation of factors important for favoring conspecific pollen or inhibiting heterospecific pollination (Kandasamy et al., 1994).
Ovules are known to emit signals that attract pollen tubes, and the ability to respond to these signals declines so rapidly with phylogenetic distance that even A. arenosa ovules are noticeably less capable of attracting A. thaliana pollen tubes than are A. thaliana ovules (Palanivelu and Preuss, 2006). Recent work in Torenia fournieri has shed some light on the molecular identity of a highly species-specific pollen tube attractant produced by the synergid cells of the ovules (Higashiyama et al., 2006). This signal consists of a cocktail of secreted cysteine-rich proteins named LUREs, which are related to a diverse class of proteins called defensins (Okuda et al., 2009). Female gametophytes of Arabidopsis and maize also express large numbers of diverse defensin-related proteins (Cordts et al., 2001; Jones-Rhoades et al., 2007; Punwani et al., 2007; Yang et al., 2006), suggesting that this type of signal is widely utilized, and that the particular mixture produced may confer high specificity. In the Arabidopsis genus the ease of genetic manipulation will provide ample opportunity to test the roles that genetic diversity of these defensin-like molecules may play in mediating the specificity of pollination.
Another gene that has been suggested to confer species-specificity in the final stages of pollination of A. thaliana is a receptor-like kinase, FERONIA (FER). Plants mutant for FER show a phenotype of pollen tube overgrowth and fertilization failure that is phenotypically similar to that observed in interspecies crosses onto A. thaliana (Escobar-Restrepo et al., 2007). The extracellular domain of FER appears to be rapidly evolving; it is highly divergent between A. thaliana and A. lyrata (Escobar-Restrepo et al., 2007) as well as among populations of A. lyrata (Gos and Wright, 2008). That sequence divergence of FER plays a role in interspecies barriers is certainly plausible from these findings, but remains to be formally demonstrated.
These patterns suggest that pollination barriers may evolve sufficiently rapidly to play a crucial role in species-barriers in the Arabidopsis genus. The degree to which the multiple steps of the pollen recognition process play a role in gene flow in Arabidopsis in the wild has not, to our knowledge, been quantified.
POSTZYGOTIC FACTORS
The Role of Chromosomal Differences in Hybrid Sterility and Inviability
Polyploidy: Polyploidy is thought to be one of the major mechanisms of speciation in plants, and one of the few by which reproductive isolation may be essentially instant, since interploidy crosses often fail or produce hybrids that are sterile due to meiotic aberrations (Comai, 2005; Coyne and Orr, 2004; Levin, 2002). Though meiotic problems associated with interploidy crosses do present a strong barrier to gene flow, many species vary in ploidy without being considered separate. This is true for at least three species within the Arabidopsis genus — A. thaliana, A. lyrata and A. arenosa all show natural variation in ploidy without taxonomic recognition as separate species (Al-Shehbaz and O'Kane, 2002). Arabidopsis thaliana and its relatives have provided excellent systems for studying the immediate and longer-term consequences of polyploidy and interploidy crosses both within and between species (Comai et al., 2000; Comai, 2005).
Tetraploid A. thaliana plants are fully viable, fertile and barely distinguishable from diploids (Koornneef et al., 2003) as is also true of A. lyrata and A. arenosa tetraploids. Diploid and tetraploid A. thaliana can be crossed to produce viable triploids (Scott et al., 1998), allowing quantification of the degree to which ploidy changes can cause gene flow barriers via triploid sterility or other mechanisms. Triploids in A. thaliana have reduced fertility, as predicted, but they are not completely sterile (Henry et al., 2005). When selfed, they spawn aneuploid swarms with a wide range of phenotypes, variable fertility and genomic instabilities (Henry et al., 2005; Huettel et al., 2008). After several generations, descendant populations resolve into stable diploid and tetraploid groups (Henry et al., 2005). Discrete genetic loci are associated with aneuploid propagation success, implying that some genotypes are more likely than others to contribute to gene flow across ploidy barriers (Dilkes et al., 2008; Henry et al., 2007; Henry et al., 2009). This work demonstrates that ploidy barriers are not impenetrable to gene flow.
Much is known about why interploidy crosses sometimes fail to produce viable progeny in Arabidopsis. Reciprocal interploidy crosses have different effects on endosperm development: An excess of the maternal genome results in an underdeveloped endosperm in the hybrid seed, while an excess of the paternal genome causes the endosperm to over-proliferate, the severity of which correlates with relative parental genomic doses (Scott et al., 1998). DNA methylation has been implicated as a major factor in this type of hybrid failure - the effect of paternal genome excess can be phenocopied if the maternal (but not the paternal) genome is hypomethylated (Adams et al., 2000).
Barriers to allopolyploid formation: In interspecies crosses differing in chromosome number (due either to differences in ploidy or a different base chromosome number), very similar patterns of seed failure due to endosperm imbalance are often observed (Haig and Westoby, 1991). For studying the mechanisms underlying this, work in the Arabidopsis genus has again provided many insights. Work in this area has focused on hybrids between A. thaliana (n=5) and A. arenosa (n=8 or 16). Though diploid A. thaliana can be readily fertilized by tetraploid A. arenosa, most of the resulting seeds abort (Comai et al., 2000). As was observed within A. thaliana, hybrid seed lethality in these interspecies crosses worsens if the maternal (A. thaliana) genome is hypomethylated, suggesting a role for epigenetic regulation in this type of hybrid failure (Bushell et al., 2003).
In crosses between A. thaliana and A. arenosa, an increase in expression of maternal (A. thaliana) genes that are normally expressed only from paternal copies is correlated with an increase in relative paternal (A. arenosa) genome dosage (Josefsson et al., 2006; Walia et al., 2009). Seed viability is increased when the A. thaliana parent is homozygous for a mutation in one of these genes, PHERES1 (PHE1), demonstrating that this mis-regulation of imprinted genes plays a causal role in this type of incompatibility (Josefsson et al., 2006). Normally, maternal PHE1 expression is suppressed in A. thaliana by a maternally expressed polycomb group protein MEDEA (MEA) (Gagliardini et al., 2005; Köhler et al., 2003; Makarevich et al., 2006). It has been proposed that in hybrids excess paternal genome sequesters much of the maternally expressed MEA with the result that an insufficient amount remains to suppress maternal PHE1 expression. This is consistent with the “Dosage-Dependent Induction (DDI)” model, which suggests that regulatory molecules are produced in proportion to genome size or copy number and that hybrids can be compromised if one parental genome cannot suppress deleterious expression of genes from the other parent due to insufficient production of regulatory factors (Erilova et al., 2009; Josefsson et al., 2006). A recent genome-wide study of DNA methylation in the endosperm of A. thaliana helped uncover five additional imprinted genes and about 50 candidates (Gehring et al., 2009). It will be interesting to see whether any of these genes are also misregulated to ill effect during hybrid seed development. Consistent with a possible role for epigenetic factors regulating seed development in hybridization barriers, MEA appears to be evolving under diversifying selection (Miyake et al., 2009). This fits with the idea that imprinting can arise from parental conflicts, which drive rapid gene evolution (Haig and Westoby, 1991; Wilkins and Haig, 2003). Self-fertilization is expected to affect selection for imprinting (Brandvain and Haig, 2005), thus it would be especially interesting in this context to examine whether imprinting-related incompatibility affects hybrids involving A. thaliana differently from hybrids among outcrossing members of the genus.
Short interfering RNAs (siRNAs) may also play an important role in hybrid seed failure. Many siRNAs produced from transposons and other repetitive sequences are important in their silencing (Matzke et al., 2009). Recently, it has been demonstrated that many siRNAs are expressed only from the maternal genome in the endosperm (Mosher et al., 2009). Plants defective in the production of these siRNAs show transcription of some transposons, though within A. thaliana this leads to no obvious defects (Mosher et al., 2009). Paternally derived transposon siRNAs of a different size class are produced in pollen grains (Slotkin et al., 2009) and it is thought that they may function in coordination with maternally-derived siRNAs to suppress transposon expression (Martienssen, 2010). The combination of maternal and paternal transposon-derived siRNAs may not function together correctly in interspecies hybrids, or may not be produced in adequate quantities to silence transposons from both parental genomes. These possibilities are intriguing, as improper silencing of transposons has been shown to negatively impact interspecies hybrids. For example, in Drosophila, highly deleterious activation of paternal transposons occurs in hybrids among strains in which only the father carries P-element transposons, because the mother lacks the transposon siRNAs necessary to silence them in the fertilized egg and developing embryo (Blumenstiel and Hartl, 2005). Interspecies hybrids in rice and evening primrose similarly have high levels of transposon activity, which leads to a high mutation rate and varied morphological defects in subsequent progeny (Wang et al., 2009). In interspecies hybrids in both banana and petunia, inadequate silencing in hybrids can lead to activation of integrated pararetroviruses, which can generate active infections (Harper et al., 1999; Ndowora et al., 1999; Richert-Poggeler et al., 2003). In A. thaliana × A. arenosa hybrid seeds, expression of paternal ATHILA transposons is elevated proportional to the severity of genome imbalance and correlates with seed failure (Josefsson et al., 2006; Madlung et al., 2005). The A. arenosa genome contains more ATHIILA elements than the A. thaliana genome and thus the amount of maternally produced ATHILA repressing elements (probably siRNAs) may not suffice to silence the A. arenosa elements in hybrid seeds (Josefsson et al., 2006).
Problems can also arise later in allopolyploid hybrid plants that have survived the initial seed development hurdle. Hybrid A. thaliana × A. arenosa plants often exhibit reduced fitness, low fertility and developmental abnormalities. Differences in gene and siRNA expression among hybrids, and between hybrids and the parent plants, help to explain the reduced fitness as well as phenotypic variation (Ha et al., 2009; Wang et al., 2006b). Multiple chromosomal rearrangements and segmental losses observed in A. thaliana × A. arenosa hybrids may be a main cause of meiotic abnormalities, the prevalence of which in turn is inversely proportional to pollen viability (Madlung et al., 2005; Pontes et al., 2004). It has been suggested that increased transposon activity in allopolyploids causes chromosomal breaks, which lead to joining of homeologous chromosome segments (Pontes et al., 2004). Homeologous recombination could also be an explanation for the observed fusion of homeologous chromosomes, but this remains to be demonstrated (Madlung et al., 2005).
Allopolyploidy as an opportunity: In some cases, crosses between species with different chromosome numbers can give rise to stable allopolyploid hybrids, with a full genome complement from each parent (Hegarty and Hiscock, 2005; Mallet, 2007; Rieseberg and Willis, 2007). Allopolyploidy is commonly observed in nature and may be a common mode of speciation in plants (Leitch and Leitch, 2008). Arabidopsis thaliana × A. arenosa hybrids, replicating the cross that gave rise to A. suecica, can easily be produced in the laboratory. Though first generation hybrids often have reduced viability and low fertility, stable, fertile lines can be generated and have provided useful models for studying hybridization and early events following allopolyploidy (Comai et al., 2000).
Nonadditive gene expression observed in resynthesised A. suecica can confer novel traits and provide the nascent allopolyploid hybrid species with novel habitat and help isolate it and protect it from parental lineages. For example, interaction of a flowering time regulator from an A. thaliana accession with one from an A. arenosa accession causes significantly delayed flowering in inter-species hybrid plants relative to either parent (Wang et al., 2006a, 2006b). Such a change in flowering time could impose a significant prezygotic gene flow barrier. The allotetraploid species A. kamchatica also enjoys a far broader range of climates than its parents (Hoffmann, 2005) allowing it to exploit more habitats and develop geographic isolation from its parents. That hybrid species may have novel adaptive potential that can provide the nascent species a novel niche and an easy route to limiting gene flow with parental populations is well known, especially from wild sunflower species (Rieseberg et al., 2003; Schwarzbach et al., 2001; Ungerer et al., 1998;). The Arabidopsis genus has already been providing the opportunity to uncover molecular details of this speciation mechanism.
The potential role of centromeres in hybrid sterility: Evolutionary divergence of centromere sequences and associated proteins could cause aberrant chromosome segregation in hybrids, and thus, meiotic instability and hybrid sterility. The centromeric repeats and associated proteins make up part of the kinetochore (a protein-based structure that connects the centromere to the spindles), which is essential for proper meiotic and mitotic cell division. A complex interaction exists between the sequence of the repeats, the chromatic state, and the kinetochore proteins of the centromere (Dawe and Henikoff, 2006). Hybrids of species with diverged centromeres and centromeric proteins may exhibit reduced fitness due to unequal behavior of centromeres during cell division, which could lead to nondisjunction (Henikoff et al., 2001). In Drosophila species, rapid evolution of heterochromatic repeats and associated proteins can contribute to hybrid sterility by disrupting chromosome segregation (Bayes and Malik, 2009; Ferree and Barbash, 2009). The role that centromeres and associated, perhaps adaptively evolving proteins might play in speciation is an intriguing question and one that certainly merits further exploration.
A main component of most plant and animal centromeres is a repeated sequence usually around 180 base pairs in length. Analysis of the centromeric repeat sequences of different Arabidopsis species demonstrated that each species has its own unique repeat sequence or a characteristic combination of different repeats (Heslop-Harrison et al., 2003; Kamm et al., 1995; Kawabe and Nasuda, 2005). Proteins specifically associated with the centromeric repeats are surprisingly divergent between A. thaliana and A. arenosa and there is evidence that they are rapidly evolving (Talbert et al., 2002; Talbert et al., 2004). Furthermore, there has been a burst in the number of LTR retrotransposons in A. lyrata and A. halleri compared to A. thaliana and most of these are found in the centromeres (Tsukahara et al., 2009). Centromere transposon complements have been shown to vary considerably in abundance and type even between closely related species (Hawkins et al., 2006; Vitte and Bennetzen, 2006). Thus the idea that centromere evolution can contribute to gene flow barriers among species in the Arabidopsis genus is enticing, though a formal demonstration that this contributes to speciation in this genus is still lacking.
Genic Causes of Hybrid Sterility or Inviability
Segregation distortion: Segregation distortion (SD), also known as transmission ratio distortion, has been used throughout the speciation literature as a way of identifying genomic regions that might contribute to genetic isolation between populations or species (e.g. Bradshaw et al., 1998; Fishman et al., 2008; Fishman et al., 2001; Hall and Willis, 2005; McDaniel et al., 2007; Myburg et al., 2004; Orr and Irving, 2005; Yin et al., 2004). SD is conceptually simple - a deviation from expected Mendelian segregation ratios in a particular region of the genome - but in practice, interpreting SD and its causes can be complex. SD may be caused by diverse factors, including segregation of genetic incompatibilities, chromosomal rearrangements, nuclear cytoplasmic incompatibility, meiotic drive, fitness differences that lead to differential survival, and/or inadvertent human selection during line generation (especially in approaches such as recombinant inbred line construction where multiple generations are successively propagated). SD can arise due to selection during the haploid (gametic) phase (e.g. through differences in fertilization success) or in the diploid (zygote) stage (e.g. through genetic incompatibilities or maladaptation).
Progeny of crosses within A. thaliana commonly show SD (e.g. Balasubramanian et al., 2009; Clerkx et al., 2004; el-Lithy et al., 2006; Liu et al., 1996; McKay et al., 2008; Törjek et al., 2006; Wilson et al., 2001). This has been primarily a nuisance factor in mapping experiments and thus only a few studies addressed the underlying causes. However, SD among strains within a species may be of interest in speciation research, because it could indicate early-arising incompatibilities. To date all SD cases within A. thaliana for which causes have been identified result from genetic incompatibilities. In crosses between the Col and C24 accessions, for example, two unlinked loci interact epistatically to cause severely reduced male fertility in complementary homozygotes (Törjek et al., 2006). The causal genes remain to be identified, but will provide an interesting insight into how male sterility, a common and probably early-arising type of between-species incompatibility (Coyne and Orr, 2004), may originate as within-species variation. In another case, segregation distortion results from the loss of alternate copies of a duplicated essential gene leading to embryo lethality in complementary double homozygotes (Bikard et al., 2009), supporting the idea that loss of alternate copies of a duplicated essential gene in different lineages could lead to incompatibility (Lynch and Force, 2000). Some cases of segregation distortion in A. lyrata appear to arise from linkage of deleterious mutations that affect pollen performance to the self-incompatibility (SI) locus. Since at the SI locus rare alleles have a mating advantage, haplotypes carrying linked deleterious alleles are difficult to purge from the population and can result in gamete-level SD and long-term balancing selection (Bechsgaard et al., 2004; Leppälä et al., 2008). Similar hitchhiking of deleterious mutations possibly leading to balancing selection has been suggested to occur at MHC locus alleles in humans — the high diversity, heterozygosity and low recombination, all features shared with the S-locus, render purifying selection inefficient for purging deleterious alleles (van Oosterhout, 2009).
As populations are genetically isolated by geography, edaphic adaptations, pollinator fidelity, incompatibility or other factors, genetic incompatibilities are expected to accumulate and further impede gene flow (Rieseberg and Willis, 2007). This indeed appears to be the pattern in numerous taxa (Coyne and Orr, 2004; Orr and Turelli, 2001; Rieseberg and Willis, 2007), but we still know very little about the degree to which incompatibility accumulates during divergence in the Arabidopsis genus. However, patterns are beginning to emerge: For example, within a population of A. lyrata ssp. petraea from Iceland, SD was observed at only about 10% of markers in crosses. Most of this could be attributed to gameticlevel selection, due in part to deleterious mutations linked to the SI locus (Bechsgaard et al., 2004; Leppälä et al., 2008). Reciprocal crosses among a Swedish and Russian population on the other hand showed about twice as much SD, and much more of it attributable to zygotic-level selection (Kuittinen et al., 2004; Leppälä et al., 2008). Crosses between species show, as predicted, much more extensive distortion: hybrids between A. lyrata ssp. petraea and A. halleri ssp. halleri, have reduced vigor (Ramos-Onsins et al., 2004) and marker distortion in different intercrossing strategies ranges from 40% (Willems et al., 2007) to 76% of markers (Filatov et al., 2007). These studies indicate a general trend of increasing accumulation of genetic incompatibilities with divergence time, but more studies are needed to understand the underlying molecular causes and how this might contribute to gene flow barriers.
Hybrid necrosis: Hybrid necrosis, a phenotype involving dwarfism and extensive cell death is very common in the plant kingdom (Bomblies and Weigel, 2007b), and is also a repeated cause of failure of hybrids among A. thaliana accessions. Hybrid necrosis occurs in progeny of about 2% of crosses among accessions and can cause strong segregation distortion in recombinant inbred lines or F2 populations (Alcazar et al., 2009; Bomblies et al., 2007). While in the Arabidopsis genus hybrid necrosis is rare, in some taxa it is common and may play an important role in gene flow restriction (Brieger, 1929; Jiang et al., 2000; Kostoff, 1930; Lee, 1981; Phillips, 1977). Thus the Arabidopsis system provides a model for a phenomenon relevant to divergence and speciation in plants more generally. Several genes have been cloned in A. thaliana, showing that this type of hybrid failure is caused by deleterious epistatic interactions among diverged components of the plant immune system that interact to trigger a highly detrimental induction of pathogen responses (Alcazar et al. 2009; Bomblies et al., 2007). Hybrid necrosis has been attributed to immune induction and resistance-related genes in other species as well (Hannah et al., 2007; Jeuken et al., 2009; Khanna-Chopra et al., 1998; Krüger et al. 2002; Yamamoto et al., 2010). Defense-related genes are among the most diverse in the A. thaliana genome, and often show patterns of nucleotide polymorphism suggestive of non-neutral evolution, including patterns of diversifying and balancing selection (Bakker et al. 2006; Borevitz et al. 2007; Clark et al., 2007; Mondragón-Palomino et al. 2002). This implicates adaptation to rapidly diversifying pathogens in the accumulation of genetic incompatibilities among populations or species (Bomblies, 2009). Modeling has recently shown that under some circumstances, hybrid necrosis could contribute to the evolution of reproductively isolated populations (Ispolatov and Doebeli, 2009).
Outlook
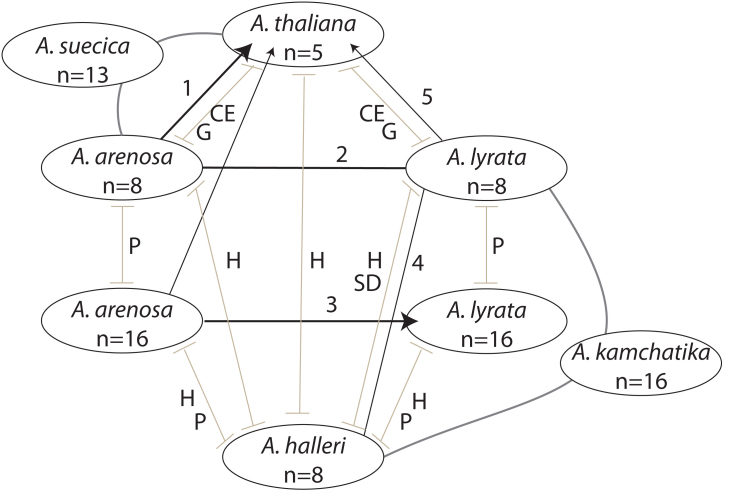
Though we know quite a lot about the molecular basis of a number of evolutionary processes in the Arabidopsis genus, we still know comparatively little about speciation, gene flow barriers and genetic divergence in this tractable group of plants. What is known so far about potential and realized gene flow, as well as barriers among species, is summarized in figure 2. Though many barriers and gene flow possibilities have been uncovered, many gaps remain in our understanding of what drove the divergence of these species, and what barriers maintain their distinctiveness.
Figure 2:
Overview of known or suspected compatibility (black lines or arrows, if unidirectional) and barriers (grey bars) between the major Arabidopsis lineages. Hybrid species are linked to their parents with dark grey bars. Chromosome numbers are indicated within the circles with the taxon names. Notes on chart: CE = chromosomal differences and epigenetic incompatibility, P = Ploidy difference, G = gamete recognition preference, H = possible habitat isolation, SD = strong segregation distortion. Specific notes: (1) Hybridization between A. arenosa (especially diploids) and A. thaliana is possible in the lab, and gave rise to allopolyploid A. suecica in nature, (2) Hybridization between A. lyrata and A. arenosa is inferred from molecular data, (3) A hybrid zone between tetraploid A. arenosa and tetraploid A. lyrata is known (see text), (4) Historical gene flow between A. lyrata and A. halleri is indicated in molecular data. Hybridization gave rise to some isolates of A. kamchatika, (5) Crosses of A. lyrata onto A. thaliana are possible in the lab (Beaulieu et al., 2009), but hybrid zones have not been documented in nature.
The Arabidopsis species, with extensive variation in habitats, evidence of local adaptation, variation in ploidy, at least two allopolyploid hybrid species, a newly discovered hybrid zone and well-established molecular models and tools, provide exciting opportunities for using this genus more fully to study speciation-related processes in molecular detail. To fully capitalize on these opportunities, we still need some groundwork: The need for a detailed, molecularly informed re-evaluation of morphology and taxonomy in the Arabidopsis genus has already been emphasized (Koch et al., 2008), but we also need to greatly improve our understanding of the present and historical patterns of local adaptation and divergence (i.e. with population genetics and evolutionary modeling approaches), especially as they might relate to gene flow barriers. We need to understand more fully why Arabidopsis species so rarely occur in the same areas. What are their specific adaptations and to what degree do these limit contact? Attempting to correlate this information with genome sequence divergence will be informative for understanding which barriers may have been particularly significant over the long term. While studies that attempt to characterize all gene flow barriers acting between two species or populations still have some limitations (for example, they do not necessarily allow us to infer the temporal order in which isolating barriers arose, or whether distinct barriers are genetically independent), they can provide an extremely valuable starting point for investigating what factors might have been important in speciation. Such studies of gene flow barriers (see for examples Lowry et al., 2008a) would be very useful in Arabidopsis and will allow us to more effectively capitalize on molecular and genomic tools to ask ecologically informed questions and benefit from the power of this genus to uncover mechanisms relevant to speciation and adaptation. In addition, it will be very informative to begin including the outcrossing species in more detailed studies of species barriers — for example, why do diploid A. arenosa and A. halleri, whose ranges overlap, not seem to hybridize in the wild, or why do A. lyrata and A. halleri, whose hybrid is viable in the lab, and whose ranges also overlap in Europe, not form hybrid zones? Are hybrid zones such as that observed with A. lyrata and A. arenosa tetraploids in Austria also observed among diploids? There are many remaining questions about the causes and consequences of divergence in the Arabidopsis genus, and the molecular tools available for a growing number of species provide exciting opportunities for understanding the molecular basis of speciation in this model genus.
ACKNOWLEDGEMENTS
We are grateful to Levi Yant, Marcus Koch, Roswitha Schmickl, Jesse Hollister and Brian Arnold for discussions and helpful comments on the manuscript. The authors were supported by setup funds from Harvard University and a fellowship from the John D. and Catherine T. MacArthur Foundation (KB).
Footnotes
Citation: Hunter B., and Bomblies K. (2010) Progress and Promise in using Arabidopsis to Study Adaptation, Divergence, and Speciation. The Arabidopsis Book 8:e0138. doi:10.1199/tab.0138
elocation-id: e0138
First published on September 23, 2010
REFERENCES
- Adams S., Vinkenoog R., Spielman M., Dickinson H.G., Scott R.J. Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development. 2000;1278(1):2493–2502. doi: 10.1242/dev.127.11.2493. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz I.A., O'Kane S.L.J. Taxonomoy and Phylogeny of Arabidopsis (Brassicaceae). The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar R., Garcia A.V., Parker J.E., Reymond M. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. USA. 2009;1068(1):334–339. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Schwartz C., Singh A. QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS One. 2009;48(1):e4318. doi: 10.1371/journal.pone.0004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E.G., Toomajian C., Kreitman M., Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;188(1):1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N.H., Hewitt G.M. Adaptation, speciation and hybrid zones. Nature. 1989;3418(1):497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Baxter I. Ionomics: studying the social network of mineral nutrients. Curr. Op. Plant Biol. 2009;128(1):381–386. doi: 10.1016/j.pbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes J.J., Malik H.S. Heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;3268(1):1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J., Jean M., Belzile F. The allotetraploid Arabidopsis thaliana-Arabidopsis lyrata subsp. petraea as an alternative model system for the study of polyploidy in plants. Mol. Genet. Genomics. 2009;2818(1):421–435. doi: 10.1007/s00438-008-0421-7. [DOI] [PubMed] [Google Scholar]
- Bechsgaard J., Bataillon T., Schierup M.H. Uneven segregation of sporophytic self-incompatibility alleles in Arabidopsis lyrata. J. Evol. Biol. 2004;178(1):554–561. doi: 10.1111/j.1420-9101.2004.00699.x. [DOI] [PubMed] [Google Scholar]
- Bechsgaard J.S., Castric V., Charlesworth D., Vekemans X., Schierup M.H. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 2006;238(1):1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- Bernard C., Roosens N., Czernic P., Lebrun M., Verbruggen N. A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens. FEBS Lett. 2004;5698(1):140–148. doi: 10.1016/j.febslet.2004.05.036. [DOI] [PubMed] [Google Scholar]
- Bert V., Bonnin I., Saumitou-Laprade P., de Laguerie P., Petit D. Do Arabidopsis halleri from nonmetallicolous population accumulate zinc and cadmium more effectively than those from metallicolous population? New Phytol. 2002;1558(1):47–57. doi: 10.1046/j.1469-8137.2002.00432.x. [DOI] [PubMed] [Google Scholar]
- Bikard D., Patel D., Le Mette C., Giorgi V., Camilleri C., Bennett M.J., Loudet O. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;3238(1):623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Blumenstiel J.P., Hartl D.L. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc. Natl. Acad. Sci. USA. 2005;1028(1):15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N.A., Dwyer K.G., Nasrallah M.E., Nasrallah J.B. In vivo detection of residues required for ligand-selective activation of the S-locus receptor in Arabidopsis. Curr. Biol. 2009;198(1):786–791. doi: 10.1016/j.cub.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K. Too much of a good thing? Hybrid necrosis as a byproduct of plant immune system diversification. Botany. 2009;878(1):1013–1022. [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., Dangl J.L., Weigel D. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;58(1):1962–1972. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. Arabidopsis: a model genus for speciation. Curr. Opin. Genet. Dev. 2007a;178(1):500–504. doi: 10.1016/j.gde.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. Hybrid Necrosis: Autoimmunity as a common barrier to gene flow in plants. Nat. Rev. Genet. 2007b;88(1):382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. Arabidopsis and relatives as models for the study of genetic and genomic incompatibilities. Phil. Trans. R. Soc. B. 2010;3658(1):1815–1823. doi: 10.1098/rstb.2009.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Hazen S.P., Michael T.P., Morris G.P., Baxter I.R., Hu T.T., Chen H., Werner J.D., Nordborg M., Salt D.E. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;1048(1):12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boublík K., Petřík P., Sádlo J., Hédl R., Willner W., Černý T., Kolbek J. Calcicolous beech forests and related vegetation in the Czech Republic: a comparison of formalized classifications. Preslia. 2007;798(1):141–161. [Google Scholar]
- Bradshaw H.D., Jr., Otto K.G., Frewen B.E., McKay J.K., Schemske D.W. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics. 1998;1498(1):367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y., Haig D. Divergent mating systems and parental conflict as a barrier to hybridization in flowering plants. Am. Nat. 2005;1668(1):330–338. doi: 10.1086/432036. [DOI] [PubMed] [Google Scholar]
- Brieger F. Vererbung bei Artbastarden unter besonderer berücksichtigung der Gattung Nicotiana. Der Züchter. 1929;18(1):140–152. [Google Scholar]
- Buerkle C.A., Morris R.J., Asmussen M.A., Rieseberg L. The likelihood of homoploid hybrid speciation. Heredity. 2000;848(1):441–451. doi: 10.1046/j.1365-2540.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- Bushell C., Spielman M., Scott R.J. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell. 2003;158(1):1430–1442. doi: 10.1105/tpc.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černý T., Petřík P., Boublík K., Kolbek J. Habitat requirements of Cardaminopsis petraea—Rare and relict species of the Czech Republic. Biologia. 2006;618(1):51–61. [Google Scholar]
- Chini A., Grant J.J., Seki M., Shinozaki K., Loake G.J. Drought tolerance established by enhanced expression of the CC-NBSLRR gene, ADR1. requires salicylic acid, EDS1 and ABI1. Plant J. 2004;388(1):810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- Clark N.L., Aagaard J.E., Swanson W.J. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;1318(1):11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Clark R.M., Schweikert G., Toomajian C. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;3178(1):338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Clauss M.J., Koch M.A. Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci. 2006;118(1):449–459. doi: 10.1016/j.tplants.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Clerkx E.J., El-Lithy M.E., Vierling E., Ruys G.J., Blankestijn-De Vries H., Groot S.P., Vreugdenhil D., Koornneef M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004;1358(1):432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;68(1):836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Comai L., Madlung A., Josefsson C., Tyagi A. Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;3588(1):1149–1155. doi: 10.1098/rstb.2003.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tyagi A.P., Winter K., Holmes-Davis R., Reynolds S.H., Stevens Y., Byers B. Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell. 2000;128(1):1551–1568. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S., Bantin J., Wittich P.E., Kranz E., Lorz H., Dresselhaus T. ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 2001;258(1):103–114. doi: 10.1046/j.0960-7412.2000.00944.x. [DOI] [PubMed] [Google Scholar]
- Courbot M., Willems G., Motte P., Arvidsson S., Roosens N., Saumitou-Laprade P., Verbruggen N. A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4: a gene encoding a heavy metal ATPase. Plant Physiol. 2007;1448(1):1052–1065. doi: 10.1104/pp.106.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J.A., Orr H.A. Speciation. Sunderland, Massachusetts, USA: Sinauer; 2004. [Google Scholar]
- Crosby M.A, Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., The FlyBase Consortium FlyBase: genomes by the dozen. Nucl. Acids Res. 2007;88(1)(1):35. D486–491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart S., Kron P., Mable B.K. Characterizing polyploidy in Arabidopsis lyrata using chromosome counts and flow cytometry. Botany. 2004;828(1):185–197. [Google Scholar]
- Dawe R.K., Henikoff S. Centromeres put epigenetics in the driver's seat. Trends Biochem. Sci. 2006;318(1):662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Dilkes B.P., Spielman M., Weizbauer R., Watson B., Burkart-Waco D., Scott R.J., Comai L. The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol. 2008;68(1):2707–2720. doi: 10.1371/journal.pbio.0060308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger D.B., Desbrosses-Fonrouge A.G., Krach C., Chardonnens A.N., Meyer R.C., Saumitou-Laprade P., Kramer U. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins cosegregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 2004;398(1):425–439. doi: 10.1111/j.1365-313X.2004.02143.x. [DOI] [PubMed] [Google Scholar]
- el-Lithy M.E., Bentsink L., Hanhart C.J., Ruys G.J., Rovito D., Broekhof J.L., van der Poel H.J., van Eijk M.J., Vreugdenhil D., Koornneef M. New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics. 2006;1728(1):1867–1876. doi: 10.1534/genetics.105.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erilova A., Brownfield L., Exner V., Rosa M., Twell D., Mittelsten Scheid O., Hennig L., Kohler C. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 2009;58(1):e1000663. doi: 10.1371/journal.pgen.1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo J.M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.C., Grossniklaus U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;3178(1):656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- Ferree P.M., Barbash D.A. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009;78(1):e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov V., Dowdle J., Smirnoff N., Ford-Lloyd B., Newbury H.J., Macnair M.R. A quantitative trait loci analysis of zinc hyperaccumulation in Arabidopsis halleri. New Phytol. 2007;1748(1):580–590. doi: 10.1111/j.1469-8137.2007.02036.x. [DOI] [PubMed] [Google Scholar]
- Fishman L., Aagaard J., Tuthill J.C. Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution. 2008;628(1):2958–2970. doi: 10.1111/j.1558-5646.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Fishman L., Kelly A.J., Morgan E., Willis J.H. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics. 2001;1598(1):1701–1716. doi: 10.1093/genetics/159.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxe J.P., Slotte T., Stahl E., Neuffer B., Hurka H., Wright S.I. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA. 2009;1068(1):5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.L., Garcia D., Kim D., Hopf A., Salt D.E. Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol. 2005;1378(1):1082–1091. doi: 10.1104/pp.104.055293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardini V., Grossniklaus U., Page D.R., Kohler C. The Arabidopsis thaliana MEDEA polycomb group protein controls expression of PHERES1 by parental imprinting. Nature Genet. 2005;378(1):28–30. doi: 10.1038/ng1495. [DOI] [PubMed] [Google Scholar]
- Gavrilets S., Vose A. Case studies and mathematical models of ecological speciation. 2. Palms on an oceanic island. Mol. Ecol. 2007;168(1):2910–2921. doi: 10.1111/j.1365-294X.2007.03304.x. [DOI] [PubMed] [Google Scholar]
- Gazzani S., Gendall A.R., Lister C., Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;1328(1):1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Bubb K.L., Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;3248(1):1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gos G., Wright S.I. Conditional neutrality at two adjacent NBS-LRR disease resistance loci in natural populations of Arabidopsis lyrata. Mol. Ecol. 2008;178(1):4953–4962. doi: 10.1111/j.1365-294X.2008.03968.x. [DOI] [PubMed] [Google Scholar]
- Grant V. Plant Speciation. Columbia University Press; New York, NY: 1971. [Google Scholar]
- Guo Y.-L., Bechsgaard J.S., Slotte T., Neuffer B., Lascoux M., Weigel D., Schierup M.H. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA. 2009;1068(1):5246–5251. doi: 10.1073/pnas.0808012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K.D., Chapman E.J., Carrington J.C., Chen X., Wang X.J., Chen Z.J. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA. 2009;1068(1):17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D., Westoby M. Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Phil. Trans. R. Soc. Lond. B Biol. Sci. 1991;3338(1):1–13. [Google Scholar]
- Hall M.C., Willis J.H. Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus: implcations for genomic divergence. Genetics. 2005;1708(1):375–386. doi: 10.1534/genetics.104.038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.C., Willis J.H. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;608(1):2466–2477. [PubMed] [Google Scholar]
- Hanikenne M., Talke I.N., Haydon M.J., Lanz C., Nolte A., Motte P., Kroymann J., Weigel D., Krämer U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;4538(1):391–395. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- Hannah M.A., Kramer K.M., Geffroy V. Hybrid weakness controlled by the dosage-dependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytol. 2007;1768(1):537–549. doi: 10.1111/j.1469-8137.2007.02215.x. [DOI] [PubMed] [Google Scholar]
- Harper G., Osuji J.O., Heslop-Harrison J.S., Hull R. Integration of Banana Streak Badnavirus into the Musa genome: Molecular and cytogenetic evidence. Virology. 1999;2558(1):207–213. doi: 10.1006/viro.1998.9581. [DOI] [PubMed] [Google Scholar]
- Hawkins J.S., Kim H.R., Nason J.D., Wing R.A., Wendel J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;168(1):1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Tang R.H., Hao Y. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;3058(1):1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- Hegarty M.J., Hiscock S.J. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 2005;1658(1):411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H.S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;2938(1):1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Comai L. Genetic basis for dosage sensitivity in Arabidopsis thaliana. PLoS Genet. 2007;38(1):e70. doi: 10.1371/journal.pgen.0030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Young K., Watson B., Wu H., Comai L. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics. 2005;1708(1):1979–1988. doi: 10.1534/genetics.104.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Tyagi A.P., Lin H.Y., Comai L. Dosage and parent-of-origin effects shaping aneuploid swarms in A. thaliana. Heredity. 2009;1038(1):458–468. doi: 10.1038/hdy.2009.81. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J.S., Brandes A., Schwarzacher T. Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res. 2003;118(1):241–253. doi: 10.1023/a:1022998709969. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Inatsugi R., Sakamoto S., Sasaki N., Mori T., Kuroiwa H., Nakada T., Nozaki H., Kuroiwa T., Nakano A. Species preferentiality of the pollen tube attractant derived from the synergid cell of Torenia fournieri. Plant Physiol. 2006;1428(1):481–491. doi: 10.1104/pp.106.083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.H. Evolution of the realized climatic niche in the genus Arabidopsis (Brassicaceae). Evolution. 2005;598(1):1425–1436. [PubMed] [Google Scholar]
- Howard D.J. Conspecific sperm and pollen precedence and speciation. Ann. Rev. Ecol. Syst. 1999;308(1):109–132. [Google Scholar]
- Huettel B., Kreil D.P., Matzke M., Matzke A.J. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 2008;48(1):e1000226. doi: 10.1371/journal.pgen.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husová M. Azidophile Eichenwälder auf Quartziten im Tribec-Gebirge, Slowakei. Folia Geobotanica & Phytotaxonomica. 1967;28(1):121–136. [Google Scholar]
- Hylander N. Cardaminopsis suecica (Fr.) Hiit., a northern amphidiploid species. Bulletin du Jardin Botanique de l'Etat. 1957;278(1):591–604. [Google Scholar]
- Ispolatov I., Doebeli M. Speciation due to hybrid necrosis in plant-pathogen models. Evolution. 2009;638(1):3076–3084. doi: 10.1111/j.1558-5646.2009.00800.x. [DOI] [PubMed] [Google Scholar]
- Jain S. The evolution of inbreeding in plants. Annu. Rev. Ecol. Syst. 1976;78(1):469–495. [Google Scholar]
- Jakobsson M., Hagenblad J., Tavaré S., Säll T., Halldén C., Lind-Halldén C., Nordborg M. A unique recent origin of the allotetraploid species Arabidopsis suecica: Evidence from nuclear DNA markers. Mol. Biol. Evol. 2006;238(1):1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- Jeuken M.J.W., Zhang N.W., McHale L.K., Pelgrom K., den Boer E., Lindhout P., Michelmore R.W., Visser R.G.F., Niks R.E. Rin4 Causes Hybrid Necrosis and Race-Specific Resistance in an Interspecific Lettuce Hybrid. Plant Cell. 2009;218(1):3368–3378. doi: 10.1105/tpc.109.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C.X., Chee P.W., Draye X., Morrell P.L., Smith C.W., Paterson A.H. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution Int. J. Org. Evolution. 2000;548(1):798–814. doi: 10.1111/j.0014-3820.2000.tb00081.x. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Borevitz J.O., Preuss D. Genomewide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 2007;38(1):1848–1861. doi: 10.1371/journal.pgen.0030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson C., Dilkes B., Comai L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol. 2006;168(1):1322–1328. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Juenger T.E., Sen S., Stowe K.A., Simms E.L. Epistasis and genotype-environment interaction for quantitative trait loci affecting flowering time in Arabidopsis thaliana. Genetica. 2005;1238(1):87–105. doi: 10.1007/s10709-003-2717-1. [DOI] [PubMed] [Google Scholar]
- Justin C. Über bemerkenswerte Vorkommen ausgewählter Pflanzensippen auf Serpentinstandorten Österreichs, Sloweniens sowie der Tschechischen Republik. Linzer Biol. Beitr. 1993;258(1):1033–1099. [Google Scholar]
- Kamm A., Galasso I., Schmidt T., Heslop-Harrison J.S. Analysis of a repetitive DNA family from Arabidopsis arenosa and relationships between Arabidopsis species. Plant Mol. Biol. 1995;278(1):853–862. doi: 10.1007/BF00037014. [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Nasrallah J.B., Nasrallah M.E. Pollenpistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development. 1994;1208(1):3405–3418. [Google Scholar]
- Kawabe A., Nasuda S. Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol. Genet. Genomics. 2005;2728(1):593–602. doi: 10.1007/s00438-004-1081-x. [DOI] [PubMed] [Google Scholar]
- Khanna-Chopra R., Dalai M., Kumar G.P., Laloraya M. A genetic system involving superoxide causes F1 necrosis in wheat (T. aestivum L.). Biochem. Biophys. Res. Comm. 1998;2488(1):712–715. doi: 10.1006/bbrc.1998.9043. [DOI] [PubMed] [Google Scholar]
- Koch M.A., Matschinger M. Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;1048(1):6272–6277. doi: 10.1073/pnas.0701338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Wernisch M., Schmickl R. Arabidopsis thaliana's wild relatives: an updated overview on systematics, taxonomy and evolution. Taxon. 2008;578(1):933–943. [Google Scholar]
- Köhler C., Hennig L., Spillane C., Pien S., Gruissem W., Grossniklaus U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Devel. 2003;178(1):1540. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolník M., Marhold K. Distribution, chromosome numbers and nomenclature conspect of Arabidopsis halleri (Brassicaceae) in the Carpathians. Biologia. 2006;618(1):41–50. [Google Scholar]
- Koornneef M., Alonso-Blanco C., Peeters A.J.M., Soppe W. Genetic control of flowering time in Arabidopsis. Ann. Rev. Plant Phys. Plant Mol. Biol. 1998;498(1):345–370. doi: 10.1146/annurev.arplant.49.1.345. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Fransz P., de Jong H. Cytogenetic tools for Arabidopsis thaliana. Chromosome Res. 2003;118(1):183–194. doi: 10.1023/a:1022827624082. [DOI] [PubMed] [Google Scholar]
- Korves T.M., Bergelson J. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 2003;1338(1):339–347. doi: 10.1104/pp.103.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostoff D. Ontogeny, Genetics, and Cytology of Nicotiana Hybrids. Genetica. 1930;128(1):33–139. [Google Scholar]
- Krüger J., Thomas C.M., Golstein C., Dixon M.S., Smoker M., Tang S., Mulder L., Jones J.D.G. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science. 2002;2968(1):744–747. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Sas-Nowosielska A., Małkowski E., Japenga J., Kuperberg J.M., Pogrzeba M., Krzyżak J. The use of indigenous plant species and calcium phosphate for the stabilization of highly metal-polluted sites in southern Poland. Plant and Soil. 2005;2738(1):291–305. [Google Scholar]
- Kuittinen H., de Haan A.A., Vogl C., Oikarinen S., Leppälä J., Koch M., Mitchell-Olds T., Langley C.H., Savolainen O. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics. 2004;1688(1):1575–1584. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawesson J.E., Mark S. pH and Ellenberg reaction values for Danish forest plants. Proc. IAVS Symp. 2000;8(1):151–153. [Google Scholar]
- Lee J.A. Genetics of D3 complementary lethality in Gossypium hirsutum and G. barbadense. J. Hered. 1981;728(1):299–300. [Google Scholar]
- Leitch A.R., Leitch I.J. Genomic plasticity and the diversity of polyploid plants. Science. 2008;3208(1):481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;18(1):109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppälä J., Bechsgaard J.S., Schierup M.H., Savolainen O. Transmission ratio distortion in Arabidopsis lyrata: effects of population divergence and the S-locus. Heredity. 2008;1008(1):71–78. doi: 10.1038/sj.hdy.6801066. [DOI] [PubMed] [Google Scholar]
- Levin D.A. The Role of Chromosomal Change in Plant Evolution. New York, NY: Oxford University Press; 2002. [Google Scholar]
- Levin D.A. Flowering-time plasticity facilitates niche shifts in adjacent populations. New Phytol. 2009;1838(1):661–666. doi: 10.1111/j.1469-8137.2009.02889.x. [DOI] [PubMed] [Google Scholar]
- Liu S.C., Kowalski S.P., Lan T.H., Feldmann K.A., Paterson A.H. Genome-wide high-resolution mapping by recurrent intermating using Arabidopsis thaliana as a model. Genetics. 1996;1428(1):247–258. doi: 10.1093/genetics/142.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D.B., Modliszewski J.L., Wright K.M., Wu CA., Willis J.H. The strength and genetic basis of reproductive isolating barriers in flowering plants. Proc. R. Soc. Lond. B Biol. Sci. 2008a;3638(1):3009–3021. doi: 10.1098/rstb.2008.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D.B., Rockwood R.C., Willis J.H. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution. 2008b;628(1):2196–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Force A.G. The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 2000;1568(1):590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- Macnair M.R. Within and between population genetic variation for zinc accumulation in Arabidopsis halleri. New Phytol. 2002;1558(1):59–66. doi: 10.1046/j.1469-8137.2002.00445.x. [DOI] [PubMed] [Google Scholar]
- Macnair M.R., Bert V., Huitson S.B., Saumitou-Laprade P., Petit D. Zinc tolerance and hyperaccumulation are genetically independent characters. Proc. R. Soc. Lond. B Biol. Sci. 1999;2668(1):2175–2179. doi: 10.1098/rspb.1999.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Tyagi A.P., Watson B., Jiang H., Kagochi T., Doerge R.W., Martienssen R., Comai L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;418(1):221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- Makarevich G., Leroy O., Akinci U., Schubert D., Clarenz O., Goodrich J., Grossniklaus U., Köhler C. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;78(1):947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;4468(1):279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Martienssen R.A. Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol. 2010;1868(1):46–53. doi: 10.1111/j.1469-8137.2010.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C., Pons E., Prats G., León J. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 2004;378(1):209–217. doi: 10.1046/j.1365-313x.2003.01954.x. [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J.M. RNA-mediated chromatin-based silencing in plants. Curr. Op. Cell Biol. 2009;218(1):367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- McDaniel S.F., Willis J.H., Shaw A.J. A linkage map reveals a complex basis for segregation distortion in an interpopulation cross in the moss Ceratodon purpureus. Genetics. 2007;1768(1):2489–2500. doi: 10.1534/genetics.107.075424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J.K., Richards J.H., Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotrpy contributes to genetic correlations among ecological traits. Mol. Ecol. 2003;128(1):1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- McKay J.K., Richards J.H., Nemali K.S., Sen S., Mitchell-Olds T., Boles S., Stahl E.A., Wayne T., Juenger T.E. Genetics of drought adaptation in Arabidopsis thaliana II. QTL analysis of a new mapping population, KAS-1 × TSU-1. Evolution. 2008;628(1):3014–3026. doi: 10.1111/j.1558-5646.2008.00474.x. [DOI] [PubMed] [Google Scholar]
- Měsíček J. The chromosome morphology of Arabidopsis thaliana (L.) Heynh. and some remarks on the problem of Hylandra suecica (Fr.) Löve. Folia Geobotanica & Phytotaxonomica. 1967;28(1):433–436. [Google Scholar]
- Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol. Evol. 2001;168(1):693–700. [Google Scholar]
- Miyake T., Takebayashi N., Wolf D.E. Possible diversifying selection in the imprinted gene, MEDEA. Arabidopsis. Mol. Biol. Evol. 2009;268(1):843–857. doi: 10.1093/molbev/msp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Meyers B.C., Michelmore R.W., Gaut B.S. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 2002;128(1):1305–1315. doi: 10.1101/gr.159402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. Uniparental expression of PollV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;4608(1):283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- Myburg A.A., Vogl C., Griffin A.R., Sederoff R.R., Whetten R.W. Genetics of postzygotic isolation in Eucalyptus: whole-genome analysis of barriers to introgression in a wide interspecific cross of Eucalyptus grandis and E. globulus. Genetics. 2004;1668(1):1405–1418. doi: 10.1534/genetics.166.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah J.B., Lui P., Sherman-Broyles S., Schmidt R., Nasrallah M.E. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics. 2007;1758(1):1965–1973. doi: 10.1534/genetics.106.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndowora T., Dahal G., LaFleur D., Harper G., Hull R., Olszewski N.E., Lockhart B. Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology. 1999;2558(1):214–220. doi: 10.1006/viro.1998.9582. [DOI] [PubMed] [Google Scholar]
- Okuda S., Tsutsui H., Shiina K. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;4588(1):357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- Orr H.A., Irving S. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics. 2005;1698(1):671–682. doi: 10.1534/genetics.104.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A., Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;558(1):1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Preuss D. Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol. 2006;68(1):7. doi: 10.1186/1471-2229-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoyan A., Kochian L.V. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 2004;1368(1):3814–3823. doi: 10.1104/pp.104.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels M., Saumitou-Laprade P., Holl A.C., Petit D., Bonnin I. Multiple origin of metallicolous populations of the pseudometallophyte Arabidopsis halleri (Brassicaceae) in central Europe: the cpDNA testimony. Mol Ecol. 2005;148(1):4403–4414. doi: 10.1111/j.1365-294X.2005.02739.x. [DOI] [PubMed] [Google Scholar]
- Phillips L.L. Interspecific incompatibility in Gossypium. IV. Temperature-conditional lethality in hybrids of G. klotzschianum. Am J Bot. 1977;648(1):914–915. [Google Scholar]
- Pigliucci M., Schlichting C.D. Reaction norms of Arabidopsis. V. Flowering time controls phenotypic architecture in response to nutrient stress. J Evol Biol. 2002;118(1):285–301. [Google Scholar]
- Polatschek A. Cytotaxonomische beiträge zur flora der Ostalpenländer, I. Österr Botan Zeitschrift. 1965;1138(1):1–46. [Google Scholar]
- Pontes O., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci USA. 2004;1018(1):18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przedpełska E., Wierzbicka M. Arabidopsis arenosa (Brassicaceae) from a lead-zinc waste heap in southern Poland—a plant with high tolerance to heavy metals. Plant and Soil. 2007;2998(1):43–53. [Google Scholar]
- Punwani J.A., Rabiger D.S., Drews G.N. MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell. 2007;198(1):2557–2568. doi: 10.1105/tpc.107.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punz W., Mucina L. Vegetation on Anthropogenic metalliferous soils in the Eastern Alps. Folia Geobotanica & Phytotaxonomica. 1997;328(1):283–295. [Google Scholar]
- Rajakaruna N. The edaphic factor in the origin of plant species. Int Geol Rev. 2004;468(1):471–478. [Google Scholar]
- Ramos-Onsins S.E., Stranger B.E., Mitchell-Olds T., Aguadé M. Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics. 2004;1668(1):373–388. doi: 10.1534/genetics.166.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert-Poggeler K.R., Noreen F., Schwarzacher T., Harper G., Hohn T. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 2003;228(1):4836–4845. doi: 10.1093/emboj/cdg443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H., Blackman B.K. Speciation genes in plants. Ann Bot. AOP. 2010. [DOI] [PMC free article] [PubMed]
- Rieseberg L.H., Willis J.H. Plant speciation. Science. 2007;3178(1):910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H., Raymond O., Rosenthal D.M., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;3018(1):1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H., Whitton J., Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;1528(1):713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riihimäki M., Savolainen O. Environmental effects on flowering differences between Northern and Southern populations of Arabidopsis lyrata (Brassicaceae). Am J Bot. 2004;918(1):1036–1045. doi: 10.3732/ajb.91.7.1036. [DOI] [PubMed] [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., Salt D.E. Natural variants of AtHKT1 enhance Na accumulation in two wild populations of Arabidopsis. PLoS Genet. 2006;28(1):e210. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säil T., Jakobsson M., Lind-Hallden C., Hallden C. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. J Evol Biol. 2003;168(1):1019–1029. doi: 10.1046/j.1420-9101.2003.00554.x. [DOI] [PubMed] [Google Scholar]
- Säil T., Lind-Halldén C., Jakobsson M., Halldén C. Mode of reproduction in Arabidopsis suecica. Hereditas. 2005;1418(1):313–317. doi: 10.1111/j.1601-5223.2004.01833.x. [DOI] [PubMed] [Google Scholar]
- Salt D.E., Baxter I., Lahner B. lonomics and the study of the plant ionome. Ann Rev Plant Biol. 2008;598(1):709–733. doi: 10.1146/annurev.arplant.59.032607.092942. [DOI] [PubMed] [Google Scholar]
- Sandring S., Agren J. Pollinator-mediated selection on floral display and flowering time in the perennial herb Arabidopsis lyrata. Evolution. 2009;638(1):1292–1300. doi: 10.1111/j.1558-5646.2009.00624.x. [DOI] [PubMed] [Google Scholar]
- Sandring S., Riihimäki M.A., Savolainen O., Ågren J. Selection on flowering time and floral display in an alpine and a lowland population of Arabidopsis lyrata. J Evol Biol. 2007;208(1):558–567. doi: 10.1111/j.1420-9101.2006.01260.x. [DOI] [PubMed] [Google Scholar]
- Savolainen V., Anstett M.C., Lexer C., Hutton I., Clarkson J.J., Norup M.V., Powell M.P., Springate D., Salamin N., Baker W.J. Sympatric speciation in palms on an oceanic island. Nature. 2006;4418(1):210–213. doi: 10.1038/nature04566. [DOI] [PubMed] [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;3238(1):737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Schluter D., Conte G.L. Genetics and ecological speciation. Proc Natl Acad Sci U S A. 2009;106 S18(1):9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R., Jorgensen M. H., Brysting A. K., Koch M. A. The evolutionary history of the Arabidopsis lyrata complex: A hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evolutionary Biology. 2010. in press. [DOI] [PMC free article] [PubMed]
- Schmickl R., Koch M.A. Introgression, hybridization and polyploid evolution in Arabidopsis lyrata and A. arenosa. Manuscript in preparation .
- Scholz H. Nomenklatorische und systematische Studien an Cardaminopsis arenosa (L.) Hayek. Willdenowia. 1962;38(1):137–149. [Google Scholar]
- Schwarzbach A.E., Donovan L.A., Rieseberg L.H. Transgressive character expression in a hybrid sunflower species. Am J Bot. 2001;888(1):270–277. [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Bailey J., Dickinson H.G. Parentof-origin effects on seed development in Arabidopsis thaliana. Development. 1998;1258(1):3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- Sherman-Broyles S., Boggs N., Farkas A., Liu P., Vrebalov J., Nasrallah M.E., Nasrallah J.B. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 2007;198(1):94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Inatsugi R., Lihova J., Iwanaga H., Kudoh H., Marhold K., Savolainen O., Watanabe K., Yakubov V.V., Shimizu K.K. The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Mol Ecol. 2009;188(1):4024–4048. doi: 10.1111/j.1365-294X.2009.04329.x. [DOI] [PubMed] [Google Scholar]
- Shimizu K.K. Ecology meets molecular genetics in Arabidopsis. Pop Ecol. 2002;448(1):221–233. [Google Scholar]
- Silvertown J., Servaes C., Biss P., Macleod D. Reinforcement of reproductive isolation between adjacent populations in the Park Grass Experiment. Heredity. 2005;958(1):198–205. doi: 10.1038/sj.hdy.6800710. [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Dean C. Arabidopsis, the Rosetta stone of flowering time? Science. 2002;2968(1):285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzic M., Becker J.D., Feijó J.A., Martienssen R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;1368(1):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaydon R.W., Davies T.M. Rapid divergence of plant populations in response to recent changes in soil conditions. Evolution. 1982;368(1):289–297. doi: 10.1111/j.1558-5646.1982.tb05044.x. [DOI] [PubMed] [Google Scholar]
- Sobel J. M., Chen G. F., Watt L. R., Schemske D. W. The biology of speciation. Evolution. 2009;648(1):295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- Stelfox A.W. The forms of Cardaminopsis petraea (L.) in Ireland. Irish Naturalists’ J. 1970;168(1):308–309. [Google Scholar]
- Stinchcombe J.R., Schmitt J. Flowering time plasticity in Arabidopsis thaliana: a reanalysis of Westerman & Lawrence (1970). J Evol Biol. 2004;178(1):197–207. doi: 10.1046/j.1420-9101.2003.00641.x. [DOI] [PubMed] [Google Scholar]
- Swanson R., Edlund A.F., Preuss D. Species specificity in pollen-pistil interactions. Ann Rev Genet. 2004;388(1):793–818. doi: 10.1146/annurev.genet.38.072902.092356. [DOI] [PubMed] [Google Scholar]
- Swanson W.J., Vacquier V.D. The rapid evolution of reproductive proteins. Nature Rev Genet. 2002;38(1):137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Bryson T.D., Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;38(1):18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Masuelli R., Tyagi A.P., Comai L., Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;148(1):1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke I.N., Hanikenne M., Kramer U. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 2006;1428(1):148–167. doi: 10.1104/pp.105.076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y.-L., Hu T.T., Clark R.M., Nasrallah J.B., Weigel D., Nordborg M. The evolution of selfing in Arabidopsis thaliana. Science. 2007;3178(1):1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- Törjek O., Witucka-Wall H., Meyer R.C., von Korff M., Kusterer B., Rautengarten C., Altmann T. Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet. 2006;1138(1):1551–1561. doi: 10.1007/s00122-006-0402-3. [DOI] [PubMed] [Google Scholar]
- Tsukahara S., Kobayashi A., Kawabe A., Mathieu O., Miura A., Kakutani T. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;4618(1):423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- Turner T.L., von Wettberg E.J., Nuzhdin S.V. Genomic analysis of differentiation between soil types reveals candidate genes for local adaptation in Arabidopsis lyrata. Plos One. 2008;38(1):e3183. doi: 10.1371/journal.pone.0003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T.L., Bourne E.C., Von Wettberg E.J., Hu T.T., Nuzhdin S.V. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet AOP. 2010. doi:10.1038/ng.515. [DOI] [PubMed]
- Ungerer M.C., Baird S.J., Pan J., Rieseberg L.H. Rapid hybrid speciation in wild sunflowers. Proc Natl Acad Sci USA. 1998;958(1):11757–11762. doi: 10.1073/pnas.95.20.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen H., Kliebenstein D.J., West M.A., Kim K., van Poecke R., Katagiri F., Michelmore R.W., Doerge R.W., St Clair D.A. Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell. 2007;198(1):2099–2110. doi: 10.1105/tpc.107.050641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterhout C. A new theory of MHC evolution: beyond selection on the immune genes. Proc R Soc B. 2009;2768(1):657–665. doi: 10.1098/rspb.2008.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C., Bennetzen J.L. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci USA. 2006;1038(1):17638. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H., Josefsson C., Dilkes B., Kirkbride R., Harada J., Comai L. Dosage-dependent deregulation of an AGAMOUS-LIKE Gene Cluster Contributes to Interspecific Incompatibility. Curr Biol. 2009;198(1):1128–1132. doi: 10.1016/j.cub.2009.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Chai Y., Chu X., Zhao Y., Wu Y., Zhao J., Ngezahayo F., Xu C., Liu B. Molecular characterization of a rice mutator-phenotype derived from an incompatible cross-pollination reveals transgenerational mobilization of multiple transposable elements and extensive epigenetic instability. BMC Plant Biol. 2009;98(1):63. doi: 10.1186/1471-2229-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S., Chen Z.J. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics. 2006a;1738(1):965–974. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006b;1728(1):507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K. Arabidopsis kamchatica (Fisch, ex DC.) K. Shimizu & Kudoh and A. kamchatica subsp. kawasakiana (Makino) K. Shimizu & Kudoh, New Combinations. Acta phytotaxonomica et geobotanica. 2005;568(1):163. [Google Scholar]
- Weigel D., Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;108(1):107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgolaski F.E. Phenological modifications in plants by various edaphic factors. Int J Biometeorol. 2001;458(1):196–202. doi: 10.1007/s004840100100. [DOI] [PubMed] [Google Scholar]
- Wilczek A.M., Roe J.L., Knapp M.C., Cooper M.D., Lopez-Gallego C., Martin L.J., Muir C.D., Sim S., Walker A., Anderson J. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;3238(1):930–934. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- Wilkins J.F., Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nature Rev Genet. 2003;48(1):359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Willems G., Drager D.B., Courbot M., Gode C., Verbruggen N., Saumitou-Laprade P. The genetic basis of zinc tolerance in the metallophyte Arabidopsis halleri ssp. halleri(Brassicaceae): an analysis of quantitative trait loci. Genetics. 2007;1768(1):659–674. doi: 10.1534/genetics.106.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.W., Schiff C.L., Hughes D.L., Somerville S.C. Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession Kashmir-1. Genetics. 2001;1588(1):1301–1309. doi: 10.1093/genetics/158.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E., Takashi T., Morinaka Y., Lin S., Wu J., Matsumoto T., Kitano H., Matsuoka M., Ashikari M. Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol Gen Genomics. 2010;2838(1):305–315. doi: 10.1007/s00438-010-0514-y. [DOI] [PubMed] [Google Scholar]
- Yang H., Kaur N., Kiriakopolos S., McCormick S. EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta. 2006;2248(1):1004–1014. doi: 10.1007/s00425-006-0283-3. [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Schmid M. Just say no: floral repressors help Arabidopsis bide the time. Curr Op Plant Biol. 2009;128(1):580–586. doi: 10.1016/j.pbi.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Yin T.M., DiFazio S.P., Gunter L.E., Riemenschneider D., Tuskan G.A. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor Appl Genet. 2004;1098(1):451–463. doi: 10.1007/s00122-004-1653-5. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S. W.-L., Chen H., Henderson I. R., Sinn P., Pellegrini M., Jacobsen S. E., Ecker J. R. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;1268(1):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]