Abstract
Treatment of cell lines with type I IFNs activates the formation of ISGF3 (STAT1/STAT2/IRF9), which induces the expression of many genes. To study this response in primary cells, we treated fresh human blood with IFN-β and used flow cytometry to analyze phosphorylated STATs1, 3 and 5 in CD4+ and CD8+ T cells, B cells, and monocytes. The activation of STAT1 was remarkably different among these leukocyte subsets. In contrast to monocytes, CD4+ and CD8+ T cells, few B cells activated STAT1 in response to IFN-β, a finding that could not be explained by decreased levels of IFNAR2 or STAT1 or enhanced levels of SOCS1 or relevant protein tyrosine phosphatases in B cells. Micro-array and real-time PCR analyses revealed the induction of STAT1-dependent pro-apoptotic mRNAs in monocytes but not in B cells. These data show that ISGF3 or STAT1 homodimers are not the main activators of gene expression in primary B cells of healthy humans. Notably, in B cells and especially in CD4+ T cells, IFN-β activated STAT5 in addition to STAT3, with biological effects often opposite from those driven by activated STAT1. These data help to explain why IFN-β increases the survival of primary human B cells and CD4+ T cells, but enhances the apoptosis of monocytes, and also to understand how leukocyte subsets are differentially affected by endogenous type I IFNs during viral or bacterial infections, and by type I IFN treatment of patients with multiple sclerosis, hepatitis or cancer.
Keywords: STATs, Cytokines, Transcription Factors, Signal Transduction
Introduction
IFNs are pleiotropic cytokines that play important roles in infection and inflammation. Three different classes of IFNs are known: Type I IFNs includes IFNs α, β, ω, τ, δ, κ and ε (1), type II is IFN-γ and type III is IFN-λ. IFNs-α and -β utilize the IFNAR1 and IFNAR2c receptor subunits to signal (2), each of which binds constitutively to a single member of the JAK family of kinases, IFNAR1 to TYK2 and IFNAR2 to JAK1. Ligand binding induces the phosphorylation of JAK1, TYK2, intracellular tyrosine residues of each receptor subunit, and STATs. Activated STATs dimerize, dissociate from the receptor and translocate to the nucleus to induce the expression of ISGs (3). Current data suggest that ISGF3 is the major transcription factor activated in response to IFN-α/β (3, 4). ISGF3, a complex of phosphorylated STAT1, STAT2 and unphosphorylated IRF-9, binds to ISREs present in the promoters of many ISGs. In response to type I IFNs, activated STAT1 can also form homodimers that bind to GAS elements in some ISG promoters (3, 5).
It is becoming clear that, in addition to ISGF3 and STAT1 homodimers, other transcription factors play important roles as cytoplasmic messengers between the receptor and the nucleus (4), helping to explain why type I interferons (IFNs), which were discovered on the basis of their potent antiviral activities, are now known to act much more broadly, as pleiotropic cytokines that regulate many different cellular functions. For example, STAT3 is activated in response to type I IFNs in most cell lines, forming STAT3 homodimers, or heterodimers with activated STAT1 (4). In contrast, activation of STAT4 and STAT5 by IFN-α/β is found mostly in NK and T cells (6–8). Interestingly, the activation of STAT6 induced by type I IFNs has been described thus far only in B cell lines (9). STAT homo- and heterodimers bind to GAS elements in the promoters of ISGs, but it is clear that different STAT dimers have different preferences for specific GAS elements (5). The differential activation of STATs 4, 5, and 6 in different cell lines suggests the possibility of cell type-specific activation of STATs by IFN-α/β in vivo. Of note, evidence for a cell type-specific response to IFN-α has been described previously with respect to differential ISG induction in human T cells and dendritic cells (10).
We have now investigated how primary human leukocytes signal in response to IFN-β. Undiluted freshly drawn human whole blood was stimulated with IFN-β in vitro in order to mimic the situation in vivo as closely as possible. Since the activation of STATs occurs only transiently, the isolation of many different leukocyte subsets after stimulation of whole blood in combination with Western analysis is not feasible because, by the time the subsets could be isolated, the optimal time point for activation of STATs would have already passed. Therefore, we employed a flow cytometry-based technique that enables the detection of intracellular phospho-tyrosine-STATs1, 3, and 5 at the single cell level, allowing cells to be fixed at the optimal time for STAT activation. IFN-β-induced activation of STATs1, 3, and 5 was chosen because these three transcription factors regulate cell survival in opposite directions (11–13). Furthermore, this approach allowed us to address whether differential activation of these STATs might explain how IFN-β enhances the survival of mature B cells and T cells (14–19) while increasing apoptosis in monocytes and many cancer cell lines (20–23). Notably, we found that IFN-β induced significant differences in the activation of STATs 1 and 5 in different leukocyte subsets and that these differences are related to the induction of pro- and anti-apoptotic genes, respectively. Our results provide important insights into the differential effects that type I IFNs may have on leukocyte subsets during infection and upon treatment with type I IFNs of multiple sclerosis, hepatitis and some cancers.
Materials and Methods
Cell culture and IFN-β stimulation
The HT cell line was acquired from the ATCC (CRL-2260, human B lymphoblast derived from a patient with diffuse mixed lymphoma) and cultured in RPMI-1640 medium supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 4500 mg/L glucose, and 10 % fetal bovine serum. This cell line was maintained in 10- or 15-cm culture dishes and always stimulated at a concentration of 1× 106 cells/ml with IFN-β1a (Avonex, 30 μg/0.5 ml Prefilled Syringe, 12×106 IU/ml, Biogen Idec Inc., Cambridge, MA, USA). Stimulated cells were subsequently fixed for flow cytometry analysis or lysed for Western analysis.
Heparinized whole blood was obtained from healthy donors according to an IRB-approved protocol (Cleveland Clinic). Within 15 min after venupuncture, undiluted whole blood was stimulated in 6- or 10-cm culture dishes or 50 ml canonical tubes (BD Biosciences) in vitro with recombinant human IFN-β1a (Biogen Idec), IFN-α1 (3×103 IU/μl), IFN-α2 (Intron A, 10×103 IU/μl, Schering-Plough, Kenilworth, NJ, USA) or IFN-γ (10×103 IU/μl, Genentech Inc, San Francisco, CA) as indicated. For each staining, 130 μl of whole blood was used. All in vitro stimulations were performed in an incubator at 37°C (Thermo Fisher Scientific Inc., Asheville, NC, USA); no cell clumping or adhesion to tissue culture plates was observed. After stimulation, whole blood was fixed and lysed for intracellular detection of PY-STATs.
Western Analysis of HT cells and isolated leukocyte subsets
After stimulation of HT cells with IFN-β1a, or after leukocyte subsets were purified from unstimulated whole blood, the cells were washed once with phosphate-buffered saline, and the cell pellets were lysed for 30 min at 4 °C in 250 μl (per 5 × 106 cells) of lysis buffer containing 50 mM HEPES, pH 7.9, 250 mM potassium chloride, 0.1% NP-40, 10% glycerol, 0.1 mM EDTA, 10 mM sodium fluoride, 5 mM sodium orthovanadate, 1 mM phenylmethane-sulfonyl fluoride, 20 μg/ml aprotinin, 20 μg/ml pepstatin, and 20 μg/ml leupeptin. Cellular debris was pelleted by centrifugation at 13,000 × g at 4 °C for 10 min. Cell extracts were fractionated by electrophoresis in 10% or 12 % SDS-PAGE and transferred to PVDF membranes (Millipore Corporation, Bedford, MA, USA). The following antibodies were used: rabbit polyclonal α-SOCS1 and rabbit polyclonal α-SH-PTP1 (clones H-93 and C-19 respectively, Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal α-TCP45 (clone CF4-1D, Calbiochem, via EMD Chemicals, Gibbstown, NJ, USA), mouse monoclonalα-N-terminal STAT1 (clone 42, BD Transduction Laboratories), rabbit polyclonal α-PY701-STAT1, rabbit polyclonal α-PY705-STAT3, rabbit polyclonal α-STAT3, rabbit polyclonalα-PY694-STAT5, rabbit polyclonal α-STAT5 (Cell Signaling Technology Inc., Danvers, MA, USA), rabbit polyclonal α-PY689-STAT2 and rabbit polyclonal α-STAT2 (Upstate Cell Signaling Solutions, Lake Placid, NY, USA), and mouse monoclonal anti-β-actin (clone AC-74, Sigma-Aldrich, St. Louis, MO). Horseradish peroxidase-coupled goat anti-rabbit or goat anti-mouse IgG (Rockland Immunochemicals Inc. Gilbertsville, PA, USA) was used for visualization, using the enhanced chemiluminescence (ECL Plus) Western analysis detection system (PerkinElmer LAS, Inc., Boston, MA, USA).
Intracellular detection of PY-STATs using flow cytometry
The published method of Chow et al. (24), developed to measure intracellular phospho-ERK in whole blood cells, was adapted slightly to measure the induction of phospho-STATs in human whole blood. A number of commercially available anti-human CD3, CD4, CD8, CD19 and CD14 antibodies were screened. The best α-CD3 and α-CD14 clones were those that performed optimally after fixation and erythrocyte lysis, but before methanol incubation. In contrast, the best α-CD8, α-CD4 and α-CD19 antibodies performed best after methanol incubation. As previously published (25), IFN-β-induced phospho-STATs were optimally detected in leukocytes that were permeabilized with 90% methanol (data not shown).
After stimulation with IFN-β1a in vitro or leaving cells untreated for the same time period, whole blood or cell lines were fixed in 4% or 2% formaldehyde, respectively, by adding 10% prewarmed methanol-free formaldehyde (Polysciences, Warington, PA, USA), followed by incubation at 37°C for 10 min. After fixation, cell lines were washed twice with 50 ml of ice-cold wash buffer (Dulbecco’s Phosphate-Buffered Saline (D-PBS), 5% FBS, 0.09% NaN3). Erythrocytes were lysed by adding 0.12% Triton X-100 (0.1% Triton X-100 final concentration; Pierce Biotechnology, Rockford, IL, USA) dissolved in 1 × D-PBS and incubating for 30 min at room temperature with rocking. Lysed erythrocytes were removed by washing 3 times with 50 ml ice-cold wash buffer, followed by spinning at 300 × g for 10 min at 4°C. One-hundred μl of fixed cells were subsequently added to each five ml Falcon FACS tube (equivalent to 100 μl HT or 130 μl of whole blood per tube), containing FITC- or pacific blue-conjugated α-CD3 (clone UCHT1, BD Biosciences, San Jose, CA, USA), α-CD14-FITC (clone RM052, Beckman Coulter, Miami, FL, USA) or α-CD14-AF700 (clone TÜK4, Invitrogen Corporation, Carlsbad, CA, USA) antibodies in the amounts recommended by the manufacturers. Cells were incubated for 30 min at room temperature in the dark and washed twice, once with 2 ml ice-cold wash buffer per tube and once with 2 ml ice-cold 1 × D-PBS. While vortexing at high speed, 1 ml 90% methanol in 1 × D-PBS was added per tube and the mixture was incubated at −20°C overnight. The next day, the contents of each tube were washed twice with 2 ml of wash buffer (with spinning at 300 × g, 4°C). For the blocking step, the cell pellets were resuspended in 50 μl of wash buffer and incubated for 10 min at room temperature in the dark. A combination of antibodies directed against human CD8 (clone B9.11, PE-Cy5 conjugated, Beckman Coulter), CD19 (clone J4.119, PE-Cy5 or PE-Cy7 conjugated, Beckman Coulter), or CD4 (clone 13B8.20, PE-Cy5 conjugated, Beckman Coulter), and the following AlexaFluor 647 (AF647)- or PE- conjugated antibodies against PY(701)-STAT1, PY(705)-STAT3, or PY(694)-STAT5 (clones 4A, 4, and 47, respectively, BD Biosciences) were added in amounts as advised by the manufacturer, followed by incubation at room temperature in the dark for 1 h. Either α-CD8 or α-CD4 antibodies were used; the alternative T cell subset was identified by selecting the CD8−CD3+ or CD4−CD3+ population, respectively. To detect total STAT1, cells were incubated with anti-STAT1-PE (N-terminus of STAT1, clone 1/Stat1, BD Biosciences) after permeabilization with 90% methanol. To detect the activation of STAT2, rabbit polyclonal α-PY(689)-STAT2 (Upstate Cell Signaling Solutions, Lake Placid, NY, USA) was added at 6.5 μg/ml. Experiments demonstrating the specificity of this anti-PY-STAT2 antibody are shown in Supplemental Figs. 1A + 1B. For the fluorochrome-conjugated antibodies, the last wash step was performed with 3 ml of wash buffer per tube (300 × g, 10 min, 4°C). To detect PY-STAT2, cells were incubated with 5 μl of a 1: 10 dilution of goat-anti-rabbit-IgG-PE (Jackson ImmunoResearch Laboratories, Inc. West Grove, PA, USA) at room temperature for 30 min. After the final washing step, each cell pellet was resuspended in 350 μl wash buffer and measured on a LSR I (Becton Dickinson) or LSRII (BD Biosciences) flow cytometer. Samples stained with a single color were used for compensation. Intact cells were gated on forward and side scatter (FCS/SSC) and 50,000 cells were measured. Flow data were analyzed with WinList (Verity Software House, Topsham, ME, USA). The percentage of IFN-β-induced phospho-STAT-positive cells were determined by subtracting the percentage of positive cells in unstimulated cells, which were set at <2%. An example of an analysis is shown in Fig. 1.
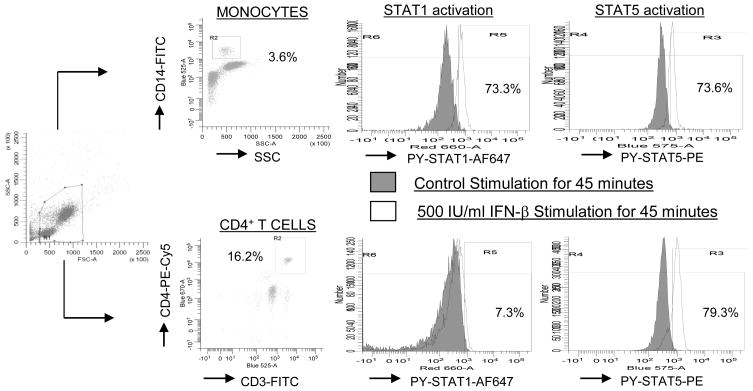
FIGURE 1. Analysis of activation of STAT1 and STAT5 in human leukocytes.
Undiluted whole blood from a healthy donor was stimulated with 500 IU/ml of IFN-β1a or was untreated for 45 min. After processing as described in Materials and Methods, the cells were analyzed by using flow cytometry. CD14+ (monocytes) and CD4+/CD3+ (CD4+ T cells) were determined within the live gate. The percentages of IFN-β-induced PY-STAT positive cells within each leukocyte subset was determined by subtracting the percentages of positive unstimulated cells, which were set at <2%.
Detection of IFNAR2 and caspase 3 activation by Flow Cytometry
Unstimulated and undiluted whole blood (150 μl) was incubated with mouse-anti-IFNAR2-PE (PBL Biomedical Laboratories via R&D Systems, clone MMHAR-2, Minneapolis, MN, USA) or control mIgG2a-PE(BD Biosciences, clone G155–178) as recommended by the manufacturer, in combination with the same anti-CD-antibodies as above-mentioned, for 30 min at 4°C. Whole blood cells were subsequently fixed and erythrocytes were lysed as above-mentioned, and the washing steps the cell pellet was resuspended in 350 μl wash buffer and measured on a LSRII (BD Biosciences) flow cytometer the same day.
To detect induction of apoptosis, whole blood that was 1:3 diluted with plain RPMI-1640 was either stimulated with 2000 IU/ml IFN-β or unstimulated for various time-periods up till 48 h at 37°C, or kept for 1 h at 50°C (positive control). Cells were then washed one time with 25 ml of 1× D-PBS, and whole blood was divided (150 μl/tube) and incubated with same anti-CD antibodies as above-mentioned for 30 min at 4°C. Whole blood cells were subsequently fixed and erythrocytes were lysed as abovementioned, and after the washing steps, each pellet was eventually resuspended in 100 μl Permeabilization Medium B (Invitrogen, Carlsbad, CA, USA) and incubated with 20 μl anti-activated caspase 3-PE antibody (= 0.25 μg) for 30 minutes at RT. Finally, after each tube was washed with 3 ml stain buffer, the cell pellet was resuspended in 350 μl stainbuffer and measured on BD Biosciences LSR II flow cytometer the same day. Induction of activated caspase 3 in leukocyte subsets by IFN-β was determined by subtracting the percentage of caspase 3-positive cells in unstimulated cells from those in IFN-β-stimulated cells.
Statistical Analysis
GraphPad InStat 3 was used. The Nonparametric Repeated Measures ANOVA for paired samples or Friedman test was used to test whether the four blood cell subsets (monocytes, B cells, CD8+ and CD4+ T cells) differed with respect to activation of STAT1, STAT3 and STAT5. When the Friedman test showed a significant difference (p<0.05), post-hoc analysis was subsequently performed using Dunn’s test in order to detect which blood subsets differed significantly from each other. When calculating the P values, the Dunn’s test takes into account the number of comparisons one is making (Bonferroni adjustment). Pearson’s Correlation Test was used to determine whether the percentages of PY-STAT-positive leukocyte subsets that were induced after stimulation with IFN-β for 45 min correlated with the percentage of activated caspase 3-positive subsets after longer periods of stimulation with IFN-β. The coefficient of determination (R2), and the two-tailed P value are shown (if significantly correlated).
Blood cell subset isolation, gene-array analysis and real-time PCR
Twenty-two ml of undiluted whole blood from each of 2 healthy donors was stimulated with 2000 IU/ml IFN-β1a (Avonex, Biogen Idec) for 3 h, and 22 ml of blood was left unstimulated for 3 h. An aliquot of blood was taken out after 45 min of stimulation with IFN-β to determine the activation of STATs1, 3 and 5 by flow cytometry. Immediately following stimulation for 3 h, 10 ml of whole blood was incubated with either 500 μl “whole blood” α-CD14 or α-CD19 microbeads (Miltenyi Biotec, Auburn, CA) for 15 min at 4°C in order to isolate monocytes and B cells, respectively. After washing with cold running buffer (Miltenyi Biotec; PBS, 2 mM EDTA, 0.5% BSA, 0.09% sodium azide) to remove unbound microbeads, and after bringing the volume of the whole blood back to the starting volume by adding cold running buffer, the cells of interest were positively selected using the AutoMACS Pro Separator (Miltenyi Biotec) and program posselWB. During the whole isolation procedure the blood cells were kept cold. The procedure is very fast (maximally 20 min), which helps to preserve the quality of the RNA. The purity (90–99%) of the positively selected fraction and the yield were excellent, since the negative sorted fraction was totally depleted of each subset of interest. Total RNA was isolated from the isolated unstimulated (control) or IFN-βstimulated blood cell subset by dissolving the cells in TRIZOL (Invitrogen, Carlsbad, CA; 1–10 × 106 cells in 1 ml), following the protocol of the manufacturer. One μg of total RNA (100 ng is minimally needed) was sent to the Cleveland Clinic Genomics Core. A single-round of in vitro transcription (IVT) amplification was carried out using the Illumina RNA Amplification Kit, manufactured by Ambion, Inc., to amplify mRNA and thus to obtain ample amounts of cRNA to perform the whole human genome gene expression assay using the humanRef-8 v2 expression beadchips micro-array (Illumina, San Diego, CA), which has 22,184 transcript probes, representing 18,189 genes in total. The micro-array data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE23307 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23307). Expression data normalization and differential expression analysis was handled through the Illumina BeadStudio Gene Expresson module V3.2. The data were first normalized by using the Illumina background normalization algorithm and then the differential expression analyses were performed by using Illumina’s custom model. Downstream data processing and reporting was handled in R packages (http://www.r-project.org). Genes for downstream analyses were filtered to include only those with both differential expression analysis p values of less than 0.001 and fold changes greater than 2, over the unstimulated (control) stimulation. Genes in this filtered list were further grouped into those that were changed in all B cells and monocytes, those that were changed in monocytes only, and those that were changed in B cells only (both HI#1 and #2). The Gene Ontology Enrichment Analysis Software Toolkit (GOEAST: publicly available at http://omicslab.genetics.ac.cn/GOEAST/) was used to sort these groups of genes according to gene ontology, particularly apoptosis, proliferation and cell cycle regulation.
Real-time PCR (rtPCR) was used to confirm changes in gene expression obtained by micro-array analysis. rtPCR was done with RNA isolated from B cells and monocytes (present in whole blood of 6 healthy individuals and purification occurred after stimulation of whole blood as above-mentioned), which were either left untreated or were stimulated with 2000 IU/ml IFN-β for 3 h. rtPCR was thus performed with 24 samples to detect changes in 7 mRNAs. The following 7 Taqman Gene Expression Assays from Applied Biosystems were used (Gene Symbol, Gene name, Assay ID #): BAK1, BCL2-antagonist/killer 1, Hs00832876_g1; CASP3, caspase 3, Hs00263337_m1; CDKN1A, cyclin-dependent kinase inhibitor 1A (p21), Hs00355782_m1; BCL2L13, BCL2-like 13 apoptosis facilitator, Hs00209789_m1; STK3, serine/threonine kinase 3 (STE20 homolog yeast), Hs00169491_m1; IL2RA, interleukin 2 receptor, alpha, Hs00907779_m1; NAMPT, nicotinamide phosphoribosyl-transferease (= PBEF1), Hs00237184_m1. Two candidate genes were chosen for endogenous control determination based on publications about rtPCR performed with RNA from B cells and monocytes: eukaryotic 18S rRNA and human hypoxanthine phosphoribosyltransferase 1 (HPRT1). An Applied Biosystems ABI 7900HT unit with automation attachment (Foster City, CA) was used for rtPCR. This unit is capable of collecting spectral data at multiple points during a PCR run. To execute the first step and make archive cDNA, 150 ng of total RNA was reverse transcribed in a 25-μl reaction using Applied Biosystems enzymes and reagents in accordance with the manufacturer’s protocols. RNA samples were accurately quantitated using a Nanodrop Technologies ND-1000 spectrophotometer (Wilmington, DE). The cDNA reaction from above was diluted by a factor of 10. For the PCR step, 9 μl of this diluted cDNA was used for each of three replicate 15-μl reactions carried out in a 384-well plate. Standard PCR conditions were used for the Applied Biosystems assays: 50°C for 2 min, followed by 95°C for 10 min, followed by 40 cycles of 95°C for 15 s alternating with 60°C for 1 min each. The Comparative Ct method, or delta-delta Ct method, was used for relative quantitation. 18S rRNA had very little variation in expression across the sample sets and therefore was chosen as the endogenous control. For rtPCR data analysis, values of RNA abundance were normalized for each gene with respect to the endogenous control in that sample (18S), mean values for fold changes were calculated for each gene (IFN-β-stimulated over control treated). PCR confirmation of gene expression array data required that the direction of the change in expression had to be the same with rtPCR as with gene expression arrays, and be at least 2-fold increased.
Results
Exposure to low doses of IFN-β causes differential activation of STATs1, 3, and 5 in primary human blood cell subsets
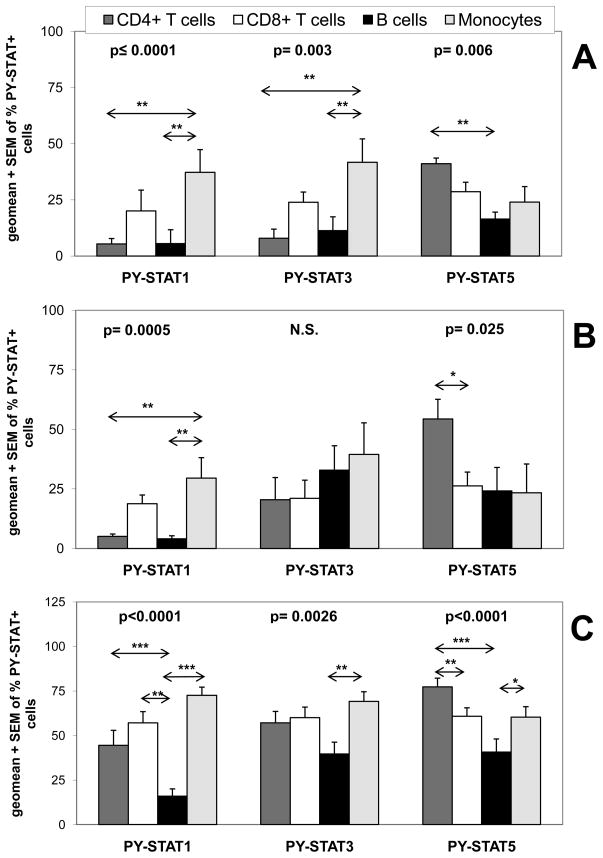
To verify antibody specificity, we compared our flow cytometry method to Western analysis, using the human leukemic cell line HT, which was stimulated with 1000 IU/ml of IFN-β (Fig. 2). The two methods yielded the same overall pattern. Of note, the highest percentage of PY-STAT-positive HT cells was found 30 min after stimulation with IFN-β, as usually found in human cell lines. The advantage of flow cytometry is that it reveals the percentage of each cell subset in which a certain STAT is activated (Figs. 1 and 2). To begin to investigate how primary human monocytes, B cells, CD4+ and CD8+ T cells respond to IFN-β, we stimulated undiluted whole blood samples from nine healthy individuals with 500 IU/ml of IFN-β for 25 min (Fig. 3A). We observed significant differences in the fractions of leukocyte subsets in which STAT1 (p≤0.0001), STAT3 (p=0.003) and STAT5 (p=0.006) was activated. Unexpectedly, remarkably few B cells and CD4+ T cells showed activation of STAT1 in comparison to monocytes. The differences in activation of STAT3 were very similar to those seen for the activation of STAT1; many fewer B cells and CD4+ T cells displayed activation of STAT3 compared to monocytes. Thus, of the blood cell subsets investigated, the highest percentage of PY-STAT1- and PY-STAT3-positive cells were found among monocytes, whereas the percentages of CD8+ T cells positive for PY-STAT1 and PY-STAT3 were intermediate (Fig. 3A). In contrast, many more CD4+ T cells than B cells showed activation of STAT5 when whole blood was stimulated with 500 IU/ml IFN-β for 25 min.
FIGURE 2. Flow cytometric and Western analyses of PY-STATs show the same pattern.
The leukemic human B cell line HT was stimulated with 1000 IU/ml of IFN-β1a. Cells were either A, lysed and subjected to Western analysis or B, fixed and stained to determine the percentage of cells with PY-STATs by flow cytometry.
FIGURE 3. In vitro stimulation of whole blood with IFN-β reveals unique STAT activation patterns in different leukocyte subsets.
Undiluted whole blood of healthy donors was left untreated or stimulated with either A, 500 IU/ml of IFN-β1a for 25 min (9 persons), B, 45 min (7 persons), or C, with 2000 of IU/ml IFN-β1a for 45 min (20 persons) and induction of PY-STATs 1, 3, and 5 was determined. The geometric means + SEM of the various donors are shown for percentages of leukocyte subsets positive for each PY-STAT. The Friedman test revealed significant differences in the activation of STATs among the four leukocyte subsets; the p values are shown in top of each Figure. The Friedman test was followed with post-hoc analysis, revealing significant differences in STAT1, 3 or 5 activation between the subsets, indicated in the figures by arrows. Significant differences were indicated as: * is p<0.05; ** is p<0.01; and *** is p<0.001. N.S. = not significantly different.
Exposure to low doses of IFN-β causes differential activation of STATs1 and 5 in primary human blood cell subsets at the optimal time for PY-STAT induction
Although 25 min of stimulation with IFN-β is optimal for STAT activation in many cell lines, it is possible that this time is not optimal in CD4+ T cells and B cells in whole blood. Therefore, blood samples from three healthy individuals were stimulated with 500 IU/ml of IFN-β for various times, up to 75 min (Fig. 4). The highest activation of STAT3 and STAT5 was detected in all blood cell subsets after 45 min of exposure to IFN-β, declining gradually thereafter. Optimal activation of STAT1 in CD8+ T cells and monocytes was also found after 45 min. Interestingly, although the IFN-β-induced activation of STAT3 in CD4+ T cells and B cells was very low after 25 min (≤ 15%), it was at least twice as high (between 22–32%) after 45 min. In contrast, the activation of STAT1 in CD4+ T cells as well as B cells remained very low for the whole time period tested (<10% positive cells).
FIGURE 4. Activation of STATs in primary human blood cells after stimulation with IFN-β is optimal after 45 min.
Undiluted whole blood from 3 healthy donors was unstimulated or stimulated with 500 IU/ml IFN-β1a for different time-periods. The geometric means + SEM of the 3 donors are shown for percentages of leukocyte subsets positive for each PY-STAT.
Since the optimal time-point for activation of STATs in whole blood cells was 45 min, we tested whether the same significant differences in PY-STAT1, 3 and 5 induction could also be found as after 25 min. To this end, whole blood from seven healthy subjects was stimulated with 500 IU/ml for 45 min (Fig. 3B). There were significant differences in the fractions of blood cell subsets that showed activation of STAT1 (p=0.0005) and STAT5 (p=0.025). Thus, even after longer stimulation with IFN-β, many fewer B cells and CD4+ T cells showed induction of PY-STAT1 than did monocytes, whereas more CD4+ T cells than CD8+ T cells showed activation of STAT5 (Fig. 3B). Although monocytes still showed the highest percentage of PY-STAT3-positive cells, the difference compared to B cells and CD4+ T cells was no longer significant. Therefore, the activation of STAT3 by IFN-β in CD4+ T cells and B cells is delayed compared to activation of STAT3 in CD8+ T cells and monocytes, in particular after 25 min (Fig. 3A), but eventually catches up after 45 min (Figs. 3A, 3B, 4).
Exposure to high-dose IFN-β also causes differential activation of STATs1, 3 and 5 in primary human blood cell subsets
Because the observed induction of PY-STATs at the optimal times occurred in no more than 50% of the cells after stimulation with 500 IU/ml IFN-β, we explored whether stimulation with higher concentrations of IFN-β would yield a higher percentage of cells positive for PY-STATs. We stimulated whole blood of 3 healthy individuals with increasing doses of IFN-β, up to 10,000 IU/ml, for 45 min (Fig. 5). At a concentration of 200 IU/ml IFN-β, virtually no activation of STATs was detected in any of the blood cell subsets tested. All four subsets responded with activation of STATs3 and 5 at 500 IU/ml IFN-β, and more cells became positive at the higher concentrations. Furthermore, the activation of STAT1 by IFN-β was also dose-responsive in CD8+ T cells and monocytes. Remarkably, it was still observed that very few B cells and CD4+ T cells (≤ 10%) showed activation of STAT1, even at the higher concentrations of IFN-β (2,000–10,000 IU/ml).
FIGURE 5. Dose responses for stimulation of PY-STATs with IFN-β in whole blood cell subsets.
Undiluted whole blood of 3 healthy donors was unstimulated or stimulated with increasing concentrations of IFN-β1a for 45 min. PY-STATs 1, 3, and 5 were determined in leukocyte subsets as indicated. Geometric means + SEM of 3 healthy donors are shown for percentage PY-STAT-positive blood cells.
To test whether the same differences in STAT1 and STAT5 activation could be observed with 2,000 IU/ml as with 500 IU/ml IFN-β, whole blood from twenty healthy subjects was stimulated for 45 min with the higher concentration (Fig. 3C). 2,000 IU/ml and not 10,000 IU/ml of IFN-β was used because the activation of STAT1 in monocytes and B cells was lower with the latter concentration (Fig. 5). At 2000 IU/ml of IFN-β, there were significant differences in the fractions of blood cell subsets that showed activation of STAT1 (p<0.0001), STAT3 (p=0.0026) and STAT5 (p<0.0001). Remarkably, in contrast to the lower dose, using this high dose of IFN-β to stimulate whole blood of many donors revealed now also high numbers of PY-STAT1-positive CD4+ T cells (Fig. 3C). However, even at such a high concentration of IFN-β, many fewer B cells than monocytes, CD4+ and CD8+ T cells activated STAT1. Also, fewer B cells than monocytes activated STAT3 and STAT5 (Fig. 3C). Of interest, the highest fraction of PY-STAT5-positive cells was still found among the CD4+ T cell subset (Fig. 3C).
In summary, irrespective of IFN-β concentration or stimulation time, peripheral blood B cells do not show appreciable activation of STAT1 in response to IFN-β, indicating that ISGF3 or STAT1 homodimers are not the main transcription factors driving ISG induction in the majority of primary human B cells, and also not in CD4+ T cells at lower IFN-β concentrations. In contrast, these subsets show activation of STAT3 and STAT5, and the highest activation of STAT5 occurs in CD4+ T cells.
Differential activation of STAT1, 3 and 5 in leukocyte subsets by IFN-β is associated with differences in apoptosis induction
Activation of STAT1 usually leads to induction of cell-cycle arrest and apoptosis, whereas enhanced survival and proliferation results from PY-STAT3 and PY-STAT5 induction. We therefore explored whether the differential activation of these STATs could be responsible for previously unexplained differences in induction of apoptosis in the various primary human leukocyte subsets by IFN-β. To this end, whole blood from four healthy subjects was stimulated with IFN-β for 4, 6, 8, 10, 12, 24, or 48 h and apoptosis induction in the various subsets was determined by the activation of caspase 3 (Fig. 6A). Stimulation with IFN-β induces the most induction of apoptosis in monocytes, much less in CD8+ T cells and the least in B cells and CD4+ T cells (Fig. 6A), in agreement with previous reports. Using the blood of the same donors, the induction of PY-STAT1, 3 and 5 was also determined after stimulation with IFN-β for 45 min in vitro. Interestingly, in none of the leukocyte subsets could we observe a correlation between the total percentages of PY-STAT1 positive cells after 45 min and the percentages of activated caspase 3-positive cells at any of the later time-points (data not shown). This result might be explained by the fact that many of the PY-STAT1 positive cells are actually doubly positive for PY-STAT3 and PY-STAT5, resulting in opposing effects on apoptosis induction at an individual cell level. Therefore, a double staining was also performed with anti-PY-STAT1/PY-STAT3 or anti-PY-STAT1/PY-STAT5 antibodies, to detect doubly positive cells after IFN-β stimulation. Fig. 6B shows that the induction of PY-STAT1+/PY-STAT3−, PY-STAT1+/PY-STAT3+, PY-STAT1−/PY-STAT3+ or PY-STAT1+/PY-STAT5−, PY-STAT1+/PY-STAT5+, PY-STAT1−/PY-STAT5+ positive cells between the four leukocyte subsets are strikingly different. For instance, the percentage of PY-STAT1+/PY-STAT3− cells is highest in CD4+ T cells, followed by CD8+ T cells and is lowest in B cells and monocytes (Fig. 6B). In contrast, the percentage of PY-STAT1+/PY-STAT5− cells is highest in CD8+ T cells, followed by CD4+ T cells and monocytes, but again lowest in B cells. Remarkably, the generation of PY-STAT1−/PY-STAT3+ and PY-STAT1−/PY-STAT5+ positive cells could only be observed in the B cells subset (Fig. 6B), because in this subset IFN-β induced the lowest percentage of PY-STAT1 positive cells. The monocyte subset showed the highest amount of apoptosis induction by IFN-β and Fig. 6A illustrates that after 8 h of stimulation, for the first time significant apoptosis induction could be observed in all 4 donors tested. Strikingly, the generation of PY-STAT1+/PY-STAT3− monocytes after 45 min correlated very significantly with the fraction of activated caspase 3-positive monocytes after 8 h of IFN-β stimulation (p=0.0008, Fig. 6C). Of note, after 10 or 12 h of IFN-β stimulation, the number of activated caspase 3-positive monocytes doubled or tripled, compared to 8 h of stimulation (Fig. 6A), suggesting that even in PY-STAT1+/PY-STAT3+ or PY-STAT1+/PY-STAT5+ monocytes apoptosis was induced eventually. None of the other subsets showed a correlation between induction of PY-STAT1+/PY-STAT3− orPY-STAT1+/PY-STAT5− after 45 min with apoptosis induction after 8 h (data not shown), but this could be because individual donors showed variation in the B cell, CD4+ or CD8+ T cell subsets with respect to significant activation of caspase 3. Therefore, the highest observed activation of caspase 3 (Fig. 6A, CD4+: 2× 6 h, 10 and 12 h; CD8+: 4, 6 and 2× 10 h; B cells: 2 × 10 h and 2 × 12 h) within a leukocyte subset was plotted against the % PY-STAT1+/PY-STAT3− cells, revealing a significant correlation only within the CD8+ T cell subset (Fig. 6C, p=0.0357). Due to the finding that this subset has the highest percentage of PY-STAT1+/PY-STAT5− cells, many of the PY-STAT1+/PY-STAT3− CD8+ T cells are likely to be PY-STAT1+/PY-STAT3−/PY-STAT5− and prone to apoptosis induction. In contrast, CD4+ T cells and B cells have the lowest percentages of PY-STAT1+/PY-STAT5− cells and consequently possibly the lowest amount of apoptosis induction due to the anti-apoptotic effects in PY-STAT1+/PY-STAT5+ cells.
FIGURE 6. Differences in activation of STAT1 in leukocyte subsets are associated with variation in induction of apoptosis.
A, Undiluted whole blood from four healthy donors was stimulated with 2000 IU/ml IFN-β or was unstimulated for varying times. Induction of activated caspase 3 in leukocyte subsets by IFN-β was determined by subtracting the percentage of caspase 3-positive cells in unstimulated cells from those in IFN-β-stimulated cells; results are shown for individual donors. B, Undiluted whole blood from five healthy donors was stimulated with 2000 IU/ml IFN-β or was not treated for 45 min. Geometric means + SD of 5 donors are shown for either PY-STAT1+/PY-STAT3−, PY-STAT1+/PY-STAT3+, PY-STAT1−/PY-STAT3+ or PY-STAT1+/PY-STAT5−, PY-STAT1+/PY-STAT5+, PY-STAT1−/PY-STAT5+ positive subsets. C, Data from four healthy individuals (data of fig. 6A and 6B combined) are depicted: induction of PY-STAT3−/PY-STAT1+-positive monocytes and CD8+ T cells after stimulation with IFN-β for 45 min correlated with caspase 3 activation after 8 and 4–10 h, respectively. For each leukocyte subset, R2, the coefficient of determination, and the P value (if significantly correlated) are shown.
Differential activation of STAT1 in primary human monocytes and B cells is connected to differential induction of apoptosis-promoting mRNAs by IFN-β
We investigated whether the observed differential activation of STAT1 in B cells and monocytes would result in differences in mRNA induction and, in particular, in the induction of pro-apopotic mRNAs. To this end, undiluted whole blood from 2 healthy individuals was stimulated with 2000 IU/ml of IFN-β for 3 h and pure B cells and monocytes were isolated by using magnetically labeled antibodies. An aliquot of blood was taken out after 45 min of stimulation with IFN-β to detect the activation of STATs1, 3, and 5 by flow cytometry. Supplemental Fig. 2A shows the expected differential activation of STATs in B cells and monocytes from two healthy individuals. Remarkably, mRNA induction by IFN-β was very different in primary human B cells and monocytes. Three hours of stimulation with IFN-β caused a total of 1462 different mRNAs to be up- or downregulated by at least 2-fold in monocytes or B cells (836 up and 626 down). Fig. 7A shows the Venn diagram of the 836 mRNAs that were that were increased by at least 2-fold in monocytes and B cells in response to IFN-β. Remarkably, of the mRNAs that changed in the monocytes of HI#1, 337 of 596 (57%) were increased in monocytes only, whereas 233 of 596 (39%) were shared between monocytes and B cells of HI#1, and 229 of 596 (38%) were shared with B cells of HI#2. In contrast, the responses in B cells of HI#1 and HI#2 were very similar, because 316 mRNAs out of 405 (HI#1) or 410 (HI#2) were increased by 2-fold (78% or 77% respectively).
FIGURE 7. Striking differences in gene-induction in purified B cells and monocytes after in vitro stimulation with IFN-β.
Undiluted whole blood from 2 healthy donors was stimulated with 2000 IU/ml IFN-β1a or was unstimulated for 3 h. A, After 3 h, pure B cells and monocytes were isolated by using magnetically labeled antibodies. RNA isolated from the purified subsets was used for analysis of gene expression using micro-array. The fold-increase in mRNA was calculated comparing expression in unstim ulated with stimulated subsets. The Venn diagram shows increases in mRNA by at least 2-fold and illustrates different mRNA induction patterns in B cells (CD19) and monocytes (CD14). B, Differences in expression of apoptosis-inducing mRNAs (BAK1, CASP3, CDKN1A, STK3) and the survival-promoting mRNA PBEF1 between B cells and monocytes after stimulation with IFN-β, revealed by micro-array analysis (Table 1), was confirmed by real-time PCR, using mRNA isolated from purified monocytes and B cells, normalized relative to 18S rRNA. The median values of monocytes and B cells derived from 6 healthy subjects are shown for fold-changes in mRNA expression in stimulated compared to control subsets.
The mRNAs that increased by at least 2-fold in monocytes only were sorted according to their ontology, using the GOEAST program. The following mRNAs, increased in monocytes only, are classified as “inducers of programmed cell death”: CDKN1A, BAK1, BCL2L13, CASP3, and STK3 (Table 1), and are all known to be very potent apoptosis-inducing proteins (26–29). The mRNA of cyclin-dependent kinase inhibitor 1A (CDKN1A, p21 or Cip1) was increased 3-fold in monocytes, whereas the mRNA expression in B cells of both healthy individuals remained unchanged. The induction of p21 is dependent on binding of activated STAT1 to the GAS element in the p21 promoter after IFN-γ stimulation (27). Likewise, increased expression of caspase 3 (CASP3) is dependent on STAT1 activation (28). Although it has not been conclusively shown that Bcl-2 homologous antagonist/killer (BAK1), Bcl-2-like13 (BCL2L13, BCL-RAMBO), and serine/threonine-protein kinase 3 (STK3) are dependent on the activation of STAT1 homodimers, this conclusion is very likely to be correct since all these mRNAs are induced after IFN-γ stimulation (29, www.interferome.org).
Table 1.
Fold increases of mRNAs in primary human monocytes and B cells from two healthy individuals (HI) after 3 h of stimulation with IFN-β.
| mRNA | Monocytes (HI1) | B cells (HI1) | B cells (HI2) |
|---|---|---|---|
| CDKN1A (p21)a | 3.1 | 1.0 | 0.8 |
| BAK1b | 2.5 | 0.9 | 1.1 |
| BCL2L13b | 2.4 | 1.1 | 1.4 |
| CASP3a | 2.3 | 1.5 | 1.5 |
| STK3b | 2.7 | 1.3 | 1.7 |
| IL2RAc | 1.7 | 4.7 | 5.8 |
| PBEF1d | 1.6 | 3.2 | 3.2 |
| TNFSF13 (BAFF)e | 2.8 | 10.4 | 10.0 |
| TNFSF10 (TRAIL) | 6.8 | 18.2 | 23.7 |
| FAS | 4.4 | 4.7 | 3.4 |
| IRF1 | 2.9 | 2.6 | 2.8 |
Known STAT1-dependent gene.
Probable STAT1-dependent gene.
Known STAT5-dependent gene.
Known STAT3-dependent gene.
Probable STAT3-dependent gene.
After IFN-β stimulation, some mRNAs were induced at least 2-fold both in B cells and monocytes (Table 1: BAFF, IRF-1, TRAIL, FAS). TNFS10 (TRAIL) and FAS cause apoptosis in cancer cells, but not necessarily in normal immune cells (30, 31). Remarkably, after sorting the mRNAs that were only increased by ≥ 2-fold in B cells according to their ontology, we found not one mRNA to be related to apoptosis induction or cell cycle arrest. In contrast, IL-2Rα chain (IL2RA) and pre-B cell colony enhancing factor (PBEF1) are two mRNAs that were found increased in primary human B cells only (Table 1), and B cell activating factor (BAFF) was 3.5 times more increased in B cells than in monocytes (Table I). These mRNAs are all found to be related to increased survival and proliferation (32–36). Of note, IL2RA and PBEF1 are both induced by type I IFNs only, and not by IFN-γ (www.interferome.org). In response to IL-2 or IL-6, the mRNAs of IL2RA and PBEF1 are known to be increased after binding of activated STAT5 or STAT3 (32, 35), respectively, to the promoters. Because IL-10 increases the expression of BAFF mRNA, the enhanced transcription is likely dependent on PY-STAT3 binding to the BAFF promoter (36). Because only B cells show the formation of PY-STAT1−/PY-STAT3+ and PY-STAT1−/PY-STAT5+ cells after stimulation with IFN-β (Fig. 6B), it is likely that activated STAT5 and STAT3 cause the increases of IL-2RA, PBEF1 and BAFF, respectively, in B cells only.
To confirm these micro-array data from 2 healthy individuals, the induction of specific mRNAs in monocytes and B cells after 3 h of stimulation with IFN-β was replicated in 6 healthy individuals. After 45 min of stimulation, the activation of STATs1, 3 and 5 showed the typical pattern (supplemental Fig. 2B). Induction of BCL2L13 and IL2RA mRNA was not replicated by real-time PCR, but we confirmed increased BAK1, CASP3, CDKN1A, and STK3 mRNAs (in monocytes only) and an increase in PFBEF1 mRNA (in B cells only) after IFN-β stimulation (Fig. 7B). In summary, the differential activation of STAT1 by IFN-β in primary human monocytes and B cells is associated with very significant differences in mRNA induction. High activation of STAT1 in monocytes is related to an increase in the expression of potent inducers of apoptosis, whereas poor STAT1 activation in B cells could explain why the activation of STAT3 and STAT5 leads to increased induction of certain proliferation-stimulating genes in B cells only.
IFNAR2 and STAT1 levels are similarly expressed in primary human leukocytes and STAT2 is activated normally in B cells
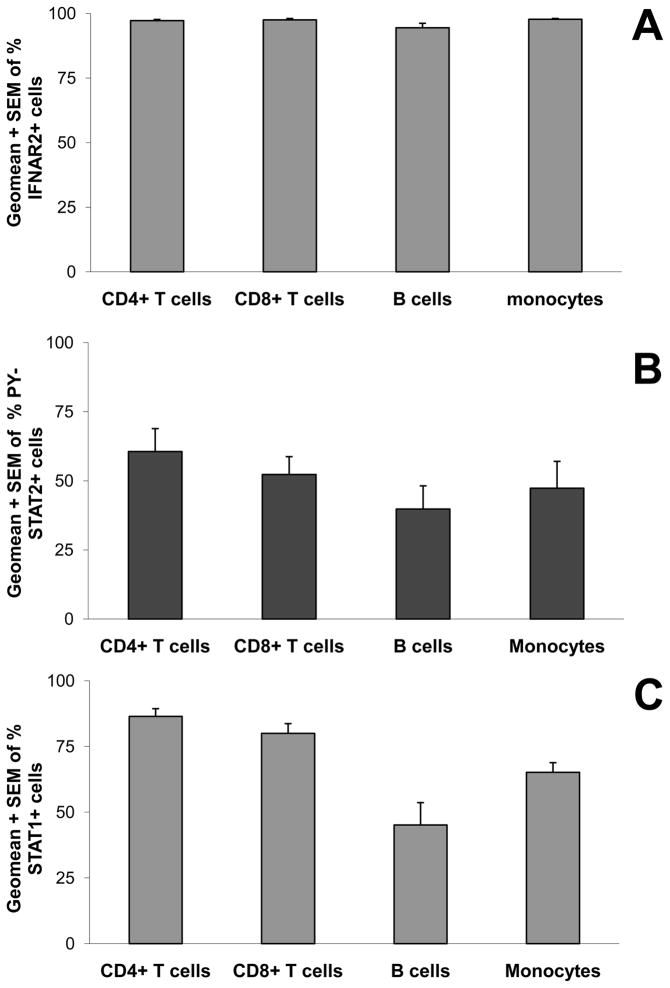
To begin to understand the mechanism that cause few B cells to activate STAT1 in response to IFN-β, we tested surface expression of IFNAR2. IFNAR1 is crucial for ligand binding, but IFNAR2 with its long cytoplasmic tail possesses two conserved tyrosine residues that are crucial for activation of STAT1, STAT2 and STAT3 (1–4). IFNAR2 expression on leukocyte subsets present in whole blood of 6 healthy individuals was studied by flow cytometry. Fig. 8A shows that 100% of monocytes, B cells, CD4+ and CD8+ T cells expressed IFNAR2, and therefore lack of its expression cannot be the reason why few B cells activated STAT1. Although IFNAR2 expression was equal, the functionality of this receptor chain might be different in B cells. We had previously found that the activation of STAT2 preceded the activation of STAT1 in fibrosarcoma cells and that STAT2-null cells are severely hampered in their capacity to activate STAT1 (37). Fig. 8B shows the percentage of cells positive for PY-STAT2 after stimulating whole blood of six healthy individuals with 2,000 IU/ml IFN-β for 45 min. Monocytes, B cells, CD4+ and CD8+ T cells all demonstrated activation of STAT2 in response to IFN-β (Fig. 8B), and therefore failure to activate STAT2 is not the cause of low STAT1 activation in B cells. Another possible explanation for the very low activation of STAT1 in B cells is that many fewer B cells express STAT1 protein. We used flow cytometry to compare the four different leukocyte subsets for percentages of cells that express total STAT1 (Fig. 8C), finding that all subsets were positive for STAT1. Although percentages of total STAT1-positive B cells were slightly lower compared to the other subsets, this difference cannot explain why usually fewer than 15% of the B cells showed activation of STAT1 in response to IFN-β. Finally, Western analysis of total STAT1 expression in isolated monocytes, B cells, CD4+ or CD8+ T cells from two different healthy individuals revealed similar levels of STAT1 expression in all subsets (data not shown).
FIGURE 8. Differences in STAT1 activation cannot be explained by differences in IFNAR2 expression levels, STAT2 activation or STAT1 levels.
A, Surface IFNAR2 expression was determined on monocytes, B cells, CD4+ T cells and CD8+ T cells present in unstimulated whole blood of six healthy individuals. B, Activation of STAT2 in leukocyte subsets was determined by stimulating undiluted whole blood of 6 healthy donors with 2000 IU/ml of IFN-β1a or unstimulated for 45 min. C, STAT1 expression was determined in monocytes, B cells, CD4+ T cells and CD8+ T cells present in unstimulated blood of four healthy subjects. Geometric means + SEM for the various donors are shown for the percentages of leukocyte subsets that are positive for surface IFNAR2 (A), PY-STAT2 (B), or intra-cellular STAT1 (C).
Type I and type II IFNs induce similar amounts of PY-STAT1-positive B cells
To test if other type I IFNs and type II IFN also stimulate few B cells to activate STAT1, undiluted whole blood of six healthy subjects was stimulated with 2000 IU/ml IFN-γ for 30 min (the optimal time for activation of STATs by IFN-γ; data not shown), and 2000 IU/ml IFN-α1, IFN-α2b, or IFN-β for 45 min. Fig. 9 shows that within all four leukocyte subsets tested (B cells, monocytes, CD4+ or CD8+ T cells), all type I IFNs induced similar percentages of cells to activate STAT1. In B cells and monocytes, type II IFN stimulated similar percentages of cells with PY-STAT1+ cells as did type I IFNs (Fig. 9). Because T cells that produce IFN-γ (such as Th1 and Tc1 cells) are unresponsive to IFN-γ due to changed type II IFNGR levels (38), CD4+ and CD8+ T cells generated lower amounts of PY-STAT1+ cells in response to IFN-γ than in response to type I IFNs (Fig. 9). Remarkably, also on an individual donor level, IFN-α1, -α2b, -β, and -γ activated equal percentages of PY-STAT1+ B cells (Fig. 9). We found that only 7% of healthy subjects (3 out of 41) showed activation of STAT1 in more than 60% of their B cells in response to IFN-β. As shown in Fig. 9, two individuals had more than 60% of their B cells positive for PY-STAT1 in response to IFN-α/β and IFN-γ. The four individuals who showed lower activation of STAT1 in B cells in response to IFN-β, about 10–20% of the cells positive for PY-STAT1, showed a similar response after stimulation with IFN-α1, -α2b and -γ. Therefore, it can be concluded that the same mechanism that influences the activation of STAT1 in B cells after ligation of the type I IFNAR, also influences its activation by the type II IFNGR.
FIGURE 9. Similar activation of STAT1 in B cells after stimulation with IFN-γ, IFN-α1, IFN-α2b and IFN-β.
Undiluted whole blood from six healthy subjects was stimulated for 30 min with 2000 IU/ml IFN-γ or for 45 min with 2000 IU/ml IFN-α1, IFN-α2b or IFN-β (optimal time-points for activation of STATs). Activation of STAT1 was determined in the various leukocyte subsets and shown for all six healthy subjects individually as percentages of PY-STAT1-positive subsets.
Suppressor of cytokine signaling 1 (SOCS1), which diminishes activation of STAT1 by the type I and type II IFN receptors by influencing the activation of the JAKs (39), and SHP1 and T-cell protein tyrosine phosphatase of 45 kDa (TCP45), two protein tyrosine phosphatases known to decrease tyrosine phosphorylation of STAT1 (40–41), are all candidates to explain the lack of PY-STAT1 induction in B cells by type I and II IFNs. We tested in isolated monocytes, B cells, CD4+ T cells and CD8+ T cells of 1 healthy individual, and isolated monocytes and B cells from another 2, whether the expression of SOCS1, SHP1 and TCP45 was enhanced in B cells compared to other leukocyte subsets. However, western analysis showed no SOCS1 expression in any of the unstimulated subsets (whereas it did in an IFN-γ-stimulated monocytic cell line), and SHP1 and TCP45 were expressed at similar levels in all subsets (data not shown).
Discussion
Mechanistic aspects of differential STAT induction in response to IFN-β
The aims of this study were to investigate how certain primary human blood cells signal in response to IFN-β and to explore how such signals might be related to apoptosis or cell survival. Up to now, non-immune cells and cell lines have mainly been used to study type I IFN signaling and to elucidate the mechanisms by which these IFNs regulate transcription. We developed a flow-cytometry-based assay to detect at the single cell level the activation of specific STATs in primary human leukocytes. IFN-α/β-induced activation of STAT1, which results mainly in the formation of the ISGF3 complex, but also leads to the formation of STAT1 homodimers in adherent and non-adherent cell lines, is a hallmark of type I IFN signaling. The human leukemic B cell line HT and the leukemic CD4+ T cell line Jurkat form abundant amounts of PY-STAT1 in response to IFN-β (Fig. 2, and unpublished results). Unexpectedly, we found that only a small fraction of primary human B cells activated STAT1 in response to IFN-β, and that this activation was independent of the concentration of IFN used (500–10,000 IU/ml) or the time of stimulation (10–75 min, Fig. 4). We tested IFN-β-induced signaling in 41 different individuals. Although most of the time we saw that a maximum of 25% of the B cells responded by activating STAT1, in 7% of these individuals, we detected >65% of the B cells positive for PY-STAT1 induction (3 out of 41). It may be that these persons have an underlying disease that has not been diagnosed or that this response is normal during a subclinical virus infection. Nevertheless, despite individual variations, we found that many fewer B cells showed activation of STAT1 compared to monocytes, CD4+ and CD8+ T cells after stimulation with 2000 IU/ml IFN-β (Fig. 3C). To begin to understand the mechanism, we studied IFNAR expression and found that 100% of primary human B cells, monocytes, CD4+ and CD8+ T cells expressed the IFNAR2 chain (Fig. 8A), which has a long cytoplasmic tail containing 2 conserved tyrosine residues that are crucial for activation of STAT1, 2 and 3 (1–4). Although IFNAR2 expression was found to be equal, the functionality of this receptor chain could still be different in B cells. An explanation for the low activation of STAT1 could be decreased activation of STAT2, because activation of STAT1 depends on the activation of STAT2 in fibrosarcoma cells and primary fibroblasts (37,42). However, STAT2-deficient peritoneal macrophages retained the ability to activate STAT1, highlighting intriguing differences in the ability of the IFNAR to activate STAT1 in fibroblasts and monocytic cells (42). Nothing is known about this function of the IFNAR in other leukocyte subsets, but we found low activation of STAT1 in primary human B cells at every IFN-β concentration and in CD4+ T cells at lower IFN-β concentrations, despite normal activation of STAT2. At the optimal time point for IFN-β-induced STAT activation, we found the highest activation of STAT5 in primary human CD4+ T cells and we also found activation of STAT5 and STAT3 in primary human B cells, suggesting that the Type I IFN receptor is functional in B cells.
It will be important to investigate whether low STAT1 activation is an intrinsic property of mature B cells and CD4+ T cells, or whether it is a result of other factors present in whole blood. Notably, our results comparing IFN-α1, IFN-α2b and IFN-γ with IFN-β showed very similar low activation of STAT1 in B cells (Fig. 9), suggesting that the same mechanism is involved. By influencing activation of the JAKs, SOCS1 can diminish the activation of STAT1 by both the type I and type II IFN receptors (39). SHP1 and TCP45 are both protein tyrosine phosphatases that decrease tyrosine phosphorylation of STAT1 (40–41). Although SOCS1, SHP1 and TCP45 are excellent candidates to explain the lack of PY-STAT1 induction in B cells by type I and II IFNs, we could not find evidence of their enhanced protein expression in B cells only. Based on our experiments we propose nevertheless that either physical properties of STAT1 protein itself are altered or that a selective negative regulator of STAT1 tyrosine phosphorylation is present in the majority of B cells. B cells are a heterogeneous population, and it will be important to characterize the minor fractions that do show activation of STAT1 by studying the expression of CD markers, chemokine receptors and adhesion molecules. Perhaps the STAT1-activating cells are immature, since type I IFNs inhibit B and T cell lymphopoiesis (43). In contrast, IFN-α induces STAT1-dependent proliferation in dormant haematopoietic stem cells (44), suggesting that the response to type I IFNs may change during the maturation of leukocyte subsets. Future experiments could address this issue by analyzing subsets separated on the basis of lineage and differentiation markers expressed on their surfaces.
Biological consequences of differential STAT activation
Type I IFNs cause apoptosis in many cancer cell lines and are used to treat several different types of tumors (22, 23). Similarly, type I IFNs induce apoptosis in primary monocytes (20, 21) but, in contrast, increase the survival and proliferation of primary B cells and T cells (14–19). In agreement, we found the highest activation of caspase 3, a hallmark of apoptosis induction, in monocytes, followed by CD8+ T cells and the least in CD4+ T cells and B cells after stimulation with IFN-β. Interestingly, during virus infection of mice, only CD8+ T cells with low STAT1 activation will proliferate in response to type I IFNs, due to lower STAT1 protein expression (45). The apoptosis-inducing capacity of type I IFNs is largely attributed to the activation of STAT1 (11,46), whereas the activation of STAT3 and STAT5 by type I IFNs is related to survival and proliferation (12,13,47). By performing double staining of IFN-β-stimulated cells with anti-PY-STAT1/PY-STAT3 or anti-PY-STAT1/PY-STAT5 antibodies, we were able to detect the activation of STAT1/STAT3 and STAT1/STAT5 together, in individual cells. Notably, the percentage of PY-STAT1+/PY-STAT3− monocytes found in response to IFN-β after 45 min correlated significantly with percentage of activated caspase 3-positive monocytes after 8 h (Fig. 6C). Enhanced STAT3 protein levels in monocytes have been demonstrated to suppress DNA-binding of STAT1 homodimers by sequestering STAT1 into STAT1/STAT3 heterodimers (48). Therefore, it is to be expected that the PY-STAT1+/PY-STAT3− monocytes would become apoptotic first, and that PY-STAT1+/PY-STAT3+ monocytes are protected from apoptosis induction only if PY-STAT3 levels are high enough to sequester activated STAT1. However, after longer stimulation with IFN-β (beyond 10 h), the percentage apoptotic cells is doubled or tripled compared to the percentage at 8 h, indicating that even PY-STAT1+/PY-STAT3+ monocytes eventually die. Very few B cells are PY-STAT1+/PY-STAT3− and, because STAT1 activation is so low in these cells, it is more likely that enhanced PY-STAT3 levels could suppress DNA-binding of STAT1 homodimers by sequestering STAT1 into STAT1/STAT3 heterodimers in PY-STAT1+/PY-STAT3+ B cells than in monocytes (48). It is interesting that CD4+ T cells that show the highest percentage of PY-STAT1+/PY-STAT3− cells in response to IFN-β after 45 min display the lowest apoptosis induction, along with B cells (Fig. 6A and 6B). Both leukocyte subsets have very low numbers of PY-STAT1+/PY-STAT5− cells and significant numbers of PY-STAT1+/PY-STAT5+ cells, suggesting that the activation of STAT5 in the majority of CD4+ T cells and B cells protects against apoptosis induction. Indeed, the anti-apoptotic and mitogenic properties of type I IFNs in mouse T cells are dependent on the activation of STATs 3 and 5 (47). Strangely, despite the fact that monocytes also generate very high numbers of PY-STAT1+/PY-STAT5+ cells in response to IFN-β, they are the most sensitive to apoptosis induction. This disparity between monocytes, B cells and CD4+ T cells might be explained by the fact that our anti-PY-STAT5 antibody recognizes both activated STAT5A and STAT5B, and human monocytic cells activate only STAT5A (49), in contrast to human T cells, which activate both STAT5A and STAT5B in response to type I IFNs (8).
In accordance with above-mentioned data, when monocytes and B cells were compared for pro-apoptotic mRNA induction by IFN-β, we only found enhancement of CDKN1A, BAK1, CASP3 and STK3 mRNA in monocytes (Table 1 and Fig. 7B). Evidence that the induction of these mRNAs depends upon phosphorylated STAT1 homodimers is as following. First, IFN-γ does not induce cyclin-dependent kinase inhibitor 1A (CDKN1A or p21) in STAT1-deficient U3A fibrosarcoma cells, but does enhance p21 expression in U3A cells in which STAT1 has been reintroduced (27), and enhancement of caspase 3 (CASP3) expression is dependent on PY-STAT1 formation (28). Secondly, because BCL-2 homologous antagonist/killer 1 (BAK1) expression is induced directly in HT-29 cells by IFN-γ (29), its induction probably depends on the formation of STAT1 homodimers. Finally, because both type I and type II IFNs enhance serine/threonine-protein kinase 3 (STK3) expression (www.interferome.org), it is likely that the induction of this mRNA occurs through activation of STAT1. CDKN1A, BAK1, CASP3 and STK3 are known to be involved at different stages of the intrinsic apoptotic pathway. For instance, increased p21 leads to cell cycle arrest in the G1 phase of fibrosarcoma and Burkitt’s lymphoma cells, and the induction of G1 arrest in Burkitt’s B cell lymphoma by type I IFNs is followed by induction of apoptosis (26). BAK1 is a member of the BCL-2 family of pro-apoptotic proteins which, upon activation by IFN-α, forms oligomers or heterodimers that interact with the mitochondria, leading to the release of cytochrome c and apoptosis induction (50, 51). Notably, it has been shown that apoptosis induction through activation of STAT1 is mediated by activation of the effector caspase 3, among others (46). Interestingly, STK3 is a direct substrate of caspase 3 and, following cleavage, translocates to the nucleus and induces chromatin condensation, followed by inter-nucleosomal DNA fragmentation (52, 53). However, increased levels of STK3 can also accelerate apoptosis induction through the activation of caspase 3 (52).
Induction by IFN-β of CDKN1A, BAK1, CASP3 and STK3, all involved in the intrinsic apoptotic pathway, seems to occur only in monocytes, whereas the induction of certain pro-apoptotic genes was not found in B cells exclusively. Nevertheless, in both monocytes and B cells, TRAIL mRNA was increased by 7–24 fold after IFN-β stimulation (Table 1). Although TRAIL expression induces apoptosis in tumor and virus-infected cells, it exhibits no apparent adverse affect on normal cells (30, 54). Moreover, TRAIL engagement on T cells can even lead to increased proliferation, but it is not known if this is also true for B cells (55). However, the expression of TRAIL on monocytic cells might still have inhibitory effects, because the rapid maturation of monocytes into short-lived dendritic cells by IFN-β is associated with TRAIL expression (21). TRAIL induction by IFN-β in fibrosarcoma cells is dependent on ISGF3 binding to the ISRE element in the promoter (56). The fact that our data suggest that TRAIL mRNA was increased in B cells by IFN-β, despite low activation of STAT1, could be explained by the binding of either STAT2dimer/IRF9 or STAT2/STAT6/IRF9 to the ISRE (4, 5), because primary human B cells can activate both STAT2 and STAT6 (Fig. 8B, and preliminary results). Alternatively, formation of STAT3, STAT5 or STAT6 homodimers (5) in response to IFN-β could be responsible for the observed increase of IRF-1 in primary human B cells (Table 1), that is, IRF1 dimers could subsequently bind to the ISRE in the TRAIL promoter (57). In addition to TRAIL induction, we found that, in both monocytes and B cells, the mRNA for the death receptor FAS was increased by 3–4 fold (Table 1). Upon FASL binding, the extrinsic apoptosis pathway could be triggered by recruitment of FAS-associated death domain protein (FADD), activation of caspase 8 and cleavage of the pro-apoptotic BCL-2 family member Bid (31,50). However, we did not observe any simultaneous increase of FASL mRNA expression in B cells or monocytes. Of note, FAS also has non-apoptotic functions (31), because individuals with homozygous caspase-8 reduction-of-function mutations display defects in FAS signaling and impaired proliferation of B, T and NK cells (58).
Very low percentages of PY-STAT1+/PY-STAT3− and PY-STAT1+/PY-STAT5− were observed in B cells, the only leukocyte subset in which the PY-STAT1−/PY-STAT3+ and PY-STAT1−/PY-STAT5+ combinations were induced upon IFN-β stimulation (Fig. 6B). These findings together are likely to be responsible for decreased induction of apoptosis and increased induction of the PY-STAT3- or PY-STAT5-dependent mRNAs that are responsible for increased survival and proliferation. Notably, the probable PY-STAT3-dependent B cell activating factor (BAFF) (36), which our preliminary data suggest to be increased 3.5-fold more in B cells compared to monocytes (Table 1), has been shown to overcome any negative effects from FAS-signaling and increase the survival of B cells (59). Another activated STAT3-dependent mRNA (35), pre-B cell colony-enhancing factor 1 (PBEF1) increased in B cells only (Table 1, Fig. 7B) and not in monocytes in response to IFN-β. Notably, PBEF inhibits the induction of apoptosis in neutrophils and epithelial cells by reducing the activity of caspases 3 and 8 (34). In addition, PBEF1 synergizes with IL-7 in pre-B cell colony formation (33). We did not observe increased BCL-2 or BCL-xL expression, which was previously suggested to be the anti-apoptotic mechanism of IFN-β in B cells (15). The promoters of both BCL-2 and BCL-xL are typical targets of activated STAT5 in response to several growth factors, and the STAT5-dependent induction of resistance to apoptosis functions through these proteins (13). Because we studied only early mRNA transcription in response to IFN-β, the induction of BCL-2 and BCL-xL in B cells might need more than 3 h of stimulation with IFN-β. We propose that the low activation of STAT1 and much higher activation of STAT3 and STAT5, and consequently the absence of induction of genes participating sequentially in the intrinsic apoptosis induction pathway (as observed in monocytes), is an important mechanism to enable primary human B cells to survive in response to IFN-β. More in-depth studies using Chip assays to determine the residence of specific STATs on specific promoters are necessary to understand in detail how STATs 1, 3, and 5 activated by IFN-α/β exert their pro-apoptotic and mitogenic effects in specific immune subsets. In addition, it will be vital to unravel the mechanism of low STAT1 activation in the great majority of primary human B cells.
Although to enable survival, few B cells activate STAT1 in response to type I and II IFNs, it is nevertheless important that the anti-viral effects of IFNs are preserved. mRNAs derived from virus-responsive genes such as MxA, OAS, PKR, ISG15, IFI44 and IFITM3 increased at least 2-fold in both B cells and monocytes in response to IFN-β (data not shown). It seems that only the promoter of MxA harbors a classical ISRE element (57), and increased MxA transcription in B cells despite low STAT1 activation by IFN-β could be the result of activation of similar transcription factor complexes as mentioned above for TRAIL. All the other typical virus-responsive genes are suggested to belong to a new subtype of ISRE termed ETS/IRF response elements (EIRE), which can bind either IRF dimers (similar as classic ISRE) or an ETS/IRF dimer (57). Because B cells express IRF4, IRF8, PU.1 and other ETS family members (57), an IRF/ETS dimer could possibly bind to these promoters in primary B cells after IFN-β stimulation. Therefore, the inability of most B cells to activate STAT1 does not lead to a deficient anti-viral response in B cells, but the mechanism has still to be elucidated. These results have important implications for understanding more fully the influence of IFN-α/β on leukocyte subsets during virus infection in humans, as well as the effects of treatment with IFN-α/β on these subsets in patients with multiple sclerosis, hepatitis and cancer.
Supplementary Material
Acknowledgments
We thank Dr. Ian M. Kerr for many helpful suggestions and Dr. Ganes C. Sen for critically reviewing this manuscript. In addition, we thank Mike Sramkoski from the Case Comprehensive Cancer Center Flow Cytometry Core of Case Western Reserve University and Cathy Shemo and Sage O’Bryant of the Flow Cytometry Core in the Lerner Research Institute of the Cleveland Clinic for advice and technical assistance. We thank Dr. Pieter Faber of the Genomics Core Facility at the Cleveland Clinic for micro-array analyses. Moreover, we thank Vai Pathak and Dr. Patrick Leahy from the Gene Expression and Genotyping Core Facility, Case Comprehensive Cancer Center at Case Western Reserve University for assistance with the real-time PCR experiments. Finally, we thank Drs. Thomas Hamilton and Ernest Borden for their kind gifts of human recombinant IFN-γ, IFN-α1 and IFN-α2b.
Abbreviations used in this paper
- ISGF3
IFN-stimulated gene factor 3
- ISG
IFN-stimulated gene
- TYK2
tyrosine kinase 2
- JAK1
Janus kinase 1
- GAS
gamma-activated sequence
- ISRE
IFN-stimulated response elements
- D-PBS
Dulbecco’s Phosphate-Buffered Saline
- SEM
standard error of mean
- PY-STAT
phospho-tyrosine-STAT
Footnotes
This work was supported by Pilot Grant PP1086 and Career Transition Fellowship Award TA3032A1/1 from the National Multiple Sclerosis Society, and by P01 CA062220 and P30 CA43703 from the NIH. This research was also supported by the Gene Expression and Genotyping Facility of the Case Comprehensive Cancer Center (P30 CA43703).
“This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the United States National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.”
References
- 1.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 2.Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 3.Stark GR, I, Kerr M, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 4.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides witha major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 6.Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, O’Shea JJ. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol. 1996;157:4781–4789. [PubMed] [Google Scholar]
- 7.Yu CR, Lin JX, Fink DW, Akira S, Bloom ET, Yamauchi A. Differential utilization of Janus kinase-signal transducer activator of transcription signaling pathways in the stimulation of human natural killer cells by IL-2, IL-12, and IFN-alpha. J Immunol. 1996;157:126–137. [PubMed] [Google Scholar]
- 8.Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I. Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–1991. [PubMed] [Google Scholar]
- 9.Gupta S, Jiang M, Pernis AB. IFN-alpha activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J Immunol. 1999;163:3834–3841. [PubMed] [Google Scholar]
- 10.Schlaak JF, Hilkens CM, Costa-Pereira AP, Strobl B, Aberger F, Frischauf AM, Kerr IM. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J Biol Chem. 2002;277:49428–49437. doi: 10.1074/jbc.M205571200. [DOI] [PubMed] [Google Scholar]
- 11.Kim HS, Lee MS. STAT1 as a key modulator of cell death. Cell Signal. 2007;19:454–465. doi: 10.1016/j.cellsig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 13.Debierre-Grockiego F. Anti-apoptotic role of STAT5 in haematopoietic cells and in the pathogenesis of malignancies. Apoptosis. 2004;9:717–728. doi: 10.1023/B:APPT.0000045785.65546.a2. [DOI] [PubMed] [Google Scholar]
- 14.Morikawa K, Kubagawa H, Suzuki T, Cooper MD. Recombinant interferon-alpha, -beta, and -gamma enhance the proliferative response of human B cells. J Immunol. 1987;139(7):61–766. [PubMed] [Google Scholar]
- 15.Su L, David M. Inhibition of B cell receptor-mediated apoptosis by IFN. J Immunol. 1999;162:6317–6321. [PubMed] [Google Scholar]
- 16.Hibbert L, Foster GR. Human type I interferons differ greatly in their effects on the proliferation of primary B cells. J Interferon Cytokine Res. 1999;19:309–318. doi: 10.1089/107999099314009. [DOI] [PubMed] [Google Scholar]
- 17.Tough DF, Borrow P, Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- 18.Marrack P, Kappler J, Mitchell T. Type I IFNs keep activated T cells alive. J Exp Med. 1999;189:521–529. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheel-Toellner D, Pilling D, Akbar AN, Hardie D, Lombardi G, Salmon M, Lord JM. Inhibition of T cell apoptosis by IFN-beta rapidly reverses nuclear translocation of protein kinase C-delta. Eur J Immunol. 1999;29:2603–2612. doi: 10.1002/(SICI)1521-4141(199908)29:08<2603::AID-IMMU2603>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 20.Moore RN, Larsen HS, Horohov DW, Rouse BT. Endogenous regulation of macrophage proliferative expansion by colony-stimulating factor-induced interferon. Science. 1984;233:178–181. doi: 10.1126/science.6606850. [DOI] [PubMed] [Google Scholar]
- 21.Santini SM, Lapenta C, Logozzi M, Parlato S, Spada M, Di Pucchio T, Belardelli F. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J Exp Med. 2000;191:1777–1788. doi: 10.1084/jem.191.10.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeffer LM, Dinarello CA, Herberman RB, Williams BR, Borden EC, Bordens R, Walter MR, Nagabhushan TL, Trotta PP, Pestka S. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 1998;58:2489–2499. [PubMed] [Google Scholar]
- 23.Clemens MJ. Interferons and apoptosis. J Interferon Cytokine Res. 2003;23:277–292. doi: 10.1089/107999003766628124. [DOI] [PubMed] [Google Scholar]
- 24.Chow S, Hedley DW, Grom P, Magari R, Jacobberger JW, Shankey TV. Whole blood fixation and permeabilization protocol with red blood cell lysis for flow cytometry of intracellular phosphorylated epitopes in leukocyte sub-populations. Cytometry A. 2005;67:4–17. doi: 10.1002/cyto.a.20167. [DOI] [PubMed] [Google Scholar]
- 25.Krutzik PO, Irish JM, Nolan GP, Perez OD. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol. 2004;10:206–221. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Subramaniam PS, Cruz PE, Hobeika AC, Johnson HM. Type I interferon induction of the Cdk-inhibitor p21WAF1 is accompanied by ordered G1 arrest, differentiation and apoptosis of the Daudi B-cell line. Oncogene. 1998;16:1885–1890. doi: 10.1038/sj.onc.1201712. [DOI] [PubMed] [Google Scholar]
- 27.Chin YE, Kitagawa M, Su WS, You Z-H, Yoshiki I, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 28.Huang YQ, Li JJ, Karpatkin S. Thrombin inhibits tumor cell growth in association with up-regulation of p21(waf/cip1) and caspases via a p53-independent, STAT-1-dependent pathway. J Biol Chem. 2000;275:6462–6468. doi: 10.1074/jbc.275.9.6462. [DOI] [PubMed] [Google Scholar]
- 29.Ossina NK, Cannas A, Powers VC, Fitzpatrick PA, Knight JD, Gilbert JR, Shekhtman EM, Tomei LD, Umansky SR, Kiefer MC. Interferon-gamma modulates a p53-independent apoptotic pathway and apoptosis-related gene expression. J Biol Chem. 1997;272:16351–16357. doi: 10.1074/jbc.272.26.16351. [DOI] [PubMed] [Google Scholar]
- 30.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 31.Peter ME, Budd RC, Desbarats J, Hedrick SM, Hueber A-O, Newell MK, Owen LB, Pope RM, Tschopp J, Wajant H, Wallach D, Wiltrout RH, Zörnig M, Lynch DH. The CD95 receptor: Apoptosis revisited. Cell. 2007;129:447–450. doi: 10.1016/j.cell.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 32.Rusterholz C, Henrioud PC, Nabholz M. Interleukin-2 (IL-2) regulates the accessibility of the IL-2-responsive enhancer in the IL-2 receptor alpha gene to transcription factors. Mol Cell Biol. 1999;19:2681–2689. doi: 10.1128/mcb.19.4.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowell MA, Richards PJ, Fielding CA, Ognjanovic S, Topley N, Williams AS, Bryant-Greenwood G, Jones SA. Regulation of pre-B cell colony-enhancing factor by STAT-3-dependent interleukin-6 trans-signaling: implications in the pathogenesis of rheumatoid arthritis. Arthritis Rheum. 2006;54:2084–2095. doi: 10.1002/art.21942. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Zhong R, Hao W, Wang H, Fan X, Zhang L, Mi Q. Interleukin-10 and interferon-gamma up-regulate the expression of B-cell activating factor in cultured human promyelocytic leukemia cells. Exp Mol Pathol. 2009;87:54–58. doi: 10.1016/j.yexmp.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Leung S, Kerr IM, Stark GR. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17:2048–2056. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bach EA, Szabo SJ, Dighe AS, Ashkenazi A, Aguet M, Murphy KM, Schreiber RD. Ligand-induced autoregulation of IFN-gamma receptor beta chain expression in T helper cell subsets. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- 39.Alexander WS. Suppressors of cytokine signaling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 40.David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krämer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Gührs KH, Stauber RH, Böhmer FD, Heinzel T. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13(7):95–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 43.Lin Q, Dong C, Cooper MD. Impairment of T and B cell development by treatment with a type I interferon. J Exp Med. 1998;187(7):9–87. doi: 10.1084/jem.187.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 45.Gil MP, Salomon R, Louten J, Biron CA. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood. 2006;107:987–993. doi: 10.1182/blood-2005-07-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sironi JJ, Ouchi T. STAT1-induced apoptosis is mediated by caspase 2, 3 and 7. J Biol Chem. 2004;279:4066–4074. doi: 10.1074/jbc.M307774200. [DOI] [PubMed] [Google Scholar]
- 47.Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David MJ. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol. 2005;174:609–613. doi: 10.4049/jimmunol.174.2.609. [DOI] [PubMed] [Google Scholar]
- 48.Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem. 2006;281:14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]
- 49.Meinke A, Barahmand-Pour F, Wöhrl S, Stoiber D, Decker T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol Cell Biol. 1996;16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 51.Panaretakis T, Pokrovskaja K, Shoshan M, Grandér D. Interferon -induced apoptosis in U266 cells is associated with activation of the proapoptotic Bcl-2 family members Bak and Bax. Oncogene. 2003;22:4543–4556. doi: 10.1038/sj.onc.1206503. [DOI] [PubMed] [Google Scholar]
- 52.Lee KK, Ohyama T, Yajima N, Tsubuki S, Yonehara S. MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J Biol Chem. 2001;276:19276–19285. doi: 10.1074/jbc.M005109200. [DOI] [PubMed] [Google Scholar]
- 53.Kakeya H, Onose R, Osada H. Caspase-mediated activation of a 36-kDa myelin basic protein kinase during anticancer drug-induced apoptosis. Cancer Res. 1998;58:4888–4894. [PubMed] [Google Scholar]
- 54.Griffith TS, Wiley SR, Kubin MZ, Sedger LM, Maliszewski CR, Fanger NA. Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J Exp Med. 1999;189:1343–1354. doi: 10.1084/jem.189.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou AH, Tsai HF, Lin LL, Hsieh SL, Hsu PI, Hsu PN. Enhanced proliferation and increased IFN-gamma production in T cells by signal transduced through TNF-related apoptosis-inducing ligand. J Immunol. 2001;167:1347–1352. doi: 10.4049/jimmunol.167.3.1347. [DOI] [PubMed] [Google Scholar]
- 56.Rani MR, Pandalai S, Shrock J, Almasan A, Ransohoff RM. Requirement of catalytically active Tyk2 and accessory signals for the induction of TRAIL mRNA by IFN-beta. J Interferon Cytokine Res. 2007;27:767–779. doi: 10.1089/jir.2007.0005. [DOI] [PubMed] [Google Scholar]
- 57.Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25(7):70–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- 58.Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, Skoda-Smith S, Atkinson T, Straus S, Lenardo MJ. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 59.Hancz A, Hérincs Z, Neer Z, Sármay G, Koncz G. Integration of signals mediated by B-cell receptor, B-cell activating factor of the tumor necrosis factor family (BAFF) and Fas (CD95) Immunol Lett. 2008;116:211–217. doi: 10.1016/j.imlet.2007.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.