Abstract
The MetaCyc database (http://metacyc.org/) provides a comprehensive and freely accessible resource for metabolic pathways and enzymes from all domains of life. The pathways in MetaCyc are experimentally determined, small-molecule metabolic pathways and are curated from the primary scientific literature. MetaCyc contains more than 1800 pathways derived from more than 30 000 publications, and is the largest curated collection of metabolic pathways currently available. Most reactions in MetaCyc pathways are linked to one or more well-characterized enzymes, and both pathways and enzymes are annotated with reviews, evidence codes and literature citations. BioCyc (http://biocyc.org/) is a collection of more than 1700 organism-specific Pathway/Genome Databases (PGDBs). Each BioCyc PGDB contains the full genome and predicted metabolic network of one organism. The network, which is predicted by the Pathway Tools software using MetaCyc as a reference database, consists of metabolites, enzymes, reactions and metabolic pathways. BioCyc PGDBs contain additional features, including predicted operons, transport systems and pathway-hole fillers. The BioCyc website and Pathway Tools software offer many tools for querying and analysis of PGDBs, including Omics Viewers and comparative analysis. New developments include a zoomable web interface for diagrams; flux-balance analysis model generation from PGDBs; web services; and a new tool called Web Groups.
INTRODUCTION
MetaCyc (http://metacyc.org/) is a highly curated, non-redundant reference database of small-molecule metabolism. It contains metabolic pathway and enzyme data experimentally demonstrated in the scientific literature (1). Because MetaCyc contains only experimentally determined pathways and enzymes, and due to its tight integration of data and references, MetaCyc is a uniquely valuable resource in fields including genome analysis, metabolism, and metabolic engineering. The metabolic pathways and enzymes in MetaCyc are derived from organisms representing all domains of life.
In conjunction with its role as a general reference on metabolism, MetaCyc is used as a reference database for the PathoLogic component of the Pathway Tools software (2) to computationally predict the metabolic network of any organism having a sequenced and annotated genome (3). In this automated process, a predicted metabolic network is created in the form of a Pathway/Genome Database (PGDB). In addition to the automated creation of PGDBs, the editing capabilities of Pathway Tools enable scientists to improve and update these computationally generated PGDBs by manual curation. MetaCyc has been used by SRI to create more than 1700 PGDBs (as of October 2011), which are available through the BioCyc (http://biocyc.org/) website. Interested scientists may adopt and curate any of these PGDBs through the BioCyc website (http://biocyc.org/intro.shtml#adoption).
In addition, MetaCyc is used by other scientists to create additional PGDBs, many of which are available to the general public through the scientists’ own websites. Together with BioCyc, these PGDBs form the MetaCyc family of databases (4).
More than 100 groups have used Pathway Tools and MetaCyc to create PGDBs for their organisms of interest, including important model organisms such as Saccharomyces cerevisiae (5), Arabidopsis thaliana (6), Oryza sativa (7), Mus musculus (8), Bos taurus (9), Medicago truncatula (10), Populus trichocarpa (11), Dictyostelium discoideum (12), Leishmania major (13), Chlamydomonas reinhardtii (14), several Solanaceae species (15), bioenergy-related organisms (BeoCyc) and many pathogenic bacteria (16) (see http://biocyc.org/otherpgdbs.shtml for a more complete list). A few examples of organisms that were studied within the last year using Pathway Tools include Bacillus acidocaldarius, B. circulans, B. filicolonicus, B. laterosporus, B. licheniformis and B. stearothermophilus (17), Clostridium difficile (18), C. thermocellum (19), Corynebacterium pseudotuberculosis (20), Ignicoccus hospitalis and Nanoarchaeum equitans (21), Mycobacterium species (22,23), Rhizobium etli CFN42 (24), Rhodococcus erythropolis (25), R. opacus PD630 (26), S. cerevisiae and Pichia pastoris (27), Serratia symbiotica (28), Shewanella species (29), Vibrio vulnificus (30), Xanthomonas axonopodis (31) and X. citri (32). Pathway Tools can also generate PGDBs from metagenomic data sets (33).
A web server included in Pathway Tools enables the publishing of PGDBs through either the Internet or an internal network. The Navigator component of Pathway Tools allows the browsing and analysis of PGDBs either locally or over the Internet. A detailed description of Pathway Tools can be found in (34).
PGDBs generated by Pathway Tools and MetaCyc are an excellent platform for the integration of genome information with many other types of data regarding metabolism, regulation and genetics. They provide powerful tools for analyzing omics data sets from experiments related to gene transcription, metabolomics, proteomics, ChIP-chip analysis and so on. During the past 2 years, we again significantly expanded the data content of MetaCyc and BioCyc. We also added supporting enhancements to the Pathway Tools software and BioCyc website, as described in the following sections.
METACYC ENHANCEMENTS
Expansion of MetaCyc
All pathways in MetaCyc are curated from the experimental literature. Since the last Nucleic Acids Research publication (2 years ago) (1), we added 413 new base pathways (pathways comprised of reactions only, where no portion of the pathway is designated as a subpathway) and 40 superpathways (pathways composed of at least one base pathway plus additional reactions or pathways), and updated 107 existing pathways, for a total of 560 new and revised pathways. The total number of base pathways grew by 28%, from 1399 (version 13.5) to 1790 (version 15.5) (the total increase is less than 413 pathways because some existing pathways were deleted from the database during this period) while the total number of superpathways grew by 17%, from 235 (version 13.5) to 275 (version 15.5).
Along with the increase in pathway number, the number of enzymes, reactions, chemical compounds and citations in the database grew by 30, 19, 13 and 49%, respectively; the number of referenced organisms increased by 23% (currently at 2216).
New pathway classes defined in MetaCyc
The pathways in MetaCyc are classified by an ontology developed at SRI that is constantly updated to reflect curation needs. Recently, we added two new top-level classes to that ontology: Activation/Inactivation/Interconversion and Metabolic Clusters.
The Activation/Inactivation/Interconversion class was added to describe certain pathways that did not fit well into any other classes, and, as its name implies, includes the three subclasses: Activation, Inactivation and Interconversion. In contrast to a standard ‘biosynthesis’ pathway in which a biologically active compound is synthesized from precursor molecules, activation pathways involve relatively minor chemical modifications to existing compounds that result in a substantial increase in their biological activity. An example activation pathway is sulfate activation for sulfonation.
Similarly, inactivation pathways involve relatively minor chemical modifications to existing biologically active compounds that result in a substantial decrease in their biological activity. This is in contrast to standard ‘degradation’ pathways in which a more complex compound is broken down into a set of simple metabolites. An example inactivation pathway is gibberellin inactivation II (methylation).
Interconversion pathways describe the bidirectional conversion of a bio-molecule to a different form, where the forward and backward conversions often prompt significant changes in the biological activity of the compound, resulting in its activation and deactivation, respectively. For an example, see medicarpin conjugates interconversion.
The Metabolic Clusters class was added to classify metabolic diagrams that do not describe the classical notion of a pathway. In pathways, all reactions are connected to one another, whereas metabolic clusters comprise a collection of non-connected but related reactions that together describe a common phenomenon. For example, see tRNA methylation (yeast), which describes a collection of tRNA methyltransferase-catalyzed reactions in yeast.
Ontology distribution of MetaCyc pathways
The six top-level categories (or classes) of the MetaCyc pathway ontology are Biosynthesis, Degradation/Utilization/Assimilation, Generation of Precursor Metabolites and Energy, Detoxification, Activation/Inactivation/Interconversion and Metabolic Clusters.
In version 15.5, the largest top-level class is Biosynthesis, with 1143 base pathways. Its main subclasses are Secondary Metabolites Biosynthesis (447); Cofactors, Prosthetic Groups, and Electron Carriers Biosynthesis (186); Amino Acids Biosynthesis (110); and Fatty Acids and Lipids Biosynthesis (124).
The second-largest top-level class is Degradation/Utilization/Assimilation, with 793 base pathways. Within this group, the largest subclasses are Aromatic Compounds Degradation (167), Amino Acids Degradation (117), Inorganic Nutrients Metabolism (94), Secondary Metabolites Degradation (85) and Carbohydrates Degradation (84).
The third-largest top-level class, Generation of Precursor Metabolites and Energy, contains 158 base pathways. Its largest subclasses are Fermentation (46), Respiration (28), Chemoautotrophic Energy Metabolism (15), Methanogenesis (13) and Electron Transfer (13).
The other three top-level classes are much smaller. The Detoxification class doubled in size and now contains 32 base pathways, and the new Activation/Inactivation/Interconversion and Metabolic Clusters classes contain 22 and 19 pathways, respectively.
During the previous 2 years, the number of metazoan pathways in MetaCyc increased by 42%, from 174 to 247 pathways. Plant pathways increased by 22% to 784, and archaeal pathways increased by 17% to 126. The number of pathways classified as bacterial actually decreased by 12%, as a result of a more accurate taxonomic classification of pathways.
Table 1 lists the species with the largest number of experimentally elucidated pathways in MetaCyc (meaning that there is experimental evidence for the occurrence of these pathways in the organism), while Table 2 describes the distribution of pathways in MetaCyc based on the taxonomic classification of associated species. The list of pathways added to MetaCyc since the last NAR publication is too long to specify here. For a complete report, see the MetaCyc Release Notes history at http://metacyc.org/release-notes.shtml.
Table 1.
List of species with 18 or more experimentally elucidated pathways represented in MetaCyc (meaning that there is experimental evidence for the occurrence of these pathways in the organism)
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Escherichia coli | 276 | Arabidopsis thaliana | 311 | Methanosarcina barkeri | 20 |
| Pseudomonas aeruginosa | 66 | Homo sapiens | 186 | Methanocaldococcus jannaschii | 21 |
| Bacillus subtilis | 57 | Saccharomyces cerevisiae | 134 | Methanosarcina thermophila | 18 |
| Pseudomonas putida | 49 | Rattus norvegicus | 77 | Sulfolobus solfataricus | 18 |
| Salmonella typhimurium | 36 | Glycine max | 67 | ||
| Pseudomonas fluorescens | 30 | Mus musculus | 50 | ||
| Mycobacterium tuberculosis | 26 | Solanum lycopersicum | 48 | ||
| Agrobacterium tumefaciens | 25 | Pisum sativum | 47 | ||
| Enterobacter aerogenes | 25 | Zea mays | 46 | ||
| Klebsiella pneumoniae | 21 | Solanum tuberosum | 41 | ||
| Mycobacterium smegmatis | 18 | Nicotiana tabacum | 39 | ||
| Delftia acidovorans | 18 | Oryza sativa | 35 | ||
| Hordeum vulgare | 31 | ||||
| Spinacia oleraca | 25 | ||||
| Triticum aestivum | 23 | ||||
| Bos taurus | 22 | ||||
| Sus scrofa | 18 | ||||
| Petunia × hybrida | 18 | ||||
The species are grouped by taxonomic domain and are ordered within each domain based on the number of pathways (number following species name) to which the given species was assigned. Some pathways may be labeled with a higher-level taxon, such as genus, if all the species within that genus are thought to have the given pathway. However, such higher-level taxa are not included in this table.
Table 2.
The distribution of pathways in MetaCyc based on the taxonomic classification of associated species
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Proteobacteria | 900 | Viridiplantae | 784 | Euryarchaeota | 125 |
| Firmicutes | 258 | Fungi | 271 | Crenarchaeota | 37 |
| Actinobacteria | 214 | Metazoa | 247 | ||
| Bacteroidetes/Chlorobi | 59 | Euglenozoa | 24 | ||
| Cyanobacteria | 48 | Alveolata | 15 | ||
| Deinococcus-Thermus | 25 | Amoebozoa | 10 | ||
| Tenericutes | 19 | Stramenopiles | 5 | ||
| Thermotogae | 19 | Fornicata | 4 | ||
| Aquificae | 13 | Rhodophyta | 4 | ||
| Spirochaetes | 12 | Haptophyceae | 3 | ||
| Chlamydiae- Verrucomicrobia | 6 | Parabasalia | 3 | ||
| Planctomycetes | 6 | ||||
| Chloroflexi | 4 | ||||
| Fusobacteria | 4 | ||||
| Nitrospirae | 2 | ||||
| Thermodesulfobacteria | 2 | ||||
| Chrysiogenetes | 1 | ||||
For example, the statement ‘Tenericutes 19’ means that there is experimental evidence for at least 19 MetaCyc pathways for their occurrence in members of this taxonomic group. Major Taxonomic groups are grouped by domain and are ordered within each domain based on the number of pathways (number following taxon name) associated with the taxon. A pathway may be associated with multiple organisms.
Curation of bioenergy pathways
Bioenergy is a rapidly growing area of research that focuses primarily on biomass conversion and biofuels production. To address the needs of the bioenergy research community we have made a priority of curating bioenergy-related pathways and enzymes in MetaCyc, starting with version 15.1 (released June 2011). Fields that receive attention are hydrogen production, cellulosic biomass biosynthesis and degradation, and algal oil production. So far we have created seven different hydrogen biosynthesis pathways, provided upgraded structures and commentary to many of the cellulosic biomass components, such as cellulose, hemicelluloses, xylan, arabinan, arabinogalactan, arabinoxylan, glucuronoxylan, glucomannan, galactomannan, galactoglucomannan and rhamnogalacturonan, and curated pathways for the biosynthesis and degradation of several of these polymers by different organisms. For an example, see cellulose degradation I (cellulosome).
Curation of engineered pathways
Since its inception, MetaCyc included only natural pathways that occur in unmodified organisms. However, over the years users indicated to us that it would be useful to include genetically engineered pathways in the database. Version 15.5 of MetaCyc (released October 2011) is the first to include such engineered pathways. To avoid confusion, engineered pathways are clearly indicated by the title ‘MetaCyc Engineered Pathway’ next to the pathway name. A text line above the summary indicates ‘Note: This is an engineered pathway. It does not occur naturally in any known organism, and has been constructed in a living cell by metabolic engineering.’
In addition, the organisms that contributed enzymes to the pathway are listed under the description ‘The enzymes catalyzing the steps of this pathway have been assembled from the following organisms’. Engineered pathways are excluded by our PathoLogic software when predicting the presence of pathways in organism-specific PGDBs.
For an example of an engineered pathway, see pyruvate fermentation to hexanol.
Chimeric and conspecific pathways
Users of MetaCyc are familiar with the concept of superpathways, which are constructed in PGDBs by combining multiple elements (at least one base pathway or superpathway, along with additional pathways or reactions) to show relationships between them and depict a larger portion of the metabolic network within a single diagram. Although most MetaCyc superpathways consist of pathways known to occur in the same organism, we sometimes find it useful to construct superpathways from pathways that are known to occur in different organisms. Combining such pathways into a single superpathway can provide an overview of a metabolic field. For example, combining all the known pathways for aerobic degradation of aromatic compounds into a single diagram provides a useful overview of this topic [see superpathway of aromatic compound degradation (aerobic)].
To distinguish such pathways from those that occur in their entirety in a single organism, we defined the terms ‘conspecific pathways’ and ‘chimeric pathways’.
While a conspecific pathway comprises a set of reactions that are expected to be found within each organism that has the pathway, a chimeric pathway comprises reactions from multiple organisms, and most commonly does not occur in its entirety in a single organism. Only sections of chimeric pathways are likely to occur in their entirety in single organisms. The two types of pathways are treated differently by the PathoLogic program during the creation of new PGDBs. When PathoLogic predicts a conspecific pathway to occur in another organism, the pathway will be transferred to that organism in its entirety. In the near future we will enhance PathoLogic so that when it predicts a chimeric pathway to occur in an organism-specific PGDB, it will remove extraneous reactions from the pathway to produce a conspecific version of the pathway. Conspecific pathways can be either base pathways or superpathways, while chimeric pathways are always superpathways.
To alert the user to the fact that a pathway is chimeric the following note appears above the summary section: ‘This is a chimeric pathway, comprising reactions from multiple organisms, and typically will not occur in its entirety in a single organism. The taxa listed here are likely to catalyze only subsets of the reactions depicted in this pathway.’ In addition, the pathway's title states ‘MetaCyc Chimeric Pathway’.
Kinetic data in PGDBs
We have recently more than doubled the number of types of enzyme kinetic data that can be captured in Pathway Tools PGDBs. When available, the following types of data are now collected in newly curated MetaCyc enzymes: Vmax, Kcat, Specific activity, optimal temperature, optimal pH, Ki values for inhibitors and Km values for substrates.
Interactions with other databases
IUBMB
MetaCyc is regularly updated with data from the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB), which includes new and modified EC entries. The last supplement incorporated is supplement 17, and the data was retrieved from the ExplorEnz database (35). In addition, starting with release 15.0, the EC entries at ExplorEnz are linked to MetaCyc reaction pages and vice versa.
NCBI taxonomy
The full NCBI Taxonomy database (36) is integrated into Pathway Tools, enabling specification of taxa using NCBI Taxonomy, and allowing taxonomic querying of MetaCyc pathways and enzymes. We continue to update the taxonomy entries with each major release of MetaCyc.
Gene ontology
The mapping between MetaCyc reactions and Gene Ontology (GO) process and function terms (37) is being continuously maintained by the GO Editorial Office at the EBI. An updated file is at http://www.geneontology.org/external2go/metacyc2go.
Links to other databases
During the last 2 years we have added extensive links from MetaCyc to PubChem and to KEGG. In version 15.5 of MetaCyc there are 4014 reactions that contain links to KEGG reactions. MetaCyc compounds contain 4449 links to KEGG compounds, 8814 links to PubChem compounds and 3800 links to ChEBI compounds.
EXPANSION OF BIOCYC
The BioCyc databases are organized into three tiers.
Tier 1 PGDBs have received at least 1 year of manual curation. While some Tier 1 PGDBs (e.g. MetaCyc and EcoCyc) received decades of manual curation and are updated continuously, others are less well curated and are still in need of significant curation.
Tier 2 PGDBs have received moderate amounts of review (<1 year), and may or may not be updated on an ongoing basis and
Tier 3 PGDBs were created computationally, and received no subsequent manual review or updating.
During the past 2 years, the number of BioCyc PGDBs increased from 508 (version 13.1) to 1129 (version 15.1). Version 15.5, to be released in October 2011, will include >1700 PGDBs. The PGDBs AraCyc (A. thaliana col, curated by PMN) and YeastCyc (S. cerevisiae, curated by SGD) have been promoted from Tier 2 to Tier 1 status, and the PGDB HumanCyc (Homo sapiens, curated by SRI) will be upgraded to Tier 1 starting with release 15.5, bringing the total of Tier 1 PGDBs to five (along with EcoCyc and MetaCyc). As of version 15.1, Tier 2 includes 32 PGDBs, and Tier 3 includes 1093 PGDBs. Some Tier 2 PGDBs were provided by groups outside SRI. Database authors are identified on the database summary page (Tools → Reports → Summary Statistics).
SOFTWARE AND WEBSITE ENHANCEMENTS
The following paragraphs describe significant enhancements to Pathway Tools and to the BioCyc website during the past 2 years.
Web groups—sharing and analysis of object groups
Starting in July 2011, BioCyc includes a new feature called Web Groups, that extends the web-based interface to allow end users to create, share and compute with collections of Pathway Tools objects (Figures 1–3). Web Groups are a step in the direction of making Pathway Tools a platform for collaborative computing and knowledge sharing.
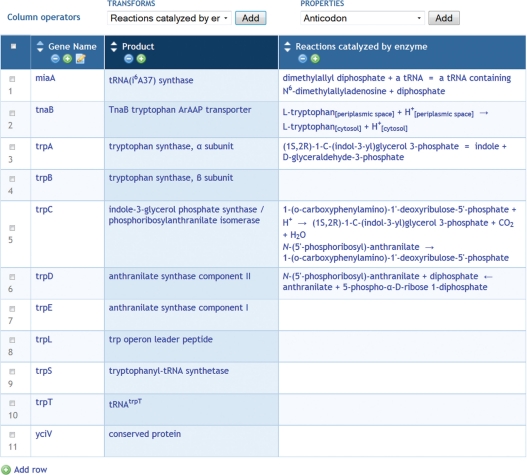
Figure 1.
An object group was created from the results of a search of the EcoCyc PGDB for genes containing the text string ‘trp’. After deleting a few rows of the table, two more columns were added by several transformations performed on the gene group, including the transformation ‘Products of gene’ and the transformation ‘Reaction of gene’.
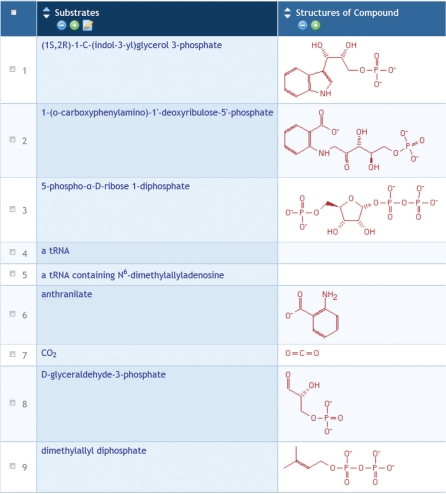
Figure 2.
An object group created by several transformations performed on the group shown in Figure 1. The first column contains all substrates that are included in the ‘Reaction’ column of that table, and the second column shows the structures of these compounds. These columns were generated using the transformations ‘Substrates of reaction’ and ‘Structures of compound’.
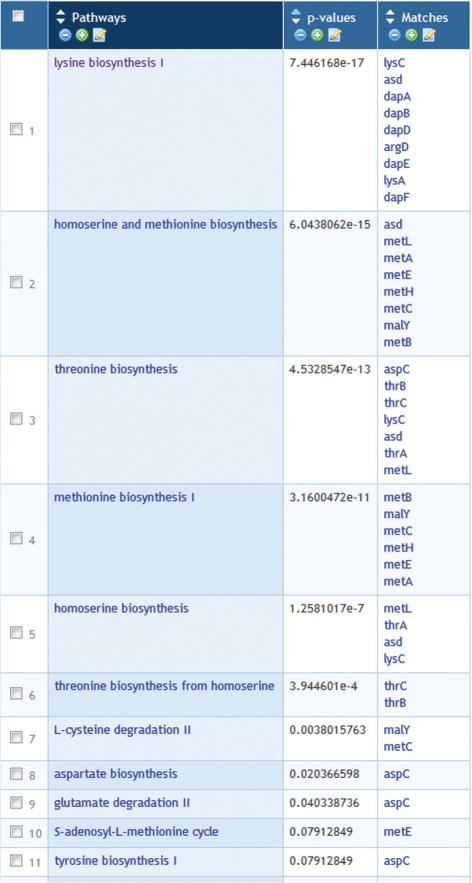
Figure 3.
Enrichment analysis of Web Groups objects. A group of Escherichia coli genes was analyzed for enrichment of the genes in pathways. The resulting table includes a list of pathways, the P-value for each pathway and the subgroup of genes from the original group that participate in each pathway. The table has been modified by removing some rows that represented pathway classes and superpathways, leaving only base pathways.
A Web Group is a spreadsheet-like structure that can contain both Pathway Tools objects and other values such as numbers or strings. Like a spreadsheet, it is organized by rows and columns. The typical group contains a set of Pathway Tools objects in the first column (e.g. a set of genes generated by a search). The other columns contain properties of the object (e.g. the chromosome position of each gene), or the result of a transformation (e.g. the reactions catalyzed by the gene products, or the corresponding genes from a different organism). The system provides 35 built-in transformations, each of which applies to a specific type of object. Example transformations include: transform a group of genes into the group of pathways containing that gene, or into the group of all genes that regulate the expression of those genes; transform a group of pathways into a group of all metabolites that are substrates within the pathway. The transformations can be applied to columns other than the first, creating a spreadsheet-like cascade.
Web Groups can be created from search result sets, by importing data from external spreadsheets or text files, and by adding objects individually from either their web pages or from the group itself. They can be exported to spreadsheets, and group columns of the appropriate types can be exported to the cellular overview. Web Groups can be shared publicly, or with selected other users.
The Web Groups interface also allows users to apply an enrichment/depletion analysis to the contents of a group (Figure 3). Enrichment/depletion analysis enables users to evaluate over- or under-representation of certain qualities or traits within an object group—for example, determining which genes out of a specified gene group are involved in one or more Gene Ontology categories. To enable this type of analysis, Pathway Tools includes a statistical analysis engine that can be applied to the content of groups. Performing enrichment analysis on a group results in creation of a new group that contains the analysis results.
Example use cases for Groups:
Users are interested in genes of the trp operon. They perform a search for genes containing the string ‘trp’, and turn the results into a group. Some of the gene names do not seem to contain that string, so the users add a column for the gene synonyms to see why they matched. After doing that, the users can see that some do not belong (e.g. the ribB gene matched because of the synonym ‘htrP’), so they delete that row from the group table. They then use a transformation from genes to their products, adding a column with the gene products; a second transformation adds a column containing the reactions that the products catalyze (Figure 1). Next they use additional transformations to obtain the substrates involved in those reactions, to create a new group from those substrates, and to add the molecular structures (Figure 2).
The users have obtained an essential gene list from experimental investigations. They can define a Web Group containing those essential genes, and use group operations to highlight the genes on the cellular overview to view its metabolic pathway distribution, or use enrichment analysis to determine over-represented GO categories.
The users have obtained a set of metabolites of interest from a metabolomics experiment. They can perform an enrichment analysis to determine over-represented metabolic pathways in that group.
New web cellular overview
We have re-engineered the web-based metabolic map diagrams available via Pathway Tools (38). As for the desktop version of Pathway Tools, the new web versions of these diagrams are organism specific, capturing the unique metabolic pathway complement of each organism, and are created by automatic layout algorithms (Figure 4). The diagrams are zoomable and queryable; users can search for metabolic entities (e.g. metabolites, enzymes and pathways) by various criteria such as by name and by EC number. Search results are highlighted on the diagram to indicate their locations. An omics viewer mode allows the diagram to be painted with large-scale data sets such as gene-expression, metabolomics and reaction flux data. Such displays can be animated (for data sets containing multiple time points), and are still zoomable. Omics data can be painted programmatically using web services (38), and bookmarks can be generated to save highlighting patterns for later use. Extensive tooltips are provided to identify metabolites, reactions and pathways within the diagram on mouse rollover.
Figure 4.
The new Web Cellular Omics Viewer. This figure, showing a Cellular Omics Viewer for the bacterium E. coli, depicts the overlay of a gene expression data set (39). The level of transcription is indicated by the color of the reactions that are catalyzed by the enzymes encoded by the specific genes. The legend for mapping colors to data values is not shown in the figure. By hovering the mouse cursor over a compound or a reaction, the user can create popup windows that provide information and enable navigation to the relevant compound page or to a pathway display.
Generation of flux-balance models from PGDBs
Pathway Tools now has the ability to generate genome-scale flux-balance analysis (FBA) models from PGDBs. Our goals for this effort were to accelerate FBA model development, and to streamline the interpretation of modeling results. We achieved those goals in several ways.
In our approach, the PGDB is both a database and an executable model. Therefore, the user can query, browse and edit the metabolic model within the PGDB using the many interactive features of Pathway Tools (such as reaction and pathway editors). The user programmatically generates from the PGDB the set of linear equations that comprise the FBA model, and Pathway Tools invokes the SCIP (40) linear solver to solve those equations, and then obtains the results via the SCIP API.
Since the FBA modeling is tightly integrated with Pathway Tools, the user does not need to directly invoke the linear solver, nor inspect its output files; Pathway Tools can paint the resulting fluxes onto the Cellular Overview for visual analysis. In addition, Pathway Tools guides the user in producing a complete functional model that produces all metabolites in the biomass equation.
We have developed special capabilities within Pathway Tools for accelerating the development of FBA models using a multiple-gap-filling approach. Using past techniques, FBA models typically had development times on the order of 1 year because metabolic network models are always incomplete at the start of the model development process, and it is very time consuming to determine how to extend the model to become functional. Using the new Pathway Tools functionality, we were able to build FBA models for the EcoCyc and HumanCyc PGDBs in ∼1 month each. Pathway Tools uses a meta-optimization approach to simultaneously suggest a minimal number of alternative types of model modifications to optimize the number of metabolites in the biomass equation that the FBA model is able to produce. The software suggests new reactions to add to the model from MetaCyc, proposes reactions within the model whose directions should be reversed, and suggests additional nutrients and secreted compounds that can be added to the model. Furthermore, in contrast to other existing tools, when metabolites cannot be produced by the model, Pathway Tools identifies those compounds, allowing the user to focus model debugging efforts on specific metabolites.
The Pathway Tools FBA module also supports evaluation of single and multiple gene and reaction knock-outs; genes or reactions whose removal prevents production of any biomass component are judged to be essential. The FBA module is available only in the desktop mode of Pathway Tools, and is not accessible via Pathway Tools based websites.
Dead-end metabolite finder
The ability to identify dead-end metabolites is a valuable method for identifying errors and incompleteness in a metabolic network, for FBA modeling and other applications. Dead-end metabolites are compounds that are only produced by, or only consumed by, the metabolic network of an organism. Although such situations sometimes reflect the correct biology, they usually indicate errors in the metabolic model. A tool for identifying dead-end metabolites is available in both web (Tools → Dead End Metabolites) and desktop modes.
Choke-point finder
Metabolic choke points are metabolites that are either produced by only a single reaction in the metabolic network, or are consumed by only one reaction in the network, and were found to be enriched for anti-microbial drug targets (41). A tool for identifying metabolic choke points is available in both web (Tools → Chokepoint Reactions) and desktop modes.
Web services
Web services allow programs to query structured data from websites, and invoke web computations. Starting with version 14.5 (Fall 2010) Pathway Tools based websites provide a number of web services (see http://biocyc.org/web-services.shtml) including
Retrieving XML-structured information about individual genes, pathways, reactions, metabolites and so on,
Performing targeted queries that return XML results, such as retrieving all of the genes or metabolites within a metabolic pathway,
Executing queries in the BioVelo (42) language against a PGDB,
Highlighting sets of objects in the Cellular Overview and
Displaying omics data on the Cellular Overview, on a table of pathways, and on individual pathways.
BioCyc ortholog data
BioCyc makes extensive use of ortholog data. Examples for ortholog use in BioCyc include local alignment of a chromosome region in a multi-genome browser, an option to show the ortholog of a gene or a protein in another organism by selecting the command ‘Gene (Protein) → Show This Gene (Protein) in Another Database’, and an editor that allows propagation of annotations from one PGDB to another, across multiple genes, based on orthology. Starting with version 15.0, BioCyc ortholog information is computed in house by running NCBI BLAST pair-wise searches between all proteomes of all PGDBs. We consider orthologs as genes that are likely to be counterparts of one another in two different organisms because they are the most closely related in this pair of organisms, and we define two proteins as orthologs if they are the bi-directional best BLAST hits of one another.
Combined gene/protein/RNA pages
BioCyc and other Pathway Tools based websites previously generated separate information pages for genes and their products. However, we merged these two pages into a single page because it was confusing to users to remember which information was contained in which page, and some users never realized that both types of pages existed. Thus, a single page now provides information about genes and their protein or RNA products.
New monoisotopic mass data and search
To facilitate analysis of metabolomics data in BioCyc, we augmented the compound search form on our website to allow searching for a list of monoisotopic molecular weight values, of the type produced by high-resolution mass spectrometry (starting with release 14.5). The search can be accessed from the menu item Search->Compounds (Figure 5) and allows changing the tolerance in ppm increments The search results are presented in a table that allows easy linking to compound pages, to simplify the identification of plausible candidates for each weight value.
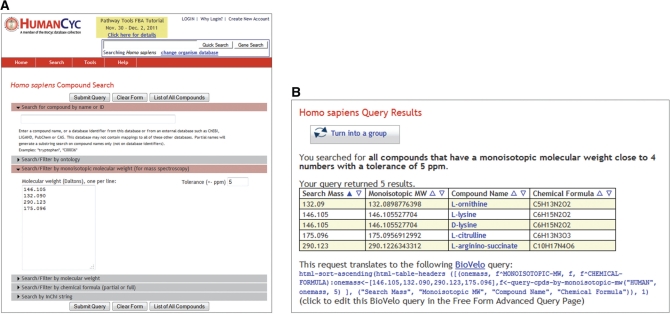
Figure 5.
(A) Searching HumanCyc for several monoisotopic molecular weights, with specified tolerance of 5 ppm. This type of search is useful for analysis of compounds identified by mass spectroscopy, enabling researchers to find candidate compounds known to exist in the organism, and to learn about their roles in the metabolic network. (B) The result of the search is a table that includes matching compounds, their monoisotopic mass, the query mass they match and their chemical formula. The compound name is a hyperlink to the compound’s page, enabling users to quickly learn about the reactions and pathways in which the compound participates in this organism.
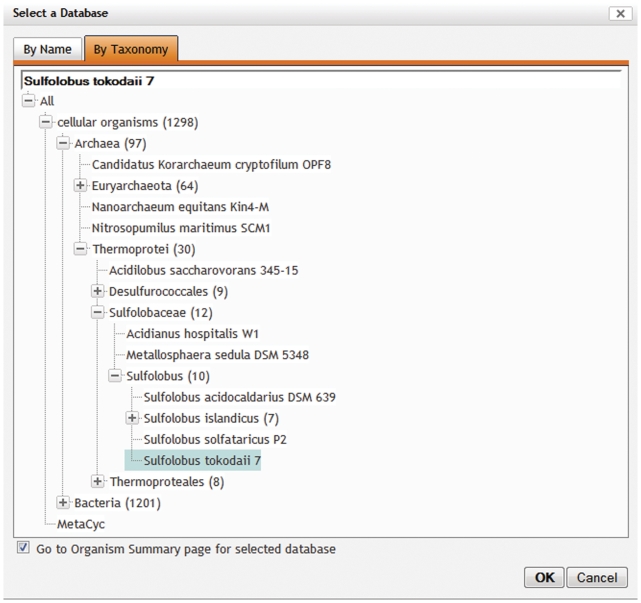
Organism selection by taxonomy
One of the challenges in designing the BioCyc website was to enable easy selection of a PGDB of interest from the large number of available databases. Previously the only selection mechanism was based on the name of the desired organism. Starting with version 15.5, it is possible to select a PGDB from BioCyc by browsing the organism taxonomy (Figure 6). In addition, the new selector window contains an option to display the Organism Summary page upon PGDB selection. This page provides background information about the PGDB such as an author list, the source for the sequence, the number and type of replicons that were used for creating the PGDB, the taxonomic lineage of the organism and relevant publications, as well as some statistics about the content of the database.
Figure 6.
The new database selector lets the user select a PGDB either by typing a name of an organism or by browsing the organism taxonomy. If the ‘Go to Organism Summary page for selected database’ box at the bottom of the selector window is checked, the software will display that page upon selection, providing background information and statistics about that database.
Ports to 64-bit Windows and 64-bit Macintosh platforms
We have ported Pathway Tools to the 64-bit Windows and Macintosh platforms. Henceforth, 32-bit versions of Pathway Tools will not be available for those platforms.
Miscellaneous enhancements
We have made many improvements to the Pathway Tools pathway layout algorithms to improve the aesthetics of pathway layouts. We have changed the color scales used in the omics viewers to improve them from a human factors perspective. We added a signaling pathway editor to Pathway Tools. We have made many performance improvements to the web mode of Pathway Tools.
How to learn more about MetaCyc and BioCyc
The BioCyc.org and MetaCyc.org websites provide several informational resources, including an online BioCyc guided tour (http://biocyc.org/samples.shtml), a guide to the BioCyc database collection (http://biocyc.org/BioCycUserGuide.shtml), a guide for MetaCyc (http://www.metacyc.org/MetaCycUserGuide.shtml), a guide for EcoCyc (http://biocyc.org/ecocyc/EcoCycUserGuide.shtml), a Pathway/Genome Database Concepts Guide (http://biocyc.org/PGDBConceptsGuide.shtml) and many webinar videos that combine narration with online demonstration of different topics (http://biocyc.org/webinar.shtml). We routinely host workshops and tutorials (on site and at conferences) that provide training and in-depth discussion of our software for beginning and advanced users. To stay informed about recent changes and enhancements to our software, join the BioCyc mailing list at http://biocyc.org/subscribe.shtml. A list of our publications is available online (http://biocyc.org/publications.shtml).
FUTURE PLANS
A variety of additional enhancements are planned. We are currently working on adding reaction atom mappings to MetaCyc and other PGDBs. Plans include the addition of many more genomes to BioCyc, including those from the Human Microbiome Project, and the addition of more types of data to BioCyc PGDBs, such as predicted GO terms and protein localizations.
DATABASE AVAILABILITY
The MetaCyc and BioCyc databases are freely and openly available to all. See http://biocyc.org/download.shtml for download information. New versions of the downloadable data files and of the BioCyc and MetaCyc websites are released four times per year.
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health (grants GM080746, GM077678, GM088849 and GM075742); Department of Energy (bioenergy-related pathway curation, grant DE-SC0004878); National Science Foundation (MetaCyc curation performed by the Plant Metabolic Network, grants IOS-1026003 and DBI-0640769). Funding for open access charge: A grant from the National Institute of General Medical Sciences of the National Institutes of Health (NIH).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This article was prepared as an account of work sponsored by an agency of the US Government. Neither the US Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe on privately owned rights. Reference herein to any specific commercial product, process or service by trade name, trademark, manufacturer or otherwise does not necessarily constitute or imply its endorsement, recommendation or favoring by the US government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the US Government or any agency thereof.
REFERENCES
- 1.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dale JM, Popescu L, Karp PD. Machine learning methods for metabolic pathway prediction. BMC Bioinformatics. 2010;11:15. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karp PD, Caspi R. A survey of metabolic databases emphasizing the MetaCyc family. Arch. Toxicol. 2011;85:1015–1033. doi: 10.1007/s00204-011-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evsikov AV, Dolan ME, Genrich MP, Patek E, Bult CJ. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo S, Lewin HA. Reconstruction of metabolic pathways for the cattle genome. BMC Syst. Biol. 2009;3:33. doi: 10.1186/1752-0509-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urbanczyk-Wochniak E, Sumner LW. MedicCyc: a biochemical pathway database for Medicago truncatula. Bioinformatics. 2007;23:1418–1423. doi: 10.1093/bioinformatics/btm040. [DOI] [PubMed] [Google Scholar]
- 11.Zhang P, Dreher K, Karthikeyan A, Chi A, Pujar A, Caspi R, Karp P, Kirkup V, Latendresse M, Lee C, et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153:1479–1491. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, et al. dictyBase - a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle MA, MacRae JI, De Souza DP, Saunders EC, McConville MJ, Likic VA. LeishCyc: a biochemical pathways database for Leishmania major. BMC Syst. Biol. 2009;3:57. doi: 10.1186/1752-0509-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May P, Christian JO, Kempa S, Walther D. ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics. 2009;10:209. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, Yoo H, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cibis E, Ryznar-Luty A, Krzywonos M, Lutoslawski K, Miskiewicz T. Betaine removal during thermo- and mesophilic aerobic batch biodegradation of beet molasses vinasse: influence of temperature and pH on the progress and efficiency of the process. J. Environ. Manage. 2011;92:1733–1739. doi: 10.1016/j.jenvman.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Scaria J, Janvilisri T, Fubini S, Gleed RD, McDonough SP, Chang YF. Clostridium difficile transcriptome analysis using pig ligated loop model reveals modulation of pathways not modulated in vitro. J. Infect. Dis. 2011;203:1613–1620. doi: 10.1093/infdis/jir112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M, Jr, et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc. Natl Acad. Sci. U.S.A. 2011;108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz JC, D’Afonseca V, Silva A, Ali A, Pinto AC, Santos AR, Rocha AA, Lopes DO, Dorella FA, Pacheco LG, et al. Evidence for reductive genome evolution and lateral acquisition of virulence functions in two Corynebacterium pseudotuberculosis strains. PLoS One. 2011;6:e18551. doi: 10.1371/journal.pone.0018551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannone RJ, Huber H, Karpinets T, Heimerl T, Kuper U, Rachel R, Keller M, Hettich RL, Podar M. Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans-Ignicoccus hospitalis relationship. PLoS One. 2011;6:e22942. doi: 10.1371/journal.pone.0022942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee R, Vats P, Dahale S, Kasibhatla SM, Joshi R. Comparative genomics of cell envelope components in mycobacteria. PLoS One. 2011;6:e19280. doi: 10.1371/journal.pone.0019280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamichhane G, Freundlich JS, Ekins S, Wickramaratne N, Nolan ST, Bishai WR. Essential metabolites of Mycobacterium tuberculosis and their mimics. MBio. 2011;2:e00301–e00310. doi: 10.1128/mBio.00301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landeta C, Davalos A, Cevallos MA, Geiger O, Brom S, Romero D. Plasmids with a chromosome-like role in rhizobia. J. Bacteriol. 2011;193:1317–1326. doi: 10.1128/JB.01184-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal S, Karimi IA, Lee DY. Flux-based analysis of sulfur metabolism in desulfurizing strains of Rhodococcus erythropolis. FEMS Microbiol. Lett. 2011;315:115–121. doi: 10.1111/j.1574-6968.2010.02179.x. [DOI] [PubMed] [Google Scholar]
- 26.Holder JW, Ulrich JC, DeBono AC, Godfrey PA, Desjardins CA, Zucker J, Zeng Q, Leach ALB, Ghiviriga I, Dancel C, et al. Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development. PLoS Genet. 2011;7:e1002219. doi: 10.1371/journal.pgen.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumann K, Dato L, Graf AB, Frascotti G, Dragosits M, Porro D, Mattanovich D, Ferrer P, Branduardi P. The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P. pastoris - a comparative analysis. BMC Genomics. 2011;12:218. doi: 10.1186/1471-2164-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol. Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues JL, Serres MH, Tiedje JM. Large-scale comparative phenotypic and genomic analyses reveal ecological preferences of Shewanella species and identify metabolic pathways conserved at the genus level. Appl. Environ. Microbiol. 2011;77:5352–5360. doi: 10.1128/AEM.00097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HU, Kim SY, Jeong H, Kim TY, Kim JJ, Choy HE, Yi KY, Rhee JH, Lee SY. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol. Syst. Biol. 2011;7:460. doi: 10.1038/msb.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Wang N. Genome-wide mutagenesis of Xanthomonas axonopodis pv. citri reveals novel genetic determinants and regulation mechanisms of biofilm formation. PLoS One. 2011;6:e21804. doi: 10.1371/journal.pone.0021804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Wang N. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol. Plant Pathol. 2011;12:381–396. doi: 10.1111/j.1364-3703.2010.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaenicke S, Ander C, Bekel T, Bisdorf R, Droge M, Gartemann KH, Junemann S, Kaiser O, Krause L, Tille F, et al. Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS One. 2011;6:e14519. doi: 10.1371/journal.pone.0014519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 35.McDonald AG, Boyce S, Tipton KF. ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009;37:D593–D597. doi: 10.1093/nar/gkn582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latendresse M, Karp PD. Web-based metabolic network visualization with a zooming user interface. BMC Bioinformatics. 2011;12:176. doi: 10.1186/1471-2105-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao H, Bausch C, Richmond C, Blattner FR, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Achterberg T. SCIP: solving constraint integer programs. Math. Program. Comput. 2009;1:1–41. [Google Scholar]
- 41.Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–924. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Latendresse M, Karp PD. An advanced web query interface for biological databases. Database. 2010;2010:baq006. doi: 10.1093/database/baq006. [DOI] [PMC free article] [PubMed] [Google Scholar]