Abstract
MetaCrop is a manually curated repository of high-quality data about plant metabolism, providing different levels of detail from overview maps of primary metabolism to kinetic data of enzymes. It contains information about seven major crop plants with high agronomical importance and two model plants. MetaCrop is intended to support research aimed at the improvement of crops for both nutrition and industrial use. It can be accessed via web, web services and an add-on to the Vanted software. Here, we present several novel developments of the MetaCrop system and the extended database content. MetaCrop is now available in version 2.0 at http://metacrop.ipk-gatersleben.de.
INTRODUCTION
The importance of crop plants goes far beyond their use for nutrition. Plants are also used for renewable resources or in the chemical industry, and thus need to be improved steadily. For a continuous improvement of crop plants, detailed understanding of their metabolism is essential. MetaCrop is a resource to manage and explore manually curated high-quality data about crop plant metabolism. It contains information at different levels of detail from overview maps to pathways, to reactions, to reaction details and contains additional related data such as literature references. MetaCrop allows researchers (i) to explore metabolic information by browsing through various levels of abstraction, (ii) to integrate experimental data into metabolic pathways and (iii) to create metabolic models for simulation purposes.
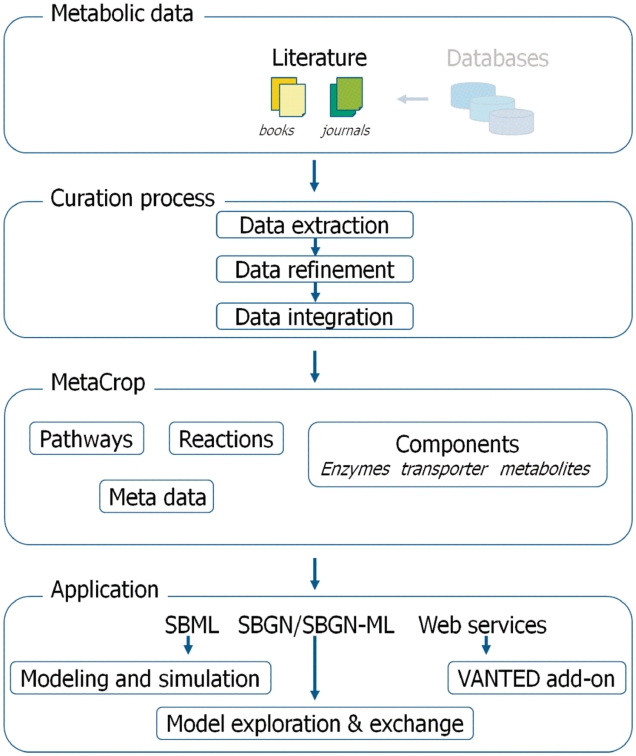
The initial system has been presented in Ref. (1), and its technical basis in Ref. (2). MetaCrop has been continuously developed in both technical aspects as well as database content over the last few years. In the following, we want to present the major improvements, which comprise a substantial extension of the content of the information system, the usage of the novel SBGN standard (3) and new ways of importing data as well as accessing the system. Figure 1 illustrates the architectural overview of the MetaCrop system including novel developments.
Figure 1.
Overview of MetaCrop, data sources, curation steps and applications.
DATABASE DESCRIPTION
Content
The data collection of MetaCrop is based on extensive manual curation. Currently, the system contains information about seven agronomically important crop plants as well as two model plants comprising both monocotyledon and dicotyledon species.
MetaCrop manages data about biochemical reactions and translocation processes, catalyzing enzymes, metabolites, macromolecules, stoichiometry, detailed locations (up to compartment level) and references. Parameters comprise, for example, names, synonym names, gene identifiers, EC and CAS numbers, chemical formulas, Gene ID, kinetic parameters and PubMed IDs.
Since the previous version was presented in Ref. (1), the database content has been almost doubled now containing information about 62 pathways, 566 reactions, 63 translocation processes and 21 compartments from >1800 scientific publications (Table 1, as of October 2011). Although MetaCrop focusses on the crop plants Hordeum vulgare (barley), Triticum aestivum (wheat), Oryza sativa (rice), Zea mays (maize), Solanum tuberosum (potato), Brassica napus (canola) and Beta vulgaris (sugar beet), and the model plants Arabidopsis thaliana and Medicago truncatula, additional data for other plants (crops and non-crops) is continuously added to the database.
Table 1.
Content of the MetaCrop database
| Organism | Pathways | Reactions | Translocations | Compartments | References |
|---|---|---|---|---|---|
| Hordeum vulgare | 54 | 362 | 44 | 9 | 454 |
| Triticum aestivum | 51 | 285 | 6 | 7 | 407 |
| Oryza sativa | 52 | 313 | 9 | 8 | 448 |
| Zea mays | 57 | 330 | 27 | 10 | 936 |
| Solanum tuberosum | 57 | 235 | 14 | 5 | 373 |
| Brassica napus | 45 | 171 | 7 | 5 | 247 |
| Beta vulgaris | 49 | 235 | - | 6 | 420 |
| Arabidopsis thalianaa | 59 | 405 | 19 | 13 | 1351 |
| Medicago truncatulaa | 49 | 247 | - | 4 | 386 |
| Totalb | 62 | 566 | 63 | 21 | 1846 |
aModel plants in life sciences research.
bDatabase object such as pathways, reactions, translocations, etc. are only listed once although they can occur in different organisms.
In addition to the extension of the data content, the database schema has been improved, in comparison to the initial MetaCrop version, in order to manage additional high-quality data. On the one hand, this comprises structures for the handling of gene identifiers, which are indispensable for data mapping and the discrimination of enzyme isoforms with different subcellular localization. On the other hand, structures for the storage of more detailed descriptions of different types of translocation processes were developed, which are important with regard to modeling and simulation of metabolic networks.
Web interface
As a point of entry to the MetaCrop database a web interface based on the Oracle Application Express technology was developed. It is intended to enable users to browse through different levels of granularity. Besides classical report tables showing, for example, detailed locations (up to compartment level) or kinetic parameters, the web interface provides clickable pathway maps with the pathways represented in the novel SBGN notation. Furthermore, an SBML exporter for the composition of individual metabolic models for analysis and simulation is available.
SBGN maps and SBGN-ML
SBGN, the Systems Biology Graphical Notation (3), has been developed as a standard for the visual representation of biochemical and cellular processes and networks. SBGN comprises three different views onto the biological system: process description (PD), entity relationship (ER) and activity flow (AF). This graphical representation helps to communicate biological knowledge in an unambiguous and easy way.
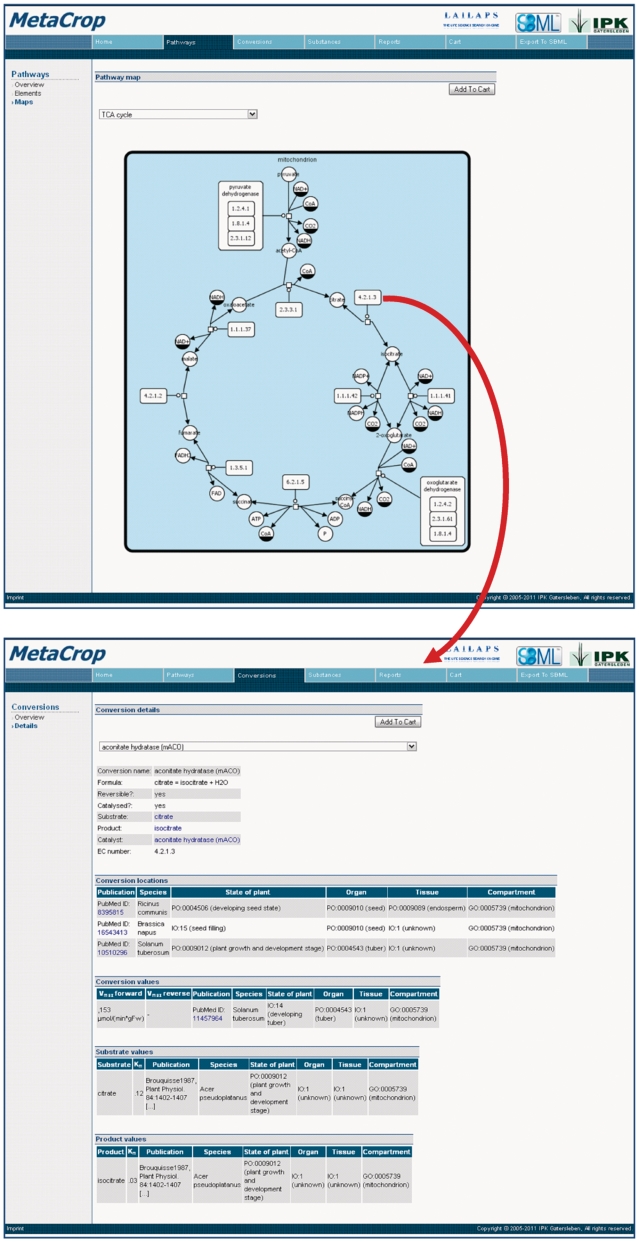
For the visualization of crop plant metabolic pathways, MetaCrop uses maps with the SBGN PD notation. Furthermore, to support the exchange of such pathway maps, they can be downloaded as SBGN Markup Language (SBGN-ML) files. Figure 2 shows an example SBGN map of a metabolic pathway as well as a corresponding report of details about one biochemical reaction of the pathway.
Figure 2.
Example from the MetaCrop web interface showing (a) an SBGN map of the TCA cycle and (b) details of a reaction chosen from this map, which could be obtained by clicking at the respective map element.
SBML exporter
In order to analyze metabolic data with stoichiometric or kinetic methods (in silico experiments), it is often necessary to construct user-specific metabolic models. For this reason, MetaCrop provides an export facility enabling the user to create models in the standardized SBML (4) format. While browsing the web interface, the user can put single elements such as reactions or substances, or even whole pathways into a kind of a shopping cart. Thereafter, the individual model can be composed, including the selection of parameter values (compartment, species, kinetic values, etc.), and finally exported as a SBML file.
Web-services
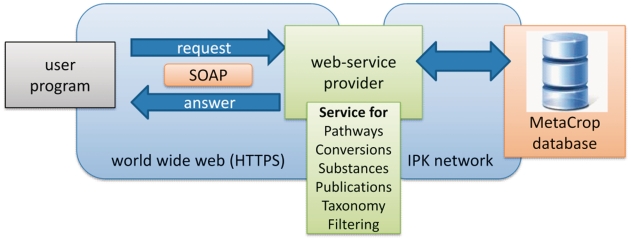
In addition to the SBML-based data exchange, SOAP-based web services were developed for interacting with external software tools, e.g. with the network visualization system Vanted (5). Web services were developed providing several methods for each of the five categories Pathway, Conversion (reaction or translocation), Substance, Publication and Taxonomy (6). The web services allow secure data transport (https) as well as filtering of data. Figure 3 illustrates the MetaCrop web service architecture.
Figure 3.
MetaCrop web services architecture.
Vanted add-on
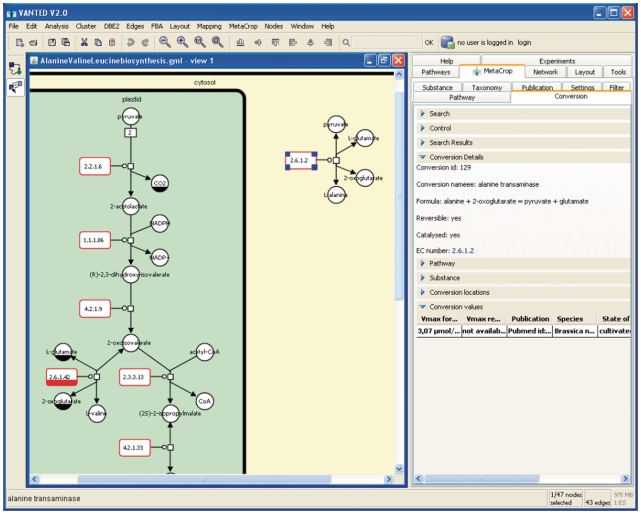
MetaCrop can easily be integrated as a data source into analysis tools. This is demonstrated by integrating MetaCrop into Vanted. An add-on for the network visualization system Vanted has been developed, which uses the web services described above. This add-on extends the search and filter capabilities of the web interface. Besides browsing of the database content, it also allows access to the graphical representations (SBGN maps) of the pathways and filtering of pathways for a species of interest. Figure 4 illustrates the user interface of the Vanted add-on.
Figure 4.
The Vanted add-on for MetaCrop which allows access to the database content using the MetaCrop web-services.
CURATION PROCESS AND CONTINUATION
MetaCrop data acquisition is performed by domain experts and is mainly based on research papers. Each record stored in the system is enriched manually by bibliography information. The main focus during the curation process is the extraction of data from scientific primary literature. In parts, meta data is extracted manually from existing databases such as BRENDA (7), ChEBI (8) and KEGG (9). The latter data is stored in MetaCrop only after extensive checks against literature. Controlled vocabulary is used to ensure high quality and to provide comparability of data, for example, by using ontology terms from Gene Ontology (10) and Plant Ontology (11).
For curators there are three possibilities for storing data in MetaCrop. First, data can be entered directly into the database using a simple curation web interface. Second, pathway data already available as a SBML file can be imported using a Java-based SBML importer. The third way includes the employment of a set of user-friendly MS-Excel templates, which can be imported by a Java application into the database.
MetaCrop is used in several projects and will be extended continuously in the future.
APPLICATION
The MetaCrop database is applicable to a broad variety of scientific questions. Exemplarily, three applications shall be mentioned here. (i) the navigation and exploration of plant metabolic pathways on different levels of detail to obtain overview and detailed knowledge concerning metabolism in plants; (ii) the analysis of–omics data to help in analyzing and understanding experimental metabolism-related–omics data such as metabolomics, transcriptomics, fluxomics and enzyme activity; and (iii) the modeling and simulation of crop plant metabolism to investigate the dynamics of the underlying biological system.
The possibility to explore plant metabolism is, for example, important in teaching. MetaCrop already supports this through its web interface, which allows a search for information about metabolites, enzymes, pathways, etc., and a click through pathway maps from overview pathways to detailed information. The Vanted add-on provides additional exploration possibilities such as the derivation of species-specific pathways. To further improve the way pathways can be explored, MetaCrop can be used in other applications using the web services. One example is the method and tool presented in Ref. (12), which introduces a new visualization approach to visualize interconnected pathways.
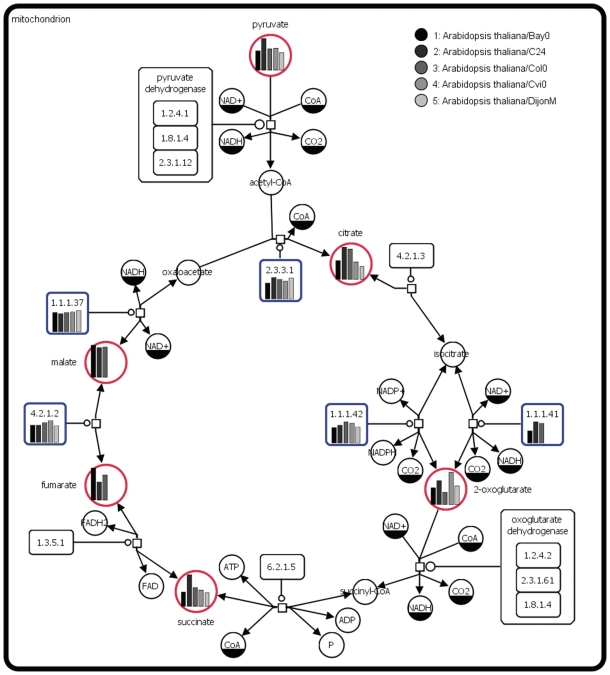
Large amounts of experimental data about metabolomes, proteomes, transcriptomes, etc. are nowadays available. MetaCrop pathways can be used to provide a context for such data and to support analyzes and understanding by mapping the data onto appropriate pathways. Figure 5 illustrates this in an example derived with the Vanted system.
Figure 5.
Metabolite concentrations and enzyme activities that were measured in several accessions of Arabidopsis thaliana (13) were mapped on a MetaCrop biochemical pathway (TCA cycle).
Another application example comprises the modeling and simulation of crop plant metabolism. Models can be built in MetaCrop and exported as SBML files. This works for stoichiometric models, which can be analyzed using constraint-based methods with tools such as FBASimVis (14) and, to some extent, for kinetic models, which can be analyzed using ODE-based methods with tools such as Copasi (15). It should be noted that the necessary kinetic values are only available for a part of the MetaCrop content as not all reactions have these parameters available in the literature. An example has been presented in Ref. (16), where a metabolic model of the primary metabolism in barley endosperm with 257 biochemical and transport reactions across four different compartments based on information in MetaCrop has been investigated using flux balance analysis.
DISCUSSION
Metabolic pathway databases contain knowledge of biochemical processes involved in the metabolism. There are a number of well-known databases for general and/or plant metabolic networks such as KEGG (9), EGENE (17), MetaCyc (18), PlantCyc (19), Arabidopsis Reactome (20) and Panther Pathways (21); for a complete list of available databases see Ref. (22). The advantage of MetaCrop is 2-fold: none of these databases covers such diverse levels of detail from overview maps to enzyme kinetics, and only some of them guarantee such high quality by manual curation and literature referencing of every database entry. MetaCrop also has its special niche by focusing on crop plants with high agronomical value.
CONCLUSION
MetaCrop is a high-quality database of metabolism in crop plants. It can be accessed in several ways and used in different application scenarios. MetaCrop will be further extended in the future.
FUNDING
German Federal Ministry of Education and Research (in part). Funding for open access charge: Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grafahrend-Belau E, Weise S, Koschützki D, Scholz U, Junker BH, Schreiber F. MetaCrop: a detailed database of crop plant metabolism. Nucleic Acids Res. 2008;36:D954–D958. doi: 10.1093/nar/gkm835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weise S, Grosse I, Klukas C, Koschützki D, Scholz U, Schreiber F, Junker BH. Meta-All: a system for managing metabolic pathway information. BMC Bioinformatics. 2006; 7:e465.1–9. doi: 10.1186/1471-2105-7-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, et al. The Systems Biology Graphical Notation. Nat. Biotech. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 4.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 5.Junker B, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:e109.1–13. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hippe K, Colmsee C, Czauderna T, Grafahrend-Belau E, Junker BH, Klukas C, Scholz U, Schreiber F, Weise S. Novel developments of the MetaCrop information system for facilitating systems biological approaches. J. Integr. Bioinform. 2010;7:e125.1–9. doi: 10.2390/biecoll-jib-2010-125. [DOI] [PubMed] [Google Scholar]
- 7.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avraham S, Tung C-W, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, et al. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008;36:D449–D454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jusufi I, Klukas C, Kerren A, Schreiber F. Guiding the interactive exploration of metabolic pathway interconnections. Information Visualization. 2011 doi:10.1177/1473871611405677. [Google Scholar]
- 13.Sulpice R, Trenkamp S, Steinfath M, Usadel B, Gibon Y, Witucka-Wall H, Pyl E-T, Tschoep H, Steinhauser MC, Guenther M, et al. Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. Plant Cell. 2010;22:2872–2893. doi: 10.1105/tpc.110.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grafahrend-Belau E, Klukas C, Junker BH, Schreiber F. FBA-SimVis: interactive visualization of constraint-based metabolic models. Bioinformatics. 2009;25:2755–2757. doi: 10.1093/bioinformatics/btp408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI—a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 16.Grafahrend-Belau E, Schreiber F, Koschützki D, Junker BH. Flux balance analysis of barley seeds: A Computational approach to Study systemic properties of central metabolism. Plant Physiol. 2009;149:585–598. doi: 10.1104/pp.108.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masoudi-Nejad A, Goto S, Jauregui R, Ito M, Kawashima S, Moriya Y, Endo TR, Kanehisa M. EGENES: Transcriptome-based plant database of genes with metabolic pathway information and expressed sequence tag indices in KEGG. Plant Physiol. 2007;144:857–866. doi: 10.1104/pp.106.095059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plant Metabolic Network (PMN). The PlantCyc database. http://plantcyc.org (4 November 2011, date last accessed) [Google Scholar]
- 20.Tsesmetzis N, Couchman M, Higgins J, Smith A, Doonan JH, Seifert GJ, Schmidt EE, Vastrik I, Birney E, Wu G, et al. Arabidopsis Reactome: a foundation knowledgebase for plant systems biology. Plant Cell. 2008;20:1426–1436. doi: 10.1105/tpc.108.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi H, Thomas P. Panther pathway: An ontology-based pathway database coupled with data analysis tools. In: Nikolsky Y, Bryant J, editors. Protein Networks and Pathway Analysis. New York: Humana Press; 2009. pp. 123–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]