Abstract
The yeast Saccharomyces cerevisiae is a prevalent system for the analysis of transcriptional networks. As a result, multiple DNA-binding sequence specificities (motifs) have been derived for most yeast transcription factors (TFs). However, motifs from different studies are often inconsistent with each other, making subsequent analyses complicated and confusing. Here, we have created YeTFaSCo (The Yeast Transcription Factor Specificity Compendium, http://yetfasco.ccbr.utoronto.ca/), an extensive collection of S. cerevisiae TF specificities. YeTFaSCo differs from related databases by being more comprehensive (including 1709 motifs for 256 proteins or protein complexes), and by evaluating the motifs using multiple objective quality metrics. The metrics include correlation between motif matches and ChIP-chip data, gene expression patterns, and GO terms, as well as motif agreement between different studies. YeTFaSCo also features an index of ‘expert-curated’ motifs, each associated with a confidence assessment. In addition, the database website features tools for motif analysis, including a sequence scanning function and precomputed genome-browser tracks of motif occurrences across the entire yeast genome. Users can also search the database for motifs that are similar to a query motif.
INTRODUCTION
The yeast Saccharomyces cerevisiae is a powerful model for the study of gene regulation, and one in which numerous computational and experimental approaches to the study of transcriptional networks have been field-tested and applied on a large scale (1–7). As a result, there has been some level of characterization of the sequence specificity of most yeast transcription factors (TFs). A TF's sequence specificity, or ‘motif’, is frequently represented as a Position Weight Matrix (PWM) whose entries represent the log-odds ratio of bases being part of the motif, relative to the background sequence, which is generally taken to represent the relative preference of the corresponding protein for that sequence (8). It is desirable to have a comprehensive collection of yeast TF motifs for use in a variety of purposes, including the computational analysis of transcriptional networks [e.g. (9)] and study of genome evolution [e.g. (10)]. However, different published motifs for the same TF often conflict and may not represent the TF's true intrinsic sequence preferences, thus potentially confounding many studies that use the motifs.
Here, we have created YeTFaSCo, a database of yeast TF sequence specificities, obtained from diverse sources. We have evaluated the motifs’ predictive power and consistency with a variety of sources, including genome-wide studies, knowledge of the types of sites that different structural classes of TFs can and cannot bind, and detailed studies from the literature. To our knowledge, no similar resources exist: UniPROBE (11) contains only Protein Binding Microarray (PBM) data, while YEASTRACT (12) does not contain any PBM data. YPA (13) and MYBS (14) have collected motifs from several different sources, but concentrate on using these motifs to predict genomic binding sites and regulatory associations. Perhaps the most commonly used index of yeast TF motifs—the MacIsaac collection (15)—contains only motifs from ChIP-chip data. While TRANSFAC (16) and JASPAR (17) compile motifs from the literature, neither these nor the aforementioned collections evaluate the motifs for predictive power. Other recent studies (18–22) have also compiled motifs, and in some cases evaluated them for consistency with external information, but do not aim to comprehensively survey the literature, or to evaluate the motifs against each other and against multiple data types. Our evaluation methods go beyond previous studies because (i) we evaluate the motifs using four independent criteria, and (ii) we include a manual curation step, producing a collection of non-redundant motifs that are annotated with expert confidence ratings. YeTFaSCo also incorporates tools for scanning new sequences for PWM matches, browsing the genome for potential binding sites, and comparing among motifs. We anticipate that, as a unique resource, YeTFaSCo will be invaluable to a wide variety of researchers.
GENERATION OF THE DATABASE
YeTFaSCo has two central tables that are related to each other. One is a table of all genes/proteins and their encoded DNA-binding domains (DBDs), if any, and the other is a table of motifs assigned to these proteins. There are often multiple motifs associated with each TF, but typically only a single TF associated with each distinct motif, unless the TF binds as a part of a complex. We considered any yeast protein with either a DBD or an associated DNA-binding motif to be a potential TF. The current version of YeTFaSCo contains 264 known and putative TFs (248 with motifs + 16 with DBDs, but no motifs yet described) and eight TF complexes (with motifs).
To assign DBDs, the union of three sets of domain predictions was taken, including those from Badis et al. (4), those from Weirauch et al. (23), and our own predictions made by scanning all yeast genes with the HMMs from the Pfam (24), SMART (25) and SUPFAM (26) databases that correspond to the Weirauch DBD set (23). Similarly, we populated the database with motifs using several approaches. First, existing databases containing motifs were used to direct the search for motifs (4,11,12,19–22,27). We re-extracted motifs in these databases from the primary literature and documented the assays used to derive them. Next, we performed general searches of the literature, looking for papers that had derived motifs for any yeast proteins (Table 1—Query Type ‘General’). Next, a reference-directed approach was taken. Here, for each popular biochemical assay from which motifs can be derived, we searched for publications that included the original publication to describe this method as a reference. This limited our search to those papers which were likely to derive motifs using these methods (Table 1—Query Type ‘Reference-directed’). These methods included: ChIP-chip (2), BIAcore (28), DIP-chip (29), MITOMI (30), PBMs (31), SELEX (32), CSI (33), CASTing (34), HT-SELEX (35), Bind-n-Seq (36), PIch (37), DNase I-seq (38), ChIP-seq (39), DamID (40). Finally, for every putative TF in the collection that had zero or one motifs derived for it, we performed a directed search, looking for motifs specifically for these factors (Table 1—Query type ‘Gene-directed’).
Table 1.
Example motif search queries
| Query type | Engine | Example query |
|---|---|---|
| General | Scholar | Yeast OR saccharomyces OR cerevisiae motif OR pfm OR logo OR pwm ‘transcription factor’ OR ‘DNA-binding’ -intitle:ahuman -intitle:drosophila |
| Scholar | Motif specificity sequence ‘transcription factor’ saccharomyces OR cerevisiae ‘DNA binding’ -intitle:arabidopsis -intitle:subtilis -intitle:drosophila -intitle:human -intitle:prokaryotic -intitle:mouse -intitle:albicans logo OR PWM OR PSSM OR PFM | |
| Scholar | Motif specificity sequence ‘transcription factor’ saccharomyces OR cerevisiae ‘DNA binding’ -intitle:arabidopsis -intitle:subtilis -intitle:drosophila -intitle:human -intitle:prokaryotic -intitle:mouse -intitle:albicans -intitle:brucei -intitle:trypanosome | |
| Scholar | ‘Transcription factor’ motif specificity cerevisiae ‘DNA-binding’ -intitle:drosophila -intitle:human -intitle:plant -intitle:mammal | |
| Pubmed | Transcription factor (motif OR specificity OR pwm OR PFM) (cerevisiae OR yeast) ‘DNA-binding’ | |
| Reference-directed | Scholar | DNA binding cerevisiae ‘transcription factor’ PWM OR PFM OR PSAM OR PSSM OR ‘sequence specificity’ motif |
| Gene-directed | Scholar | <GeneName> OR <SysName>b DNA binding cerevisiae ‘transcription factor’ motif OR PWM OR PFM OR PSAM OR PSSM OR consensus OR ‘sequence specificity’ OR ‘binding site’ |
aThe ‘-intitle:’ term is used to exclude papers with a given term in the title.
b<GeneName> and <SysName> were replaced with the gene and systematic name of the gene being searched for.
For each motif, we converted from the provided form to a position frequency matrix (PFM), the standard motif form used in this database. The entries in a PFM represent the frequency of observing each base at a given position in the motif, and thus the sum of frequencies for each position in the motif sum to 1. We chose this motif form because it is a simple, yet robust model of TF specificity, most other motif representations can be converted to PFMs with relative ease, and there are many tools developed which use either PFMs or position weight matrices (PWMs). PFMs can easily be converted to PWMs by dividing each entry by the corresponding background base frequency and changing to a log scale (8). The PWM format facilitates scanning DNA sequences by, for every possible alignment of the motif to the sequence, providing the log-odds ratio of the subsequence being an instance of the motif, versus part of the genomic background (8). Thus, scores above zero represent sequences more likely to be an instance of the motif than random DNA. As noted above, the same calculation is widely taken to represent the relative affinity of a TF to a particular sequence the same width as the PFM/PWM. Some of the motifs in the database have flanking bases with low information content. Trimming the motifs to remove low information content bases did not on average improve the motifs by our criteria, however (data not shown), so the motifs were left in their original form.
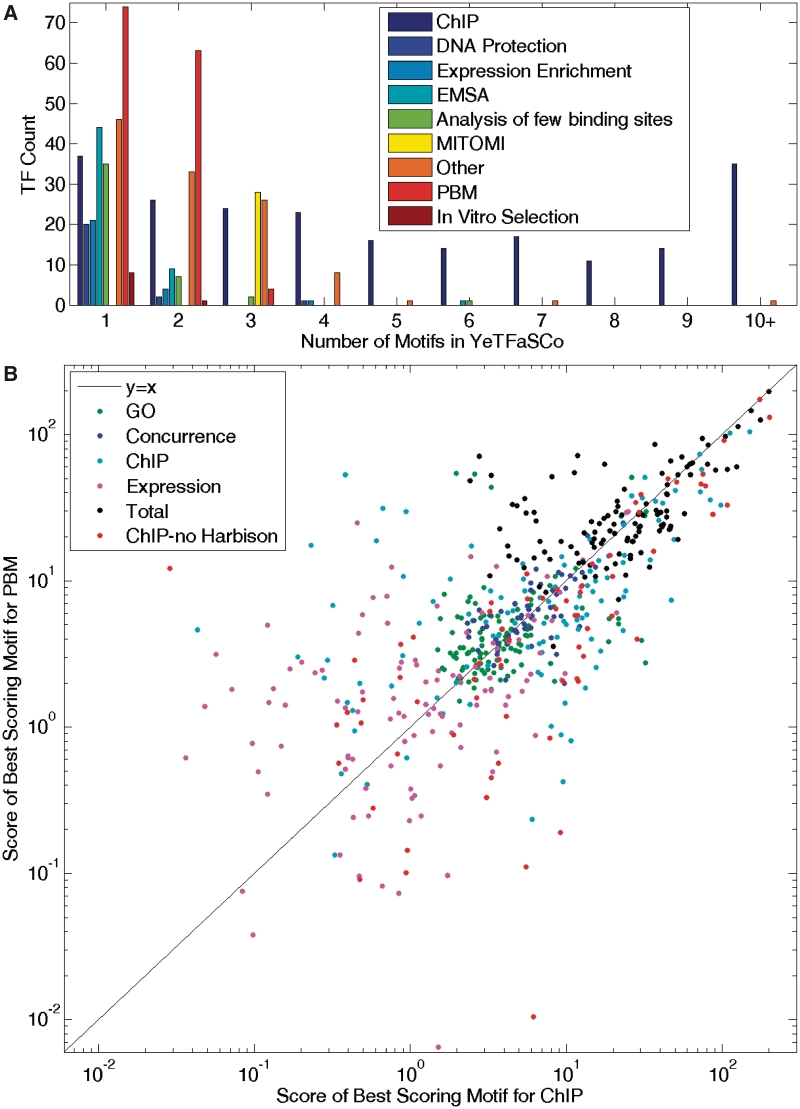
In total, YeTFaSCo currently contains 1709 motifs for 256 proteins or protein complexes, which were derived from 133 publications. As many as 445 motifs come from a single publication (19), and individual TFs have as many as 40 different associated motifs (Ste12). The motifs present in the database were derived from diverse data types, but ChIP-chip has by far the greatest number of motifs (1189), with many of these motifs being derived using different algorithms from the same source data (1), resulting in many TFs with multiple ChIP-derived motifs (Figure 1A).
Figure 1.
Comparison of ChIP and PBM-derived motifs. (A) Histogram showing the number of motifs per TF broken down by derivation method. (B) Comparison of summary scores (see text) for the highest-scoring motifs derived independently by both PBMs and ChIP-chip for 112 TFs common to both. Each point represents a single TF for one of the four scoring measures, the total score, or the average of all ChIP data, excluding the Harbison et al. (1) data, from which most of the ChIP motifs were derived.
EVALUTION OF MOTIFS: OBJECTIVE CRITERIA
We used four objective criteria to give a confidence value for the accuracy of each motif. These criteria include the correlation of predicted binding sites and ChIP-chip data, the correlation between predicted binding sites in promoters and expression changes in TF mutants, the enrichment of GO terms in genes whose promoters have binding sites, and the agreement between different studies. These criteria are described in more detail here.
ChIP-chip enrichment
For 212 proteins in the database, genome-wide chromatin immunoprecipitation data is available (1,41–43). We used these data to test the quality of the motifs by calculating the Spearman correlation coefficient between the relative probe intensities and the probability of the probe region being bound by the motif. When the ChIP experiments used entire intergenic regions as microarray probes, the ‘probe region’ to be scanned was defined as the entire probe sequence, and when the probes represented equally sized short sequences, the ‘probe region’ was defined as the maximum range of DNA around the probe that could fully hybridize to the microarray probe given the published upper size limit of the sheared ChIPed DNA (e.g. if the DNA was sheared to a maximum size of 300 bp, and the probe was 60 bp and started at x, the ‘probe region’ was taken as x − 240 to x + 300). The probability of the probe region being bound is calculated as the probability that at least one binding site in the sequence is occupied, given the motif (44). The P-value of the correlation between relative probe intensities and the probability of that probe region being bound was calculated using the Edgeworth series approximation method (45). As a summary score for each motif, we calculated the average −log(P-value) for the correlations over all ChIP-chip data sets. The distribution of these scores, in comparison to the scores for 1000 permutations of the probe intensities for each ChIP experiment, is shown in Figure 2A. Only 1% of the randomized data scored above 1.4, in contrast to 67% of the actual data. Thus, we use a cutoff of 1.4 to distinguish motifs that significantly correlate with ChIP data from those that do not, representing an empirically determined ∼1% FDR.
Figure 2.
Comparison of the distributions of actual motif evaluation scores with randomized data. For each plot, the actual data is shown in red and the permuted data is in blue with error bars (representing one SD across motifs) shown in black. (A) Mean ChIP-chip enrichment score. (B) Mean expression enrichment score. (C) GO-term enrichment score. (D) Inter-study concurrence score.
Many of the motifs in the database were derived from the same ChIP-chip data being used to evaluate it (1). This circularity would be expected to bias the analyses in favour of ChIP-chip motifs. However, comparison of the highest-scoring motif for each TF derived by ChIP-chip to the highest-scoring motif derived by PBM (the second most abundant motif derivation method in the database) revealed that, for the 112 TFs with motifs derived by both methods, PBMs perform slightly better overall; 60 of the 112 motifs have a higher total score for PBMs (Figure 1B). This is in spite of the fact that there is a much larger pool of ChIP-chip motifs from which to choose the best motif (Figure 1A). One possible explanation is that motif derivation from ChIP-chip data faces the inherent difficulty of searching for short motifs in long stretches of non-random DNA. However, we cannot exclude a bias in the evaluation criteria, or the binding model in favour of PBM-derived motifs.
Correlation with gene expression data
Several studies have used microarrays to systematically examine the effects of TF over-expression and/or deletion (46,47), and many others have analyzed one or a few TF mutants. We downloaded expression data from systematic studies (46,47) and individual studies included in the SPELL collection (48), giving us data from 58 sources that include mutant expression data for 212 of the TFs in YeTFaSCo. These data are useful for evaluating motif quality since we expect that genes with TF binding sites in their promoters will have their expression perturbed in the corresponding TF mutant. We scanned promoters (taken from −500 to +100 relative the transcription start site) using the same binding model described for the ChIP-chip Enrichment criterion to yield a probability of each promoter being bound by the TF, for each motif. Similar to the ChIP-chip metric, we then calculated the Spearman correlation coefficient between the probabilities of the TF binding each promoter and the log expression changes in the corresponding genes, with a P-value being derived as above. The summary score for this criterion is the mean of the correlation −log(P-value)s for all available mutant expression data sets. The distribution of these scores, in comparison to the scores derived from 1000 permutations of the fold expression changes for each experiment, is shown in Figure 2B. Only 1% of the randomized data scored above a threshold of 1.3, compared with 36% of the actual data.
GO term enrichment
TFs often regulate specific pathways and processes (46). To test for enrichment of binding sites in promoters of functionally related genes, we calculated the probability of the TF binding to each promoter for each motif, and performed AUROC and ranksum tests for each motif-GO slim term combination to ask whether binding probabilities differ between genes which are annotated with the GO term and those which are not. Both enriched and depleted ROCs are considered because, in addition to having certain TFs responsible for activating certain processes, it is possible that there are certain processes which specifically lack certain motifs. In general, one would expect this latter case to be uncommon. Indeed, using a 1% FDR, only 49 motifs show a significant depletion in a GO slim term, compared with 216 which are enriched. Most of these former motifs are depleted in either ‘ribosome biogenesis’ (18/49) or ‘RNA metabolic process’ (13/49), and many (5/18 and 12/13, respectively) of these are very similar to AGGGG, the ‘stress response element’ and known Msn2/Msn4 binding site (49). Since Msn2 and Msn4 are activated in stress conditions (50), it is not surprising that these sites are generally absent from genes involved in ribosome biogenesis, since these genes are generally repressed under times of environmental stress (51,52). The score for this criterion is the −log(P-value) of the ranksum test for the most significantly enriched/depleted GO term. The distribution of these scores, in comparison to 1000 permutations of the GO term labels, is shown in Figure 2C. Only 1% of this randomized data scored above 4.0, in contrast to 25% of the actual data.
Inter-study concurrence
We used the concurrence between independent studies as a measure of a motif's reliability. For instance, if two studies independently characterize the same specificity for the same protein, this is strong evidence that the motif is correct. Using Tomtom (version 4.1.0) (53), we compared each motif to the other motifs derived for the same TF (from independent data) using the Euclidean distance metric, yielding a P-value representing how likely each match is, given the similarity of the two motifs. This P-value, negated and logged, is used as the score for this metric. The distribution of these scores, in comparison to 1000 permutations of the gene labels for the motifs, is shown in Figure 2D. While 1% of the randomized data scored above a threshold of 5.3, 26% of the actual data surpassed this same threshold.
MANUAL EVALUTION OF MOTIFS: SUBJECTIVE DERIVATION OF AN ‘EXPERT CURATED’ SET
For most purposes, a library of TF motifs would ideally include the single most accurate motif for each TF. For example, in computational modelling, it is desirable to have as few features as possible, while still having a comprehensive list of relevant features. With this motivation, we created an ‘Expert Curated’ motif set, which consisted of one motif for each TF, or more than one if the TF appears to have multiple binding modes (e.g. some GAL4-class TFs appear to tolerate more than one spacing between the half-sites, and/or can bind as either monomers or dimers (54), while some bZIP proteins appear to tolerate different spacings between the half-sites, and can heterodimerize in different combinations).
We emphasize that the data available differs for each TF, and that in many cases the data is not easily comparable among TFs or even for the same TF. Consequently, no one-size-fits-all objective system can be applied automatically to all TFs; this fact is what motivated the expert curation step. Consequently, the expert curations are subjective to a degree. We did, however, employ the following procedure. First, using the Table tools described below, we manually examined all of the motifs and scores for each TF in the database. We also considered additional information in the literature (such as DNaseI footprinting and reporter assays), as well as knowledge regarding the types of sites that are normally bound by the different structural classes of DNA-binding domains. For example, monomeric GAL4-class zinc clusters typically bind sequences containing CGG, and these proteins also often bind as dimers that prefer a specific spacing and orientation of the two CGG subsites (54). We attempted to avoid selecting motifs that were likely due to cofactors (e.g. some of the motifs that have been reported for Ace2, Fhl1, Yap5, Pdr1 and Sfp1 are in fact Rap1 motifs; see Table 2). We also considered whether or not the motifs were supported by more than one criterion (e.g. if a motif was scoring highly simply because of a high correlation to ChIP-chip data, but did not perform well by any other measure), and whether the data used to derive the motif reflects the protein binding directly to the DNA. As part of our manual analysis, we gave the selected motifs a confidence score (high, medium, or low). Some putative TFs were also assigned ‘Dubious’ status in our database, due to lack of evidence that they bind DNA directly or in a sequence-specific manner; motifs for these TFs are not included in the expert curated set. The full details of the Expert Curated set are available from the ‘Expert Curation’ link on the side bar of the YeTFaSCo home page. In total, the expert curated set contains 218 motifs for 190 TFs (most of which are a 1-to-1 mapping; there are five instances of protein complexes and 28 TFs that have multiple motifs, due to evidence for multiple binding modes such as monomeric versus dimeric). 139 of the motifs are high confidence, 61 are medium confidence, and 22 are low confidence. If we consider the set of all 246 known + putative yeast TFs to include those that either have a motif in our database or contain one of the canonical eukaryotic TF DBDs (23), and are not ‘Dubious’ in our judgment, then 85% of all known + putative yeast TFs have a motif in the ‘expert curated’ set, 52% of which are high confidence.
Table 2.
Examples of potential TF cofactors
| Potential co-factor | Motif IDs | TF known to bind motif |
|---|---|---|
| Ash1 | 648, 932 | Mcm1 |
| Rlm1 | 1079 | Mcm1 |
| Ndd1 | 366 | Mcm1 |
| Arg81 | 1507 | Mcm1 |
| Arg80a | 1483 | Mcm1 |
| Fhl1 | 406, 629, 893, 1196, 1504, 1618 | Rap1 |
| Sfp1 | 357, 621, 1100, 1710 | Rap1 |
| Pdr1 | 899 | Rap1 |
| Yap5 | 896 | Rap1 |
| Ace2 | 918 | Rap1 |
| Cha4 | 1607 | Rap1 |
| YDR026Ca | 408, 696, 1160, 1581, 1921 | Reb1 |
| Stb2 | 710 | Reb1 |
| Rpn4 | 1090 | Reb1 |
| Pho2 | 1680 | Abf1 |
All motifs were derived from ChIP-chip data.
aHomologs to TF known to bind motif.
CONTENT AND INFORMATION RETRIEVAL
From the main YeTFaSCo page (http://yetfasco.ccbr.utoronto.ca), there are links to the five table views, a downloads page, a place for users to submit data, a cart, help pages, a website tour and sequence analysis tools (described below). The table views contain all the data in the database and provide links between tables for easy navigation. While navigating the database tables, the cart can be used for keeping track of specific motifs of interest, which can then be downloaded. Alternatively, the downloads page provides access to all the motifs in the database in multiple formats. These three features are described here in more detail.
Tables
The data in the database is accessible through several tables, which are the primary means of browsing the motifs. The ‘Motifs’ table displays an abbreviated version of the data in the database (Figure 3). This is the primary method of seeing which motifs are in the database and displays the TF names, motif IDs, a logo representation of the motif (55), the score for that motif, any DBDs that TF might have, the study responsible for derivation of the motif, what biochemical approach was taken to generate the motif, and the expert confidence in the motif (if it is in the ‘expert curated’ set). This page links to several other tables in the database, including the ‘Gene’ table, which shows more information about the genes, the ‘Expert Curation’ table, which shows the details of the expert curation, the ‘Reference’ table, which shows more details about the particular study from which the motif is derived, and the ‘Motifs – Details’ table which shows all the information of the ‘Motifs’ table, with additional details of how that motif scored for the various evaluation criteria. Additionally, the ‘Motifs – Details’ table provides links to the detailed breakdown of the ChIP-chip and expression enrichment scores, broken down by dataset. Each of these tables can be filtered and sorted by adding criteria to the filter bar and by clicking the header links, respectively.
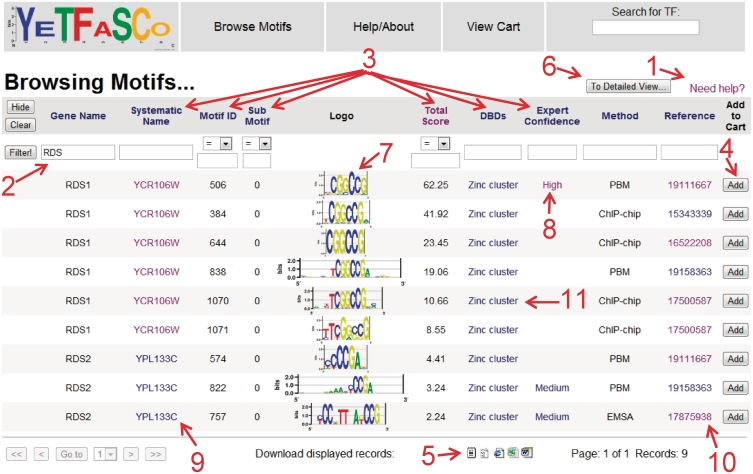
Figure 3.
The ‘Motif’ table of YeTFaSCo. For each of the database tables, there is a link to the help pages associated with that table in the upper right (1). In the help, there is an explanation of the meanings of each column. For all tables, results can be filtered by entering criteria into the filter bar (2) and pressing ‘Filter!’. The sort order can be changed by clicking on the various header links (3). For the motif table views, there is an option to add (or remove) motifs from the cart (4). There are also links provided for downloading the table data in various formats (5). The ‘To Detailed View …’ button (6) switches to the ‘Motifs – Details’ view, with the same filters applied. This view can also be reached for an individual motif entry by clicking the logo (7). The table also links to several different pages, including the expert curation (8) where additional details of the expert curation are provided, the gene entry (9) where details of the particular gene can be accessed and a link to SGD (27) is provided, and the reference entry (10) where more details of the study can be viewed, together with links to the corresponding PubMed entry. In addition, the DBDs (11) link to the corresponding entry in Interpro (65) (or other domain databases).
Cart
In the ‘Motifs’ and ‘Motifs – Details’ tables, the final column has buttons to allow the user to add motifs to their cart. This cart is useful for downloading only a subset of motifs as PFMs. The contents of the cart can be viewed through the ‘View Cart’ link in the header, which leads to a page where its contents can be edited or downloaded; in addition, users can scan a custom DNA sequence with the motifs contained in the cart.
Downloads
There are several downloads available from the downloads page, including all the motifs in the database as IUPAC, PFM, PWM (using the S. cerevisiae base content) and logo (55) representations. In addition, PFM and PWM sets are available for the ‘Expert Curated’ set, which should facilitate the use of the motifs in the database for custom sequence analysis. The download page also provides access to all microarray and ChIP-chip data used in this study, as well as the in vivo, in vitro and predicted nucleosome occupancy tracks, and conservation track present in the genome browser (see below).
ANALYSIS TOOLS
The YeTFaSCo website provides several analysis tools, which include sequence scanning with user-defined sequences, a utility for comparing a user-specified motif to the motifs in the database, and precomputed genome-wide TF binding sites available in a Genome Browser.
Sequence scanning
The sequence scanner identifies potential TF binding sites in user-defined sequences by scanning with different subsets of motifs in the database for sites that are more like the motif than like the background. This can be customized to be more or less stringent by specifying a percent of the maximum PWM score to use as a threshold (e.g. a threshold of 100% would show only perfect motif matches, while 0% shows all potential matches). By default this threshold is set to 75%. In addition, the user can also change the background base content to suit their needs.
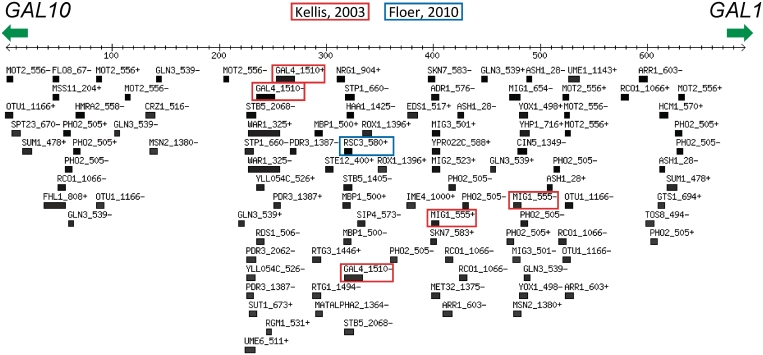
There are three ways to choose subsets of motifs with which to scan. The first is to use one of the provided motif sets (e.g. ‘all motifs’ or ‘Expert Curated’). The second is to select individual motifs by adding them to the cart, and then scanning with the cart contents. The third is to select a single TF to scan with, using all or one of the available motifs. The sequence scanner outputs a graphical representation of the motif matches along the given DNA sequence, a table containing all the hits, including details of the score, position and orientation of the match, as well as a table containing all the motifs that were searched for, but had no hits in the sequence. As an example of the sequence scanner output, we scanned the well-characterized Gal1-10 intergenic region for instances of TF motifs from the ‘Expert Curated’ set using a 75% score cutoff. The sequence scanner correctly identified the previously characterized Gal4, Mig1 and Rsc3 binding sites, as well as numerous other potential TF binding sites (Figure 4). Note that many of the motif matches are for proteins with similar binding sites to those of the known Gal1-10 regulatory factors (e.g. GAL4-class proteins, Mig2 and Mig3).
Figure 4.
Sequence scan of GAL1-10 promoter region. Only binding sites that achieve ≥75% of the maximum PWM score are shown. Previously characterized binding sites (66,67) are boxed in red and blue. The numbers next to the TF name represent the motif ID, while the ± represents the binding site orientation.
Motif similarity search
The YeTFaSCo website also has a tool for finding motifs that are similar to a user-provided motif. We anticipate this will be useful for instances where a potential regulatory motif is found, but the trans-acting factor is not known. To use this tool, users can input an IUPAC consensus motif, sequence alignment, or PFM. Using Tomtom (53) (as before), the most similar motifs are found and provided in table format in descending order of significance (until P > 0.05).
Genome browser
We scanned the yeast genome with the ‘Expert Curated’ set using an 80% of the maximum PWM score threshold (except in cases where there were fewer than 1000 or greater than 20 000 binding sites genome wide, in which cases we, respectively, repeatedly lowered or raised the thresholds by 5% until there at least 1000 or fewer than 20 000 sites were found or 0% or 100% were reached). YeTFaSCo provides these results in an implementation of the GBrowse genome browser (56). In addition to genome wide TF binding sites, YeTFaSCo provides tracks for in vivo, in vitro and predicted nucleosome occupancy (57–59). These tracks are provided as a reference because TFs are known to preferentially bind nucleosome free regions (60,61). We have also included a track representing the degree of conservation between closely related yeast species (62) because functional binding sites are more likely to be conserved (63). These data could help users to identify binding sites which are used in vivo. The YeTFaSCo genome browser uses version 64 (2011-02-03) of the S. cerevisiae genome.
FUTURE PLANS
In the process of constructing the YeTFaSCo database and manually curating the motif collection, we compiled a list of additional features and further analyses to incorporate into future versions of the database. One particularly important step will be to revisit the motif derivation steps. Browsing YeTFaSCo, it is clear that different motif-finding algorithms can yield dramatically different motifs from the same ChIP-chip data. The same may be true of motifs from PBMs and possibly also MITOMI: a recent analysis described a method for obtaining motifs that are demonstrably more accurate than those derived from previous approaches (35). More sophisticated motif evaluation methods might also yield higher correspondence between data sets; for example, correspondence between TF motifs and ChIP-chip or expression data may be higher if nucleosome occupancy over the motif match is considered, as well as the presence of General Regulatory Factor (GRF) binding sites in proximity (57,60). It is known that open chromatin is a major determinant of TF binding in vivo (61), suggesting that most TFs rely on additional cues—some of which are known and can be incorporated into computational models.
We also note that there are 16 putative TFs that still have no motif. In addition, we categorized 55 proteins that were previously annotated as known or putative TFs as ‘dubious’ and excluded them from the final manually curated list, because there is, as yet, no formal demonstration that these proteins have intrinsic sequence-specific DNA-binding activity—although there is at least some suggestion that they may. Thus, the sequence specificities of yeast TFs will, we hope, remain an active area of research, and future iterations of YeTFaSCo will incorporate emerging data. Many of these ‘dubious’ TFs with motifs assigned to them are known to be an upstream signalling component or downstream effector. For at least some such cases, the motif derived for these proteins corresponds to a known co-acting TF, suggesting that the signalling/effector protein is specific to this TF (e.g. all ChIP-chip-derived Fhl1 motifs are in fact binding sites for Rap1; see Table 2 and (64) for additional examples). It would likely be valuable for the mapping and mechanistic understanding of transcriptional networks to have, in addition to an index of TF sequence specificities, an index of which cofactors and chromatin factors are recruited by each of the individual TFs, or are involved in its recruitment.
CONCLUSIONS
As a unified and comprehensive resource of manually curated TF motifs, YeTFaSCo addresses a fundamental need in the analysis of yeast transcriptional networks. We anticipate that this database will be an extremely useful resource for the yeast community and will facilitate a greater understanding of transcriptional regulation.
FUNDING
Canadian Institutes of Health Research (Operating Grant MOP-490425 to T.R.H. and Operating Grant MOP-86705 to Corey Nislow and T.R.H.); Ontario Graduate Scholarship awards (C.G.d.B., partial). Funding for open access charge: Canadian Institutes of Health Research (Operating Grant MOP-490425).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Jack Greenblatt, Alan Moses, Harm van Bakel and Matt Weirauch for comments on the manuscript, and to Harm van Bakel, Kate Cook, Mihai Albu and Matt Weirauch for their assistance with the website.
REFERENCES
- 1.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne J-B, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 3.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 4.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant GO, Struhl K, Ptashne M. Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc. Natl Acad. Sci. USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat. Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 9.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 10.Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sá-Correia I. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang DT, Huang CY, Wu CY, Wu WS. YPA: an integrated repository of promoter features in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:D647–D652. doi: 10.1093/nar/gkq1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai HK, Chou MY, Shih CH, Huang GT, Chang TH, Li WH. MYBS: a comprehensive web server for mining transcription factor binding sites in yeast. Nucleic Acids Res. 2007;35:W221–W226. doi: 10.1093/nar/gkm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, van Nimwegen E, Rajewsky N, Siegal ML. Correlating gene expression variation with cis-regulatory polymorphism in Saccharomyces cerevisiae. Genome Biol. Evol. 2010;2:697–707. doi: 10.1093/gbe/evq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foat BC, Tepper RG, Bussemaker HJ. TransfactomeDB: a resource for exploring the nucleotide sequence specificity and condition-specific regulatory activity of trans-acting factors. Nucleic Acids Res. 2008;36:D125–D131. doi: 10.1093/nar/gkm828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morozov AV, Siggia ED. Connecting protein structure with predictions of regulatory sites. Proc. Natl Acad. Sci. USA. 2007;104:7068–7073. doi: 10.1073/pnas.0701356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- 23.Weirauch MT, Hughes TR. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell. Biochem. 2011;52:25–73. doi: 10.1007/978-90-481-9069-0_3. [DOI] [PubMed] [Google Scholar]
- 24.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandit SB, Bhadra R, Gowri VS, Balaji S, Anand B, Srinivasan N. SUPFAM: a database of sequence superfamilies of protein domains. BMC Bioinformatics. 2004;5:28. doi: 10.1186/1471-2105-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagerstam LG, Frostell-Karlsson A, Karlsson R, Persson B, Ronnberg I. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J. Chromatogr. 1992;597:397–410. doi: 10.1016/0021-9673(92)80137-j. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Noll DM, Lieb JD, Clarke ND. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 2005;15:421–427. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 33.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl Acad. Sci. USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell. Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput. Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 40.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 41.Workman CT, Mak HC, McCuine S, Tagne J-B, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG. A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc. Natl Acad. Sci. USA. 2008;105:2934–2939. doi: 10.1073/pnas.0708670105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Hughes TR, Morris Q. RankMotif++: a motif-search algorithm that accounts for relative ranks of K-mers in binding transcription factors. Bioinformatics. 2007;23:i72–i79. doi: 10.1093/bioinformatics/btm224. [DOI] [PubMed] [Google Scholar]
- 45.Best DJ, Roberts DE. Algorithm AS 89: the upper tail probabilities of Spearman's rho. J. Roy. Stat. Soc. Ser. C Appl. Stat. 1975;24:377–379. [Google Scholar]
- 46.Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl Acad. Sci. USA. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 48.Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- 49.Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 50.Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young Ra. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasch aP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta S, Stamatoyannopoulos Ja, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 60.Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 62.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cliften PF, Hillier LW, Fulton L, Graves T, Miner T, Gish WR, Waterston RH, Johnston M. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- 64.Gordan R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 67.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]