Abstract
The resolution of genome-wide association studies (GWAS) is limited by the linkage disequilibrium (LD) structure of the population being studied. Selecting the most likely causal variants within an LD block is relatively straightforward within coding sequence, but is more difficult when all variants are intergenic. Predicting functional non-coding sequence has been recently facilitated by the availability of conservation and epigenomic information. We present HaploReg, a tool for exploring annotations of the non-coding genome among the results of published GWAS or novel sets of variants. Using LD information from the 1000 Genomes Project, linked SNPs and small indels can be visualized along with their predicted chromatin state in nine cell types, conservation across mammals and their effect on regulatory motifs. Sets of SNPs, such as those resulting from GWAS, are analyzed for an enrichment of cell type-specific enhancers. HaploReg will be useful to researchers developing mechanistic hypotheses of the impact of non-coding variants on clinical phenotypes and normal variation. The HaploReg database is available at http://compbio.mit.edu/HaploReg.
INTRODUCTION
Genome-wide association studies (GWAS) are providing a flood of data associating genetic variants with common phenotypes (1). A confounding factor in such studies is linkage disequilibrium (LD), which allows many variants at the same locus to be associated with a phenotype even if only one of them is causal. Within genes, prioritizing the likely causal variant is relatively straightforward; variants are easily annotated as synonymous, missense or nonsense, changing the consensus sequence at splice sites, or residing in introns or UTRs. Often, however, GWAS associations lie far from known genes or transcribed regions, presumably in distal tissue-specific enhancers. One of the most striking examples of such a finding is the gene desert at 8q24, within which are regions specifically and independently linked to prostate, breast, ovarian, colorectal and bladder cancer. These variants have been shown to correspond to cell-type-specific distal enhancers for the MYC oncogene (2,3). Recent systematic comparisons of expression quantitative trait loci (eQTL) and GWAS suggest that the association of intergenic variants with complex phenotyes is a result of alteration of gene expression regulatory elements (4,5).
Ernst and colleagues (6) recently developed a map of chromatin states, including enhancers, promoters, insulators and heterochromatin, in nine human cell lines based on a variety of histone modifications. Using this map, it was demonstrated that these states can be used to prioritize SNPs within LD blocks associated with disease, and in some cases reveal biologically plausible enrichments for cell type-specific enhancers. Here we present a tool, HaploReg, to systematically mine these chromatin state data, along with conservation data and regulatory motif alterations.
A wide range of resources exists to make predictions about the functional consequences of variants, as well as navigating groups of linked variants using LD information. Polyphen (7), SIFT (8) and SNPS3D (9) all make predictions of the impact of missense SNPs. Algorithms such as is-rSNP (10) and RAVEN (11) use regulatory motif changes to predict SNPs that may influence transcriptional regulation. SNPinfo (12) combines missense predictions with TRANSFAC PWM disruption predictions and conservation information across 17 vertebrates for HapMap Phase III SNPs. SNAP (13) provides LD calculations using 1000 Genomes Project pilot data with information about neighboring genes and array membership for proxy/tag SNP selection, but does not currently include indels. HaploReg improves on SNAP by providing LD calculation of 1000 Genomes Project pilot indels associated with query SNPs. In addition, the features of SNPinfo are improved upon by incorporating evolutionary constraint based on two alogrithms (involving the sequences of at least 29 mammals) and considering a much larger library of PWMs.
The UCSC Genome Browser (14) and ENSEMBL Genome Browser (15,16) both allow genomic regions to be annotated with the results of cutting-edge genomic data, including chromatin state segmentations, ENCODE data, 1000 Genomes variants, evolutionary constraint, LD calculations and NHGRI catalog variants. However, the output of these browsers can be overwhelming, especially when one is interested only in a limited subset of loci (such as the variants linked to a GWAS hit.) To this end, HaploReg combines the focus on haplotype blocks provided by tools such as SNAP and SNPinfo with the breadth of genomic annotation provided by the full-featured genome browsers.
METHODS
HaploReg consists of a PHP interface to a MySQL database. The initial database table was populated using genomic coordinates and sequences for 16 151 841 biallelic SNPs and small indels from the pilot release of the 1000 Genomes Project (17). In some cases, such as novel indels, the variant call format (VCF) file from the pilot release did not have a RefSNP identifier (rsid); for the purpose of creating a unique identifier for this database, these variants were assigned the label of ‘chromosome:position’ in hg18 coordinates. To provide backward compatibility with obsolete rsids, dbSNP release 132 was checked for variants at the same position as 1000 Genomes pilot variants with multiple rsids (18). In addition, annotations of functional consequences were extracted from dbSNP.
A variety of functional annotations were then intersected with the set of variants using the BEDTools package (19), including the chromatin state segmentation of Ernst et al. (6), and conserved regions by GERP (20) and SiPhy (21,22). To obtain gene annotations, RefSeq genes (23) were downloaded from the UCSC Genome Browser and GENCODE version 7 (24) was downloaded from the project website. BEDTools was then used to calculate the proximity of each variant to a gene by either annotation, as well as the orientation (3′ or 5′) relative to the nearest end of the gene, based on the strand of the gene.
In order to annotate variants by their effect on regulatory motifs, a library of position weight matrices (PWMs) was constructed from literature sources and was scored on genomic sequences as described previously (6). Briefly, a set of PWMs was collected from TRANSFAC (25), JASPAR (26), and protein-binding microarray (PBM) experiments (27–29). The reference and alternate alleles for each of the 1000 Genomes pilot SNPs and indels were concatenated with 29 bp of genomic context on each side, using the hg18 sequence obtained from the UCSC Genome Browser (30). PWMs were then scored for instances that passed either of two thresholds, a stringent threshold of P < 4−8 and a less-stringent threshold of P < 4−7 (31). Only instances where a motif in the sequence (i) passed the stringent threshold of a PWM in either the reference or the alternate genomic sequence, and (ii) overlapped the variable nucleotide(s) (thus changing the PWM score) were considered. Then, the change in log-odds (LOD) score was calculated. In cases where the weaker match was did not pass the less-stringent threshold, an approximate minimum change of LOD score was reported, corresponding to the difference between the score of the stronger match to the score required to pass the less-stringent threshold. In cases where both allelic variants surpassed the less-stringent threshold, the exact difference in score was reported.
GWAS results were obtained from the table curated by NHGRI (32) (accessed June 29, 2011.) In cases where multiple studies were annotated as pertaining to the same phenotype, unique independent SNPs were consolidated into a single list.
LD was calculated using the phased genotype information accompanying the 1000 Genomes Project pilot release (17). VCFTools (33) was used to perform the calculation, using an LD threshold of r2 = 0.80, and a maximum distance between variants of 200 kb. Results from VCFTools were then consolidated such that for every variant in our database, a list of linked variants is accessible for each of the three populations, along with an r2 value.
To perform enhancer enrichment analysis on sets of variants, tables of common array designs were obtained from the UCSC Table Browser (34) and lists were constructed of 1000 Genomes SNPs segregating in each of the three pilot populations, as well as all SNPs in the database. Then, a background frequency of coverage was calculated for variants annotated as overlapping a strong enhancer state in each cell type. When a user submits a query list of variants, the coverage of strong enhancers in each cell type is calculated. If the coverage exceeds that of the background set selected by the user, a binomial test is performed, and enrichment is reported if it passes an uncorrected significance threshold of 0.05.
USAGE
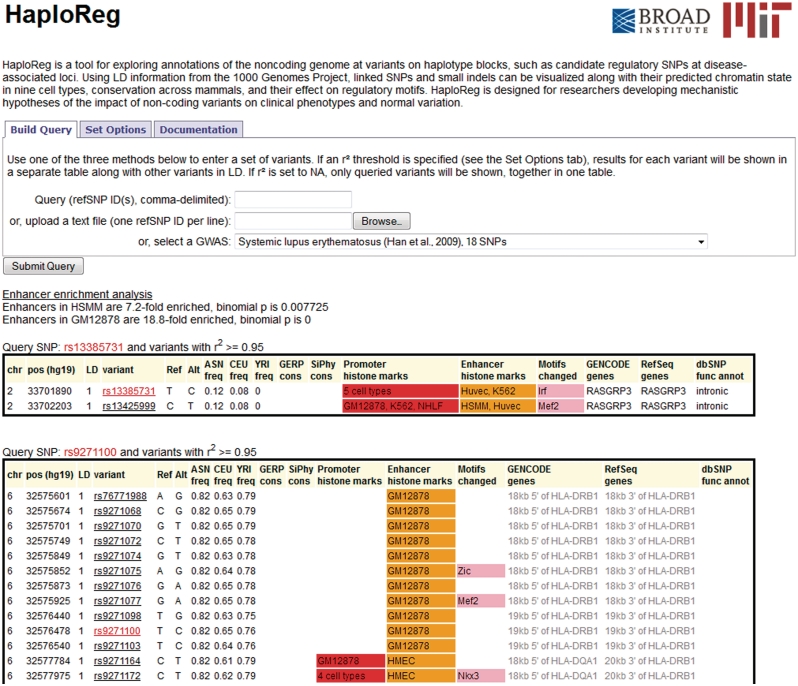
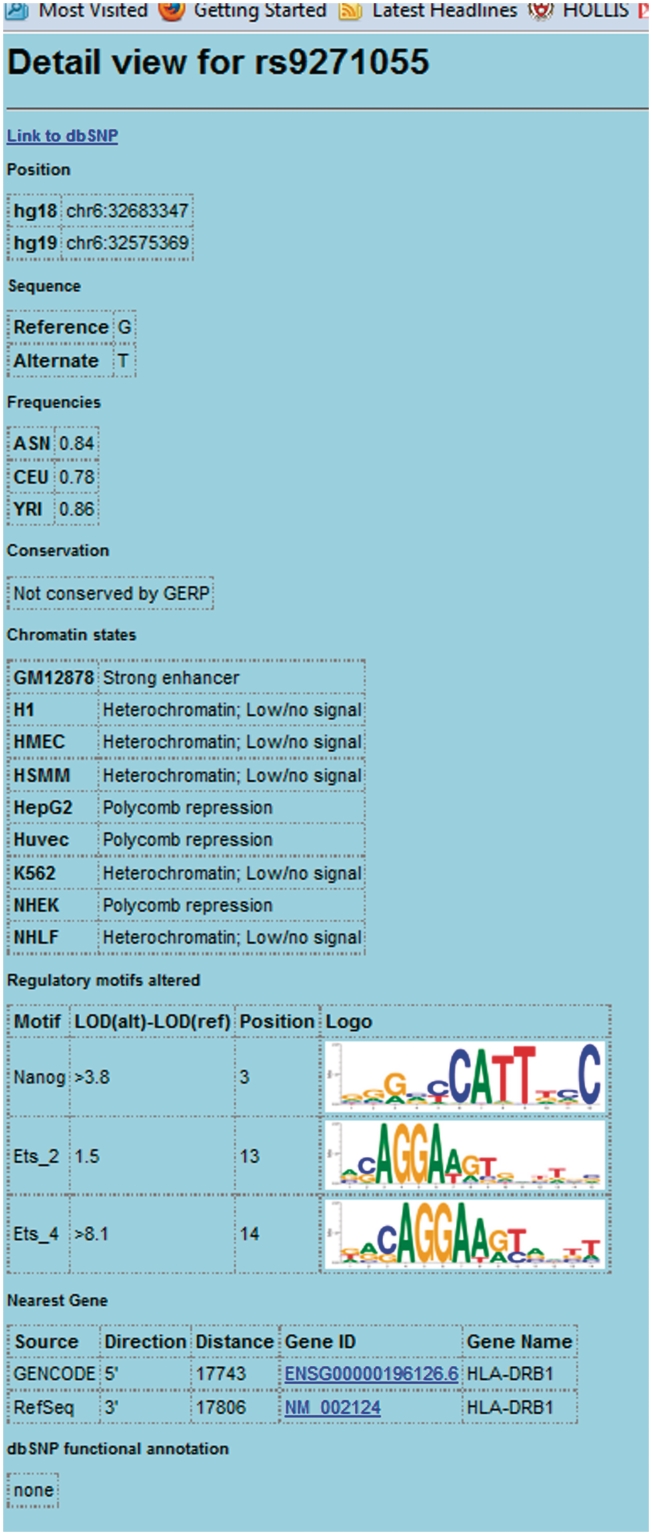
A user may submit queries in two formats: a comma-delimited list of rsids, or a one of the GWAS or traits from the NHGRI catalog. To illustrate (Figure 1), we select the lupus study by Han et al. (35). Since the study was conducted in Han Chinese, we select ASN (CHB + JPT) as the population for LD calculation, and we select all SNPs in the ASN population as the background for enhancer enrichment analysis. As was reported by Ernst et al. (6), there is a strong enrichment for GM12878 (lymphoblastoid) enhancers. To demonstrate LD blocks, we select an LD threshold of r2 = 0.95. In the LD block with lead SNP rs9271100, there is a SNP rs9271055 which affects an Ets-family binding site. Clicking on rs9271055 leads to a detail view (Figure 2) in which the complete chromatin state data are available. The positions in two literature motifs for Ets-family proteins can be seen, where the alternate T allele strengthens the predicted affinity relative to the reference G allele. In addition, links to NCBI RefSeq and ENSEMBL pages detailing the neighboring HLA-DRB1 gene are provided.
Figure 1.
HaploReg view of the SNPs from the lupus GWAS by Han et al.
Figure 2.
HaploReg detail view for the SNP rs9271055.
FUNDING
National Institutes of Health (R01-HG004037, RC1-HG005334); National Science Foundation (0644282). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Pouya Kheradpour for valuable assistance with PWM curation and scoring, and other members of the Kellis lab for helpful discussions.
REFERENCES
- 1.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl Acad. Sci. USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macintyre G, Bailey J, Haviv I, Kowalczyk A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26:i524–i530. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen MC, Engstrom PG, Lithwick S, Arenillas D, Eriksson P, Lenhard B, Wasserman WW, Odeberg J. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput. Biol. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Cunningham F, Rios D, McLaren WM, Smith J, Pritchard B, Spudich GM, Brent S, Kulesha E, Marin-Garcia P, et al. Ensembl variation resources. BMC Genomics. 2010;11:293. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011 doi: 10.1038/nature10530. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, III, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Touzet H, Varre JS. Efficient and accurate P-value computation for position weight matrices. Algorithms Mol. Biol. 2007;2:15. doi: 10.1186/1748-7188-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]