Abstract
ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/) is an integrated database that provides comprehensive information about integrative and conjugative elements (ICEs) found in bacteria. ICEs are conjugative self-transmissible elements that can integrate into and excise from a host chromosome. An ICE contains three typical modules, integration and excision, conjugation, and regulation modules, that collectively promote vertical inheritance and periodic lateral gene flow. Many ICEs carry likely virulence determinants, antibiotic-resistant factors and/or genes coding for other beneficial traits. ICEberg offers a unique, highly organized, readily explorable archive of both predicted and experimentally supported ICE-relevant data. It currently contains details of 428 ICEs found in representatives of 124 bacterial species, and a collection of >400 directly related references. A broad range of similarity search, sequence alignment, genome context browser, phylogenetic and other functional analysis tools are readily accessible via ICEberg. We propose that ICEberg will facilitate efficient, multi-disciplinary and innovative exploration of bacterial ICEs and be of particular interest to researchers in the broad fields of prokaryotic evolution, pathogenesis, biotechnology and metabolism. The ICEberg database will be maintained, updated and improved regularly to ensure its ongoing maximum utility to the research community.
INTRODUCTION
Horizontal gene transfer greatly facilitates bacterial evolution and fitness. Integrative and conjugative elements (ICEs), first proposed by Burrus et al. (1), are a diverse group of mobile elements found in both Gram-positive and Gram-negative bacteria. ICEs are defined as self-transmissible integrative elements that encode a full complement of conjugation machinery. Several well-characterized ICEs have also been shown to encode associated intricate regulatory systems that control excision of the ICE from the chromosome and, when applicable, subsequent onward conjugative transfer (2). These multi-talented entities can promote their own mobilization and thus contribute to horizontal transfer of virulence determinants, antibiotic-resistant genes and many other bacterial traits. Besides newly described ICEs, several entities discovered more than a decade ago that had previously been classified as plasmids or conjugative transposons have now been defined as ICEs (3–4). These include the ‘plasmid’ pSAM2, first reported in 1984, and the well-characterized Tn916 ‘conjugative transposon’, which was originally identified in 1995.
ICEs typically comprise three core genetic modules: (i) ICE integration and excision module; (ii) ICE conjugation module; and (iii) ICE regulation module. These functionally conserved modules are made up of a diverse range of genes that code for proteins working through disparate mechanisms. All intact ICEs contain a gene encoding an integrase (Int) that promotes site-specific integration and excision of the element, frequently into a unique site on the chromosome of the host organism (5). Members of SXT/R391 family, one of the best-studied ICE families, target the 5′-end of Vibrio spp. prfC gene, while ICEMlSym(R7A), ICEBs1, ICEHin1056, PAPI-1 and ICEclc(B13) all integrate into 3′-ends of distinct element-specific tRNA gene loci (2). However, some integrases mediate less specific integration site selection. Tn916, an 18 kb ICE first characterized in Enterococcus faecalis DS16, has a broad site preference, inserting preferentially into AT-rich sequences (3). ICE excision also requires the cognate integrase protein, thus reflecting the reversible site-specific recombination activity mediated by this class of enzyme. However, the preference between excision and integration has been shown for several ICEs to be regulated by some small, positively charged DNA-binding proteins known as ‘recombination directionality factors’ (RDFs) or excisionases (2). ICEs additionally contain genes that mediate the signature self-transmissible trait, including those that encode DNA processing, DNA replication, DNA secretion systems and/or ICE exclusion systems. Where studied, the mechanisms of DNA processing and replication appear similar to those involved in conjugative transfer of plasmids and include an origin of transfer (oriT), a relaxase, other conjugation transfer proteins and the ability to undergo rolling-circle replication, regardless of any observed autonomous extra-chromosomal replication (6). In Gram-negative bacteria, conjugation apparatuses have either been shown or are proposed to comprise type IV secretion systems (T4SSs) (7) as all such ICEs contain at least one gene encoding a type IVA secretion system homologue (2). Several ICEs also carry maintenance modules such as toxin–antitoxin systems (8) and other partition systems that ensure successful long-term vertical inheritance of these elements. Besides the functionally conserved shared ICE modules that comprise the ‘ICE backbone’, almost all identified ICEs also carry significant repertoires of accessory genes some of which have been shown or predicted to contribute towards resistance, virulence, metabolic adaptation and/or biotechnological potential (3,9). For example, the Bacteroides CTnDOT and CTnERL elements promote dissemination of antibiotic-resistant genes (10), whereas the Pseudomonas aeruginosa PAPI-1 ICE contributes to virulence in murine models of acute pneumonia and bacteremia (11). ICEs typically exhibit a number of features that are of interest to researchers in the fields of prokaryotic evolution, pathogenesis, biotechnology and metabolism. These include high levels of functional diversity, foreign and frequently patchwork origins and considerable scope for novel, functional genetics-associated discovery given the sparse availability of experimental data on the majority of these entities.
ICEs are being identified in increasing numbers as genome databases expand exponentially (4,9,12,13). Through the exploitation of hidden Markov model (HMM)-derived protein profiles of key plasmid-encoded conjugation-associated proteins, Guglielmini et al. (14) have very recently shown that ICE-like elements are abundant within sequenced bacterial chromosomes. However, given their focus on conjugative systems, these authors did not seek information on co-located integrase genes or attempt to identify ICE boundaries. Importantly, the large amount of data derived from this study was not offered as a readily accessible database. Indeed, at present only a few mobile genetic element-focused web-based recourses such as ISFinder (15), PAIDB (Pathogenicity island database) (16) and ACLAME (A Classification of Genetic Mobile Elements) (17) are available. Importantly, no well-organized and comprehensive ICE-specific dataset has been reported. We aim to progressively collate all available experimental and bioinformatics analyses data and literature about known and putative ICEs in bacteria as a PostgreSQL-based database called ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/). As its name implies, we expect that ICEberg will continue to grow from its currently visible tiny 'tip' representing presently known ICEs to a very substantial database as more and more of these entities are revealed. We envisage an ICE-specific resource to facilitate efficient investigation of large numbers of these elements, recognition of less than obvious ICE-associated patterns of sequence-, gene- and/or functional conservation and an improved understanding of the biological significance of these entities in diverse host organisms. We expect that ICEberg will prove to be of major interest to a broad community of researchers.
MATERIALS AND METHODS
As of 13 August 2011, ICEberg version 1.0 contains details of 428 ICEs found in representatives of 333 bacterial strains. These data were derived from reports of experimentally validated ICEs, published information on elements related to archetypal ICEs, computational analyses of bacterial genome sequences and GenBank entries. Over 410 references have been collected from NCBI PubMed using the search terms ‘integrative conjugative elements’, ‘integrating conjugative elements’, ‘conjugative transposon’, the names of specific ICEs, and from reference citations relating to ICEs within these sources. All references were manually screened for details of ICEs, thus identifying as of now published literature on 186 ICEs with varying levels of associated experimental evidence and papers for further 138 putative ICEs identified by bioinformatics approaches alone. We further predicted 67 new ICEs by using sequence similarity searches and manual validation for significant hits. All new ICEs identified have been named according to the method suggested by Burrus et al. (18). In addition, the database was supplemented with 37 ICEs, including unreported examples, identified by searching GenBank using terms described above and further appropriate search queries identified through manual review of the ICEberg-collated references.
Currently, 80% (344/428) of the ICEs archived within ICEberg have been assigned into 28 families, five of which have been previously described. These comprise the SXT/R391 (18), Tn916 (19), Tn4371 (13), CTnDOT/ERL (10) and ICE6013 (20) families. The remaining 23 families have been identified through the embedded WebACT-facilitated analyses and are defined based on integrase homology and synteny of other ICE backbone features. The classification of 84 ICEs remains pending, awaiting further analyses as some of them possess own integrase or unique structure features, or as some lack enough sequence information that covers the features for classification.
All annotated ICE-encoded proteins were extracted and analysed by Blastp against a number of databases, including TADB (Type 2 toxin-antitoxin loci database) (21), VFDB (Virulence factors database) (22) and ARDB (Antibiotic-resistant genes database) (23), to further enhance available ICE-relevant information.
ICEberg employs the relational database management system PostgreSQL as back-end. A customized schema was designed to organize all uploaded information including experimental and in silico analyses data and related references. ICEberg runs on a Linux platform with the Apache web server. Web interfaces were developed using HTML, CSS and JavaScript. The majority of data pipelines were developed with PHP and Perl. In addition, the following freely available components were employed: (i) Gbrowse2 genome viewer (24) and WebACT synteny browser (25); (ii) CGview circular genome visualization tool (26); (iii) MUSCLE (27) and Jalview (28) multiple sequence alignment and visualization tools, respectively. The ICEberg database is now run on a high-performance four-slot four-way server (Inspur NF8560), which had been equipped with four six-core XEON E7-4807 1.86 GHz processors and 64 GB Memory.
RESULTS AND DISCUSSION
ICEberg provides a flexible and biologist-friendly web interface. The ICEberg homepage contains the following interfaces: ‘Home’, ‘Browse’ (browse by ICE, organism or ICE family), ‘Search’ (search by species, ICE family or ICE name), ‘Tools’ [WebACT comparison tool and nucleotide/protein sequence alignment against ICEberg using Blast and HMMER3 (29)], ‘Download’ (nucleotide/protein sequences), ‘References’ (literatures relating to ICEs), ‘Introduction’ (description of ICEs and ICEberg), ‘Submission’ (Report new ICEs to ICEberg), ‘Links’ and ‘Contact’. The multiple sequence alignment tool MUSCLE and Jalview are also readily accessible to allow for user-directed analyses focused on diverse ICE-borne genes to facilitate individualized directions of research.
ICEberg browse module
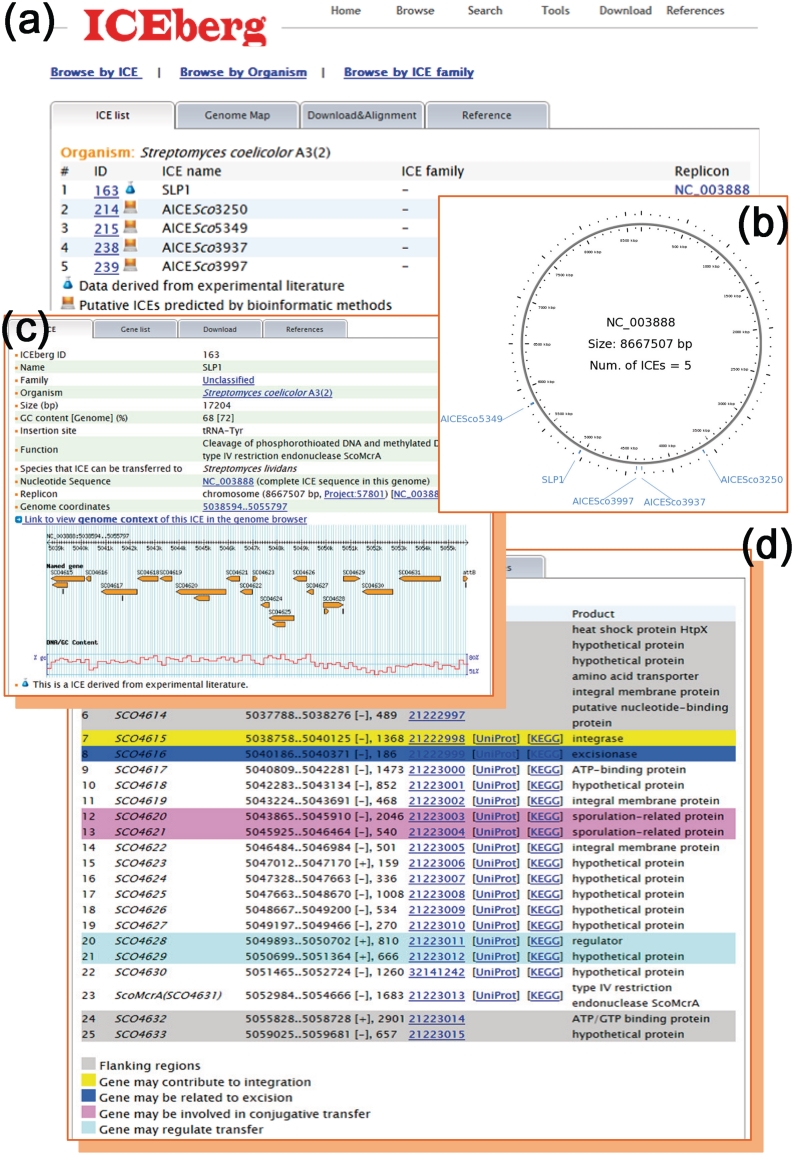
ICEberg browse module contains detailed information on all archived ICEs and the genes carried by each entity, including unique identifiers, species details and hyperlink paths to other public databases, like NCBI, UniprotKB and KEGG. The ‘Browse by ICE’ page offers accesses to all ICEs, while the ‘Browse by organism’ page provides a hyperlinked organized catalogue of bacterial strains in which ICEs have been identified. As with sequenced genomes, ICEberg allows users to view whole genome maps by using Gbrowse2 with the locations and sizes of identified ICEs flagged and hyperlinked to allow for ICE-centered zoom-in zoom-out genome-scale views. In addition, users can access individual pages dedicated to each ICE as required. The corresponding information for SLP1, an ICE in Streptomyces coelicolor A3(2), is presented as an example (Figure 1). Streptomyces coelicolor A3(2) is a soil-dwelling Gram-positive model organism used widely in studies on the production of pharmaceutically useful compounds including antibiotics, anti-tumour agents and immunosupressants. Tabulated (Figure 1a) and graphically displayed (Figure 1b) outputs of one experimentally verified (3) and four predicted ICEs (4) in the S. coelicolor A3(2) linear chromosome are as shown. The details and genomic context of a selected ICE are also highlighted (Figure 1c). SLP1 is 17.2 kb in length and exhibits a GC content of 68%, lower than the 72% genome-averaged value of its host strain. This ICE was originally found to be integrated into a tRNA-Tyr gene within the S. coelicolor A3(2) chromosome but was since shown to be transferable by conjugation to Streptomyces lividans strains. Of particular note, the three distinct modules of ICEs that mediate (i) integration and excision, (ii) conjugation and (iii) regulation of these foreign elements have been sought, examined and highlighted in the database (Figure 1d). The SLP1 genes, SCO4615 and SCO4616, encode an integrase and an excisionase, respectively. SCO4620 (traB1) and SCO4621 (traA1) encode sporulation-related proteins that are essential for intermycelial transfer and pock formation. In addition, SCO4628 and SCO4629 comprise the imp operon that regulates transfer functions and controls extra-chromosomal maintenance of SPL1 (30). Remarkably, we have recently found that the SLP1-encoded type IV restriction endonuclease ScoA3McrA (SCO4631) is able to cleave phosphorothioated and Dcm-methylated DNA and thus inferred that this is the likely molecular mechanism of the previously observed lethal zygosis resulting following mating of certain Streptomyces strains (31).
Figure 1.
An overview of ICEberg datasets and outputs using the SLP1 ICE element in Streptomyces coelicolor A3(2) as an example. (a) List of five identified ICEs in S. coelicolor A3(2), one of which has been experimentally verified (30), while the remaining four have been presently and/or previously predicted by in silico approaches (4). (b) A circular representation of the linear chromosome of S. coelicolor A3(2) generated by ICEberg-integrated CGview showing locations and sizes of ICEs within this replicon. (c) An overview of the features of SLP1: size, GC content, insertion site, known or predicted function(s), additional compatible hosts and replicon coordinates. Hyperlinks to NCBI are provided as appropriate, as is a link that allows for visualization of the gene content of the element by Gbrowse. (d) A complete gene list of SLP1 with links to NCBI, UniprotKB and KEGG. The three distinct core ICE modules that mediate integration and excision, conjugation and regulation have been sought, examined and highlighted using different colours. SCO4631 has recently been reported by us to encode a type IV restriction endonuclease, ScoA3McrA, that can cleave phosphorothioated and Dcm-methylated DNA (31).
Using the ‘Browse by ICE family’ link, users can retrieve 28 classified families that encompass 344 ICE entries that have to date been mapped to a specific family. The five previously identified families [SXT/R391 (18), Tn916 (19), Tn4371 (13), CTnDOT/ERL (10) and ICE6013 (20)] encompass nearly half the presently defined elements. Although ICEs vary in sequence and genetic organization, many share related integrase, maintenance and/or transfer genes. Hence, we have chosen to classify previously unassigned ICEs into families based firstly on integrase similarity and secondarily core structure synteny. Using this method, 23 new ICE families have been defined. A set of 230 integrases encoded by 225 ICEs were analysed for similarity and phylogeny allowing for the identification of clustered integrases (Supplementary Figure S1). In addition, broader DNA sequence comparisons and transfer gene alignments were performed to further support the classification established. Applying the same method to the previously described ICEclc family (Supplementary Figure S2), we used the well-documented ICEclc(B13) as reference (32) and confirmed that the integrase proteins encoded by all members of this family showed >60% amino acid identity to IntB13, the Int of ICEclc(B13) (Supplementary Figure S2c). Selected ICE nucleotide sequences belonging to this family were then compared to illustrate both regions of shared and variable genetic organization (Supplementary Figure S2b). We propose that a systematic and exhaustive analysis of this nature, perhaps complemented by analyses focused on other ICE-borne conserved genes or even full sets of conserved genes, and the resultant refined classification outcomes will expand our understanding of origins and functional properties of ICEs.
ICEberg search options and tools
ICEberg provides text, Blast and HMMER3 searches with varied options. Through the ‘Search’ page, users can retrieve a specific object(s) in the ICEberg by the following categories: species, ICE family or ICE name. The ‘Tools’ page allows users to search a query sequence against ICEberg to obtain and visualize potential homologous matches using WU-BLAST 2.0 (Gish,W., personal communication) and HMMER3 (29). Interestingly, the embedded WebACT provides sequence comparisons between the 231 ICEs with the entire nucleotide sequences and/or that of the user-supplied nucleotide sequence. It allows the on-line visualization of comparisons between up to 10 ICE sequences. Typically, Blastn alignment employed by WebACT is computed ‘on the fly’ in a matter of tens of seconds.
ICEberg reference module
The ICEberg reference section offers publication details of papers relating to ICEs that have been identified as described and further pertinent references relating to entries within ICEberg. Direct links to the matching PubMed entries are also provided. At present, ICEberg contains records of >400 directly relevant scientific publications. This reference collection will be updated monthly with new entries being subject to manual curation and organization in a timely manner. These references have been sorted by the following headings: experimental studies, in silico analyses, genome sequencing and reviews. ICEberg links the examined literature to relevant ICEs, ICE families and species pages as indicated by corresponding thumbnail icons. The reference collection is also searchable by ICE family, author, title, journal, year and PubMed ID, and matching abstracts can be subjected to standard word searches. This provides an easily accessible literature resource that has been subjected to both text mining, manual curation and subset categorization.
Future directions
New records of publications will be mined shortly for both experimentally verified and predicted ICEs. As current bioinformatics-based prediction and characterization of ICEs is very tedious (2), we will endeavor to establish more efficient in silico strategies to discover, characterize and define the diversity of these abundant and enigmatic, selfish ‘genetic species’. Towards this end, we will aim to tap into the complementary HMM protein profile-based ICE discovery approach recently exploited by Guglielmini et al. to more comprehensively identify co-localized relaxase, type IV coupling protein, VirB4-like ATPase, T4SS mating-pair formation and/or integrase genes. Furthermore as experimental data on the effects of ICEs on host- and self-gene expression and host fitness emerge, we will aim to systematically integrate these into ICEberg. It is likely that such data will expression patterns under diverse environmental conditions. In particular, we plan to capture data on the participation of ICE-encoded proteins and/or small RNA on host metabolic network and the specific interactions of these molecules with host DNA, RNA and proteins.
CONCLUSION
We envisage an evolving resource that captures a growing variety of ICE-related data extracted and curated from experimental literature, submitted directly by users and derived by increasingly sophisticated bioinformatics analyses of the expanding bacterial genome and metagenome database. Comprehensive utilities of this database such as similarity search, sequence alignment, genome context browser and phylogenetic tools are accessible for users to aid their research. Ultimately, we propose an ICE-specific resource to facilitate efficient investigation of large numbers of these elements, recognition of less than obvious ICE-associated patterns of sequence-, gene- and/or functional conservation, and an improved understanding of the biological significance of these potentially pleuripotent, selfish entities in diverse host organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
National Natural Science Foundation of China; the 973 program (2009CB118901, 2012CB721002) and the 863 program (2011BAD23B05-3), Ministry of Science and Technology, China; Program for New Century Excellent Talents in University, Ministry of Education, China (NCET-10-0572); Chen Xing Young Scholars Programme, Shanghai Jiaotong University; Shanghai Municipality; Action Medical Research grant (SP4255 to K.R.); Innovation Fellowship (to E.M.H, in part); East Midlands Development Agency grant (to K.R.). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Prof. Chun-Ting Zhang at Tianjin University for helpful discussions.
REFERENCES
- 1.Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 2.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 3.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 4.te Poele EM, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek. 2008;94:127–143. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd EF, Almagro-Moreno S, Parent MA. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 2009;17:47–53. doi: 10.1016/j.tim.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Lee CA, Babic A, Grossman AD. Autonomous plasmid-like replication of a conjugative transposon. Mol. Microbiol. 2010;75:268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flannery EL, Antczak SM, Mobley HL. Self-transmissibility of the integrative and conjugative element ICEPm1 between clinical isolates requires a functional integrase, relaxase, and Type IV secretion system. J. Bacteriol. 2011;193:4104–4112. doi: 10.1128/JB.05119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittle G, Shoemaker NB, Salyers AA. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell Mol. Life Sci. 2002;59:2044–2054. doi: 10.1007/s000180200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison EM, Carter ME, Luck S, Ou HY, He X, Deng Z, O'Callaghan C, Kadioglu A, Rajakumar K. Pathogenicity islands PAPI-1 and PAPI-2 contribute individually and synergistically to the virulence of Pseudomonas aeruginosa strain PA14. Infect. Immun. 2010;78:1437–1446. doi: 10.1128/IAI.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrus V, Pavlovic G, Decaris B, Guédon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
- 13.Ryan MP, Pembroke JT, Adley CC. Novel Tn4371-ICE like element in Ralstonia pickettii and genome mining for comparative elements. BMC Microbiol. 2009;9:242. doi: 10.1186/1471-2180-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon SH, Park YK, Lee S, Choi D, Oh TK, Hur CG, Kim JF. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic Acids Res. 2007;35:D395–D400. doi: 10.1093/nar/gkl790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leplae R, Lima-Mendez G, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 2010;38:D57–D61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burrus V, Marrero J, Waldor MK. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid. 2006;55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Smyth DS, Robinson DA. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J. Bacteriol. 2009;191:5964–5975. doi: 10.1128/JB.00352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011;39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Chen L, Sun L, Yu J, Jin Q. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008;36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Pop M. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. WebACT–an online companion for the Artemis Comparison Tool. Bioinformatics. 2005;21:3665–3666. doi: 10.1093/bioinformatics/bti601. [DOI] [PubMed] [Google Scholar]
- 26.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagege JM, Brasch MA, Cohen SN. Regulation of transfer functions by the imp locus of the Streptomyces coelicolor plasmidogenic element SLP1. J. Bacteriol. 1999;181:5976–5983. doi: 10.1128/jb.181.19.5976-5983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Ou HY, Wang T, Li L, Tan H, Zhou X, Rajakumar K, Deng Z, He X. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravatn R, Studer S, Zehnder AJ, van der Meer JR. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 1998;180:5505–5514. doi: 10.1128/jb.180.21.5505-5514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]