Abstract
The Molecular INTeraction Database (MINT, http://mint.bio.uniroma2.it/mint/) is a public repository for protein–protein interactions (PPI) reported in peer-reviewed journals. The database grows steadily over the years and at September 2011 contains approximately 235 000 binary interactions captured from over 4750 publications. The web interface allows the users to search, visualize and download interactions data. MINT is one of the members of the International Molecular Exchange consortium (IMEx) and adopts the Molecular Interaction Ontology of the Proteomics Standard Initiative (PSI-MI) standards for curation and data exchange. MINT data are freely accessible and downloadable at http://mint.bio.uniroma2.it/mint/download.do. We report here the growth of the database, the major changes in curation policy and a new algorithm to assign a confidence to each interaction.
INTRODUCTION
Understanding the physical and functional interactions between molecules in the cell is one of the main objectives of modern biology. Over the past decades, several powerful techniques have been developed to reveal, from different angles, the dynamics and complexity of the physiological interaction web. The retrieval, organization and analysis of these interactions are fundamental to understand the cellular machinery.
In the protein interaction field, several databases have set out to capture this information, as reported in the scientific literature, and to organize it in a structured format in order to allow users to perform automatic analysis. However, no database has sufficient resources to capture and organize all the published information and users are left with the task of querying multiple databases, wanting to interrogate the largest possible dataset.
Integrating protein interaction data from different databases has been a challenge until 2004, when the HUPO Proteomics Standards Initiative (HUPO-PSI) released the Molecular Interaction Ontology of the Proteomics Standard Initiative (PSI-MI) XML format (current version PSI-MI2.5) (1), a community standard for the representation of molecular interaction data. To date, PSI-MI formats are widely accepted and implemented by over 30 databases and supported by software tools. The detailed description of an interaction can be captured in this format, as for example, the biological and the experimental role of the interacting proteins and the kinetic parameters.
The PSI-MI standard has allowed a better cooperation between public databases, culminating in the formation of the International Molecular Exchange (IMEx) consortium (http://imex.sourceforge.net/) (2) aiming at distributing the effort of collecting large amounts of interaction data and to avoid work duplication. Scientists can now download and merge the data from several databases using a single data format. MINT (3) is an active member of IMEx consortium together with IntAct (4), DIP (5), MatrixDB (6), MPIDB (7) and InnateDB (8).
DATA GROWTH AND STATISTICS
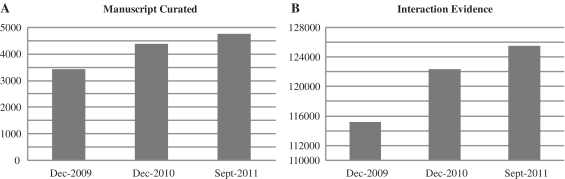
The MINT database has grown over the years as an important scientific resource. An average of 4000 queries per month are submitted to our server and thousands of PPI records are downloaded every year through our website. The current version of MINT, September 2011, contains records extracted from 4786 manually curated publications and 125 358 interaction evidences (IE) (235 635 non-redundant interaction pairs). Over the past 2 years the number of interaction evidence increased steadily and so did the number of curated publications (Figure 1).
Figure 1.
MINT growth. The bar diagrams illustrate the increase in number of MINT entries (A) and of curated manuscripts (B) since the latest update in 2009.
MINT does not specialize in selected model organisms and in the present version contains interactions between proteins from more than 30 different species such as Homo sapiens (28 283 IE), Mus musculus (4808 IE), Rattus norvegicus (2804 IE), Drosophila melanogaster (23 534 IE), Caenorhabditis elegans (7402 IE), Saccharomyces cerevisiae (48 979 IE), Escherichia coli (4188 IE) and Helicobacter pylori (1635 IE).
NEW STRUCTURAL DIGITAL ABSTRACT
Starting in 2006 MINT launched a collaboration with the FEBS Letters and FEBS Journal editorial boards aimed at developing an editorial procedure to integrate each manuscript reporting experimental evidence for protein interactions with a Structured Digital Abstract (SDA). This text appended to the traditional abstract summarizes the interaction information described in the article by using words predefined in controlled vocabularies. In addition the structured sentences are hyperlinked to relevant databases (9).
This pioneering initiative was originally conceived with the idea of involving authors in the correct annotation of their experimental evidence in database records by asking them to fill the Minimum Information required to report a Molecular Interaction Experiment (MIMIx) (10) spreadsheet. This task, however, did not prove practical because of the extra ‘burden’ that the editors felt should not be imposed on authors and because of the delay in the publication process caused by the correspondence with authors after manuscript acceptance. As a consequence, after the initial experimental phase, now authors are only asked to check and, if necessary, correct the entries made by the curators.
SDA were meant to facilitate automatic retrieval of protein interaction information by computers. At the same time we chose to maintain a human readable structure. More recently this characteristic was further enhanced by eliminating from the structured sentence the reference to the database identifiers while maintaining the hyperlinks to the database. The new structure digital abstract when compared to the initial version (Supplementary Figure S1) is more friendly to the human reader while maintaining the necessary rigor and controlled vocabulary for efficient automatic retrieval.
CURATION POLICY
The members of the IMEx consortium constantly review the annotation policy to meet the development of protein interaction technology and the evolution of the PSI-MI standards. The IMEx databases have developed and adopted a common curation manual (http://imex.sourceforge.net/doc/imex-curationManual.doc), specifying which information should be captured and how to represent it.
According to this standard all entries are annotated with richness of experimental details, such as, for example, the minimal region necessary for interaction, the mutations and the modifications that affect the interaction, the tags that are fused to the interaction partners in the experiment. The use of controlled vocabularies (CVs), mainly the PSI-MI (1) allows to capture most of the relevant experimental details and to standardize the interaction data. This standardization effort facilitates the exchange of completed records and the analysis by the users. The Controlled Vocabulary is regularly maintained and adapted via the introduction of new terms, the improvement of the description of existing terms and the upgrade of the terms hierarchy.
IMEx database curators carry out this maintenance either during annual meetings and Jamborees or by using the tracker (https://sourceforge.net/tracker/?group_id=65472&atid=612426). With the use of the tracker, it has been possible to introduce several new terms.
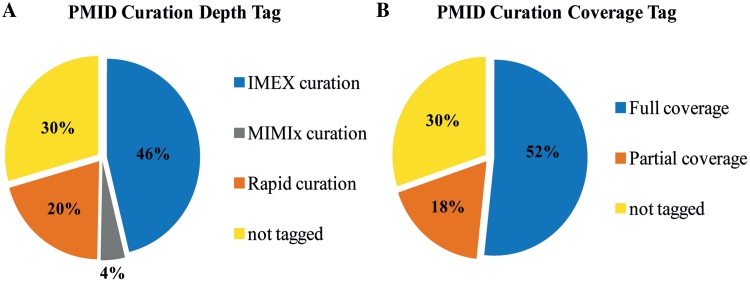
As an IMEx member, MINT has committed to regularly curate to this high standard all the articles in FEBS Letters, FEBS Journal, EMBO Journal and EMBO Reports, as they are published. Records curated to IMEx standard are easily recognizable because their publications have an IMEx ID assigned. It is important to note that MINT also contains records that are not curated to the IMEx standard that is either not all the interaction evidence or experimental details have been captured in the annotation process. These records mostly pertain to interaction relevant to topics of interest for the experimental group supporting MINT (domain peptide interactions, virus–host interaction, phosphatase interaction, etc). Most of these articles are annotated at a lower level of detail according to the MIMIx standard. For some project, records only contain information about the interaction partners and the method used to support the interaction. This shallow curation standard is labelled ‘rapid curation’. To avoid confusion, all the new entries are labelled with a tag describing the coverage (whether all the reported interactions are curated) and the depth (amount of experimental details captured) of curation. As the different tags describing the curation depth are IMEx, MIMIx or ‘rapid curation’ while those describing coverage are ‘full’ or ‘partial coverage’ (Figure 2).
Figure 2.
‘Curation-depth’ and ‘Curation-coverage’. The pie charts illustrate the fraction of MINT entries that are labelled with different tags according to curation depth (A) and curation coverage (B).
Users can choose the records that they want to utilize for their analysis by filtering according to these tags. Entries that have been curated before the advent of IMEx may still be unclassified while they are waiting to be reviewed and assigned to the correct class of curation standard
It is important to point out that the entries tagged as ‘rapid curation’ and/or ‘partial coverage’ are in principle as accurate as the IMEx entries and utilize the same controlled vocabularies recommend by the PSI-MI consortium.
NEW FEATURES
Proteomics Standards Initiative common query interface (PSICQUIC) (11) is a project aimed at standardizing the programmatic access to molecular interaction databases. MINT has implemented a PSICQUIC service. Moreover since a year any protein query on the mint search page, in addition to yielding all the interactions annotated in the MINT database also returns the results obtained by querying the PSICQUIC web services of the other IMEx databases (Supplementary Figure S2).
As discussed above, MINT contains entries curated to different annotation depth and coverage.
To make this point clear in the downloadable MITAB2.6 file we now include a ‘curation-depth’ column that can take the values ‘IMEx’, ‘MIMIX’ or ‘rapid curation’.
SCORING SYSTEM
MINT was one of the first PPI database to associate to each interaction a score estimating the reliability of the interaction, given the available experimental evidence (3,12).
The original MINT score is based on a heuristic integration of the available evidence into a ‘combined experimental evidence’ x which is then mapped in the 0–1 interval via the formula Score = 1−a−x.
x is computed by adding up all the evidence according to the formula
where i is an index iterating over all the experimental evidence, e is a coefficient that takes the value of 1 for evidence of direct interaction and 0.5 for evidence that only support and association, which may be indirect, between the two partners and d reflects the size of the experiment. Experiments are defined large scale if the article reporting them describes more than 50 interactions otherwise they are defined small scale. This coefficient is set to 1 for small scale and to 0.5 for large scale experiments. Finally n represents the number of manuscripts reporting evidence that support the interaction.
We recently decided to revise the scoring algorithm to correct some bias of the original algorithm and to include a weight that takes into account ‘community recognition/trust’.
In the new version of the score we introduced the concept of integrated supporting evidence y defined as the weighted sum of the j manuscripts supporting a given interaction.
The weight of each supporting manuscript SiRi is obtained by multiplying two coefficients each varying from 0 to 1 and reflecting the ‘validity’ of the experimental evidence (S) or estimating the recognition/trust of the scientific community (R) respectively.
To obtain S and R we first calculated s and r defined as
r = normalized citations. Where e, similarly to the same coefficient in the original score, has a different value according to the type of experiments supporting the interaction and emphasizes evidences of direct interaction (e = 1) with respect to experimental support that does not provide unequivocal evidence of direct interaction, i.e. co-ip, pull down, etc (e = 0.5) or co-localization (e = 0.1). Conversely, r is the ratio between the number of citations received by the manuscript according to Google Scholar, augmented by 20 (community trust) and the number of independent interactions reported in the manuscript. This latter normalization is made to take into account that manuscripts describing a large number of interactions have a higher number of citations, number that would be misleading to use to measure the trust for each of the high number of reported interactions.
S and R are obtained by mapping to the 0—1 interval the experimental support s and the normalized number of citations of the supporting manuscript, r. The mapping function is of type
a and b are empirically set to 1.2 . Similarly to the strategy utilized in the original MINT score the integrated supporting evidence y is mapped in the interval 0–1 according to the function
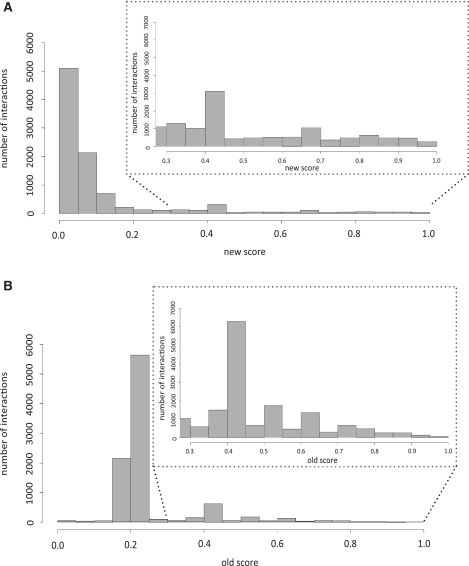
Since the normalization parameters have been set arbitrarily the absolute values of the score should not be interpreted as probabilities. However, we are planning to assemble a Golden Standard of trusted interaction to support a probabilistic approach. The density distribution of the two scoring systems is shown in Figure 3. Old and new scores are both based on the same experimental evidence and it is therefore not surprising that they are highly correlated (0.7 Pearson correlation). However, application of the two scoring systems to different interactions yields ranking lists that may substantially differ. By reviewing a number of interactions that were ranked differently by the two scores, biologists expert in the protein interaction domain, noticed that the new score tends to rank high interactions that are supported by a large number of independent evidence. In addition, as implicit in the formula, interactions reported by low throughput highly cited papers are promoted to higher levels of the ranking list. The MITAB file that can be downloaded from the website associates to each interaction both the old and new scores while the old scores are still displayed on the web site.
Figure 3.
Scoring system. The two bar diagrams illustrate the number of interactions as a function of the score according to the original algorithm (A) or to the one presented here (B).
PERSPECTIVES
The MINT curation team, in addition to the main task of looking after the four journals assigned by the IMEx consortium (FEBS Letters, FEBS Journal, EMBO Journal and EMBO Reports), will continue to cover the curation of papers reporting interactions mediated by modular domains and/or by protein phosphatases.
One limitation of the records currently annotated in MINT is that they do not capture the dynamic nature of an interaction or some other complexities such as interactions that only occur in specific context or are mediated by allosteric effects. MINT is committed to contribute to extend the PSI-MI 2.5 XML format in order to fully capture this information richness.
All MINT entries are annotated by expert curators. However, we are becoming increasingly more aware that it is unlikely that, given current funding levels, this strategy will succeed in capturing all the published PPI information. To overcome this limitation MINT has an interest in monitoring the performance of text mining methods in the automatic capture of PPI information. Over the past years MINT has participated in the BioCreAtIvE (Critical Assessment of Information Extraction systems in Biology) challenge (13) by providing manually curated datasets for the assessment exercise.
Results have demonstrated that the coverage and precision of the automatic annotation is steadily increasing (14). It is possible that in a near future MINT will contain entries corresponding to yet another curation level where automatically extracted PPI information is filtered and validated by expert curator before being stored in the database.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Funding for open access charge: Italian Association for Cancer Research (AIRC) project number 10360, Telethon GGP09243 and PSIMEx contract number 223411 within the FP7-HEALTH-2007B programme.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orchard S, Kerrien S, Jones P, Ceol A, Chatr-Aryamontri A, Salwinski L, Nerothin J, Hermjakob H. Submit your interaction data the IMEx way: a step by step guide to trouble-free deposition. Proteomics. 2007;7(Suppl 1):28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 3.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chautard E, Fatoux-Ardore M, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011;39:D235–D240. doi: 10.1093/nar/gkq830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goll J, Rajagopala SV, Shiau SC, Wu H, Lamb BT, Uetz P. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24:1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: the FEBS Letters experiment. FEBS Lett. 2008;582:1171–1177. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 10.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 11.Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatr-Aryamontri A, Ceol A, Licata L, Cesareni G. Protein interactions: integration leads to belief. Trends Biochem. Sci. 2008;33:241–242. doi: 10.1016/j.tibs.2008.04.002. author reply 242–243. [DOI] [PubMed] [Google Scholar]
- 13.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitner F, Chatr-aryamontri A, Mardis SA, Ceol A, Krallinger M, Licata L, Hirschman L, Cesareni G, Valencia A. The FEBS Letters/BioCreative II.5 experiment: making biological information accessible. Nat. Biotechnol. 28:897–899. doi: 10.1038/nbt0910-897. [DOI] [PubMed] [Google Scholar]