Abstract
The National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database is a collection of genomic, transcript and protein sequence records. These records are selected and curated from public sequence archives and represent a significant reduction in redundancy compared to the volume of data archived by the International Nucleotide Sequence Database Collaboration. The database includes over 16 000 organisms, 2.4 × 106 genomic records, 13 × 106 proteins and 2 × 106 RNA records spanning prokaryotes, eukaryotes and viruses (RefSeq release 49, September 2011). The RefSeq database is maintained by a combined approach of automated analyses, collaboration and manual curation to generate an up-to-date representation of the sequence, its features, names and cross-links to related sources of information. We report here on recent growth, the status of curating the human RefSeq data set, more extensive feature annotation and current policy for eukaryotic genome annotation via the NCBI annotation pipeline. More information about the resource is available online (see http://www.ncbi.nlm.nih.gov/RefSeq/).
INTRODUCTION
RefSeq integrates an organisms’ genomic, transcript and protein sequence with descriptive feature annotation and bibliographic information (1,2). National Center for Biotechnology Information (NCBI) builds RefSeq from sequence data available in public archival sequence databases of the International Nucleotide Sequence Database Collaboration (INSDC, including the DNA Data Bank of Japan, the European Nucleotide Archive and GenBank). Unique features of the RefSeq collection includes its broad taxonomic scope, reduced redundancy, informative cross-links between nucleic acid and protein records (both curated and computationally derived) and daily curation and maintenance. Data linkages include names, protein domains, orthologs, Enzyme Commission (E.C.) numbers, phenotypes and disease. Curation and maintenance reflect new information and enable the RefSeq collection to support numerous research directions, including associating sequence with phenotype, providing a stable and consistent coordinate system to report clinical variation, comparative genomics and evolutionary studies. The RefSeq collection is a critical element of additional resources at NCBI, including dbSNP, dbVar, Gene, Genomes, Protein Clusters and Map Viewer, enabling the integration of these resources within and among organisms.
The RefSeq database is a product of NCBI, a division of the National Library of Medicine at the US National Institutes of Health. Records are freely available by multiple methods, including Internet query, FTP downloads, BLAST or scripted query using NCBI's E-Utilities. A comprehensive FTP release is available on a bi-monthly schedule with incremental daily updates provided between releases. RefSeq records can be identified by a distinct accession format which includes an underscore (‘_’) at the third position. More information is available online (http://www.ncbi.nlm.nih.gov/books/NBK21091/).
GROWTH OF THE REFSEQ DATA SET
The comprehensive bi-monthly RefSeq release continues to grow as new genome and transcript sequence become publicly available. To support the needs of different research communities, the release is provided both comprehensively in the ‘complete’ directory and based on general taxonomic groups, mitochondrial or plastid genomes or plasmid molecules. Release 49 (September 2011) includes records from 16 248 species representing 13 137 813 protein records. Table 1 indicates an annual growth of 49.7 and 14.5% in the number of organisms and the number of accessions, respectively. Records included in the release incorporate over 200 million feature annotation links (denoted ‘db_xref=’) to 60 different Web-based resources. These links allow navigation to related information from these resources, including those within NCBI, for e.g. Gene (3), the Conserved Domain database [CDD (4)], dbSNP (5) and externally, including nomenclature groups, model organism databases, protein-focused resources and many more. Links are managed by collaboration and propagation from the INSDC records upon which the RefSeq is based.
Table 1.
Annual Growth of the RefSeq release
| Release directory | Number of organisms |
Number of records |
||||
|---|---|---|---|---|---|---|
| Release 43a | Release 49 | Increase (%) | Release 43 | Release 49 | Increase (%) | |
| Complete | 10 854 | 16 248 | 49.7 | 15 934 055 | 18 236 994 | 14.5 |
| Fungi | 280 | 301 | 7.5 | 1 178 671 | 1 319 842 | 12.0 |
| Invertebrate | 637 | 754 | 18.4 | 1 993 670 | 2 232 026 | 12.0 |
| Microbial | 5585 | 10 346 | 85.2 | 9 031 974 | 10 711 822 | 18.6 |
| mitochondrion | 2266 | 2654 | 17.1 | 34 688 | 40 664 | 17.2 |
| Plant | 182 | 229 | 25.8 | 817 648 | 842 720 | 3.1 |
| Plasmid | 952 | 1061 | 11.4 | 160 065 | 191 018 | 19.3 |
| Plastid | 186 | 233 | 25.3 | 16 908 | 21 103 | 24.8 |
| Protozoa | 134 | 146 | 9.0 | 932 990 | 956 479 | 2.5 |
| Vertebrate_mammalian | 327 | 354 | 8.3 | 1 492 157 | 1 587 895 | 6.4 |
| Vertebrate_other | 1120 | 1334 | 19.1 | 398 084 | 483 449 | 21.4 |
| Viral | 2250 | 2745 | 22.0 | 87 759 | 101 664 | 15.8 |
aRelease 43 included data available on 7 September 2010; release 49 included data available on 5 September 2011.
Microbial organisms as a group account both for the greatest number of organisms and accessions in Release 49 (Table 2) and displayed the most significant annual growth in number of organisms (85.2%; Table 1). Note, however, that the number of microbial group accessions increased by only 18.6%. This value is skewed downward relative to the growth in the number of organisms or RNA records. Release 49 actually saw a 156% increase in the number of microbial RNA records (data not shown); this reflects activity of the RefSeq Targeted Locus project (http://www.ncbi.nlm.nih.gov/genomes/static/refseqtarget.html), whose mandate is to provide a single representative 16S ribosomal RNA sequence for bacterial and archaeal genomes and strains. Release 49 included 6949 organisms with a single record, 5680 organisms with more than one but fewer than 100 accessions and 184 organisms with more than 10 000 accessions.
Table 2.
Distribution of RefSeq release 49 by ftp directory
| Release directory | Percent of total |
|
|---|---|---|
| Organisms | Accessions | |
| Fungi | 1.9 | 7.2 |
| Invertebrate | 4.6 | 12.2 |
| Microbial | 63.7 | 58.7 |
| Mitochondrion | 16.3 | 0.2 |
| Plant | 1.4 | 4.6 |
| Plasmid | 6.5 | 1.0 |
| Plastid | 1.4 | 0.1 |
| Protozoa | 0.9 | 5.2 |
| Vertebrate_mammalian | 2.2 | 8.7 |
| Vertebrate_other | 8.2 | 2.7 |
| Viral | 16.9 | 0.6 |
STATUS OF CURATING HUMAN REFSEQ RECORDS
NCBI staff actively curate several subsets of the RefSeq collection for Homo sapiens. Curation improves multiple aspects of the human RefSeq collection by (i) providing quality reference sequence records for genomic regions, transcripts and proteins; (ii) maintaining and expanding functionally relevant information integrated into both RefSeq records and NCBI's Gene database; (iii) communicating and coordinating with international curation groups to generate a unified, consistent view of human genes and their primary products (see the Consensus CDS (CCDS) project, http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi); and (iv) supporting the scientific community in response to suggestions, questions or error reports.
Genomic regions
RefSeq provides region-specific genomic region records for non-transcribed pseudogenes and for the RefSeqGene project (http://www.ncbi.nlm.nih.gov/refseq/rsg/). Pseudogene loci are defined through collaboration with the HUGO Gene Nomenclature Committee [HGNC (6)] downloaded from Pseudogene.org [http://pseudogene.org/ (7)], or defined by RefSeq curation staff when reviewing transcripts having more than one high-quality alignment to the human genome. Curation involves defining the length and location of the pseudogene locus, determining whether it is transcribed and providing a link between the pseudogene locus and a functional homolog. As an example, please see NG_002746.2, which represents a eukaryotic translation initiation factor pseudogene and the ‘General gene information’ section of its Gene record (GeneID 1986) where a link to the related functional gene (EIF5A, GeneID 1984) is provided. Additionally, putative exon regions are annotated on the pseudogene RefSeq record based on alignment to a RefSeq transcript of the functional homolog. The number of non-transcribed pseudogene records increased by 7.7% in the past year.
RefSeqGene, as part of the international Locus Reference Genomic initiative [LRG (8)], provides stable, gene-specific human genomic sequence records for reporting sequence variation in medical records and locus-specific databases (see http://www.ncbi.nlm.nih.gov/refseq/rsg/). The RefSeqGene and LRG records often represent explicitly only a subset of the known mRNA and coding regions. Identification of the sequences to use as standards depends on evaluation by the user base, but usually corresponds to the RefSeq transcript and protein records that have already been curated and reviewed by RefSeq and CCDS staff. If a question arises, review of evidence from the stakeholder, the literature and sequence evidence may result in an update to a RefSeqGene record, revision of the reference transcripts and proteins annotated on the RefSeqGene record or additional splice variants to represent in the RefSeq collection before assigning the LRG identifier. Transcript variants and protein isoforms not part of the explicit annotation are represented by alignments which can be seen in NCBI's graphical display. The number of RefSeqGene records grew by 25.8% in the past year. To request a RefSeqGene for a gene, contact rsgene@ncbi.nlm.nih.gov.
Transcripts and proteins
Transcripts and proteins are an important focus for curation at NCBI. This data set has two major categories—the ‘model’ subset generated directly by NCBI's genome annotation pipeline and the ‘known’ subset maintained independently of the genome annotation process using a combination of automated analyses and manual review. These subsets can be distinguished by the accession number prefix (models begin with ‘X’) as well as by the annotation in the COMMENT block of the record when viewed in flatfile format (see http://www.ncbi.nlm.nih.gov/books/NBK21091/ for additional details). Records in the model subset are created or updated only upon whole-genome re-annotation but may be removed from the collection following manual review between such updates. The human model set was reviewed last year, which resulted in revision of the gene type designation (e.g. protein-coding, non-coding, pseudogene, etc.), replacement of model records with known records and removal of records considered to be insufficiently supported. For example, 2068 model RefSeqs met the evidence criteria to be replaced by a known RefSeq type in the 1-year period between releases 43 and 49.
A series of status codes are annotated on the known RefSeq data set to indicate information about the level of curation (these codes are not applicable for the model record subset). Records with a status of either ‘validated’ or ‘reviewed’ are considered to be curated. As of RefSeq release 49, 92.5% of the human protein coding transcripts (and their associated proteins) are tracked with a curated status, and 57.2% of the non-coding transcripts are tracked with a curated status (Table 3). This includes curation to add or update over 7500 human transcript records between releases 43 and 49. RefSeq continues to represent protein-coding regions that are considered to be full length, and transcripts that are considered to be at least near complete. Transcripts that are obviously partial are not represented but are presented in NCBI's genome browser (Map Viewer).
Table 3.
Current status of human transcripts and proteins
| Type | Accessions in Release 49 |
||
|---|---|---|---|
| Total | Curateda | Percent curated | |
| Known protein-coding transcripts | 31 933 | 29 531 | 92.5 |
| Model protein-coding transcripts | 1118 | NA | |
| Known non-coding transcripts | 5932 | 3396 | 57.2 |
| Model non-coding transcripts | 3762 | NA | |
| Total | 42 745 | 32 927 | 77.0 |
aCurated records have a review status of ‘Validated’ or ‘Reviewed’ which is not applied to model RefSeq records.
NCBI staff coordinates closely with other major databases and curation groups to maximize consistent data representation at NCBI and other web sites. The Consensus Coding Sequence collaboration [http://www.ncbi.nlm.nih.gov/projects/CCDS/CcdsBrowse.cgi (9)] is a central hub for curation of protein-coding loci and all members must agree to updates affecting the genomic coordinates of a CDS. Ambiguous or complex cases are discussed among CCDS members in light of available supporting evidence and published reports to achieve consensus on the likely annotated protein product. CCDS review often includes communication and coordination with the HGNC, UniProt or the Genome Reference Consortium [http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/ (10)] when annotation cannot be well represented due to a concern over the sequence represented in the reference human genome assembly. The human CCDS data set was updated twice in the past year, adding 2126 CCDS IDs for 456 genes. The database currently includes 26 473 distinct human protein identifiers corresponding to 18 471 genes and is available at http://www.ncbi.nlm.nih.gov/CCDS/.
In addition to ongoing review of transcripts and proteins, several changes affecting the content of RefSeq transcript and protein records were implemented recently. These include:
new policy for management of protein names;
new policy for management of readthrough (conjoined) transcripts;
expanded representation of non-coding RNAs to include microRNAs; and
expanded feature annotation for transcript and protein records.
Protein names
RefSeq is in the process of adopting UniProtKB (11) guidelines for protein naming (http://www.uniprot.org/docs/gennameprot) for both prokaryotic and eukaryotic records. Implementation of this policy is relatively new and remains variable across taxa. For prokaryotic proteins, protein name curation occurs in conjunction with NCBI's Protein Clusters resource (http://www.ncbi.nlm.nih.gov/proteinclusters). For vertebrate proteins associated in NCBI's Gene database with a related Swiss-Prot accession number, the Swiss-Prot preferred name is used verbatim. For some vertebrate RefSeq records, a distinct protein name continues to be provided if a Swiss-Prot name is not available, or one does not adhere to the revised UniProt guidelines.
Readthrough transcripts
Transcripts representing exons from what are typically considered to be neighboring, yet distinct loci pose a particular challenge for curation. This category of data resulted in conflicting annotation, requiring extensive discussion among the CCDS collaboration members, as well as HGNC. NCBI and the CCDS collaboration recently defined an annotation policy that tracks most readthrough transcript as a distinct locus as it is not solely the product of either of the two underlying loci. This approach improves consistency of the gene extent annotated for the two smaller loci while also reflecting the transcriptional complexity of the region. RefSeq opts to annotate the readthrough transcript when it appears to be full length and there is a minimum of two independent lines of support for the readthrough event. The Gene database reports this transcriptional complexity in the ‘General gene information’ section of the record. Examples can be found using available Gene queries [e.g. ‘readthrough parent’ (properties)]. In the last year, RefSeq curators reviewed the data reported in the ConjoinG database (12) to expand representation of this type. RefSeq currently tracks 120 human loci as an instantiated readthrough locus tracked with a distinct GeneID (for example, NME1-NME2, Gene ID 654364), and 358 loci with any type of readthrough association which includes reports of readthrough transcripts that do not meet the requirements to represent in RefSeq (for example, GeneID 6728).
Non-protein-coding transcripts
RefSeq representation of non-coding RNAs grew by 30% between September 2010 and 2011. Non-coding transcripts are managed in part by downloading other publicly available data sets including that available from miRBase http://www.mirbase.org/ (13). MicroRNAs, represented in RefSeq as the stem–loop precursor product with feature annotation of the functional RNA product, currently number 6848 records, 1409 of which are for human. Other types of functional RNAs, for instance small nucleolar RNAs, may be initially defined by HGNC, or by NCBI curation staff. Long non-coding transcripts, including splice variants, have been added to the database as well. Some of these include transcripts considered unlikely to encode a protein for several reasons, including non-sense-mediated decay issues, inhibitory alternative open reading frames or an alternate splice variant for which there are concerns about significant protein truncation or numerous upstream ORFs. There are currently 6057 non-coding transcripts for 4421 human genes, including 1134 non-coding transcripts for 797 human protein-coding loci.
Expanded feature annotation
RefSeq feature annotation has expanded to indicate localization or function, and to highlight details of the sequence considered during manual review. For many years, RefSeq protein records have displayed protein annotation computed by NCBI's CDD group, including protein domains, intra- or inter-molecular binding sites and metal-binding sites. While some signal peptide, mature peptide and other features have been manually annotated by NCBI staff, these features are now also propagated from UniProtKB/Swiss-Prot records and predicted by SignalP 4.0 (14). Criteria for propagation include a high-quality alignment and confirmation that the sequence and feature length are consistently maintained. Feature types that are already provided by the CDD group are not propagated. The source of the annotated feature is indicated with a ‘/inference’ qualifier which cites SignalP4.0 or with a note ‘propagated from UniProtKB/Swiss-Prot’ and an indication of the Swiss-Prot accession number (for example, see NP_001028219.1, NP_001171622.1).
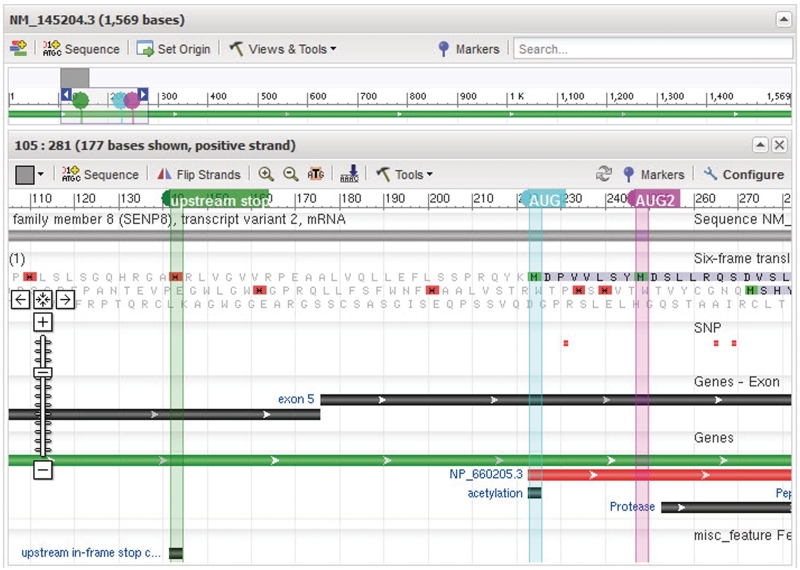
Protein-coding RefSeq transcripts now display evidence for the 5′ completeness of the annotated coding region following a computational search for an in-frame stop codon upstream of the annotated start codon. Identified stop codons are annotated with a misc_feat (see Figure 1 and NM_145204.3). Non-protein-coding transcripts, when provided for a protein-coding gene, are also computationally analysed to identify an open reading frame that shares the same start codon as a protein-coding transcript (for that gene) but that renders the transcript a candidate for non-sense-mediated mRNA (NMD) decay. Putative NMD ORFs are annotated with a misc_feat (see NR_040252.1). Misc_feat annotation is also added to a non-coding transcript if it contains an upstream ORF likely to be inhibitory to translation of the predicted ORF (see CCDS documentation at http://www.ncbi.nlm.nih.gov/CCDS/docs/CCDS-AUGguidelines.pdf and NR_003253.1 for an example).
Figure 1.
NM_145204.3 is shown in the Nucleotide Graphical display format. The display was configured to show the six-frame translation track restricted to the sense strand, and to add three markers highlighting the annotated upstream in-frame stop codon, the translation initiation codon and a second in-frame AUG codon located further downstream. The observation of a stop codon upstream of, and in the same reading frame, suggests the annotated CDS is 5′ complete.
GENOME ANNOTATION POLICY
Assembled genome sequence data are selected for inclusion in RefSeq based on several considerations including quality and completeness of a sequencing project, phylogenetic distance, model organism status, impact on disease and health studies, and identified utility to targeted research projects. Over the last several years, NCBI has developed robust whole genome annotation pipelines for both prokaryotes and eukaryotes. The prokaryotic pipeline has matured to the point that it is routinely offered as a service to submitters if genome sequences are submitted to GenBank without annotation. RefSeq genome representation for prokaryotes is currently managed by propagating annotation from the primary genome data in GenBank, calculating annotation for RefSeq when annotation is not available in GenBank within 6 months of submission of the genome, supplemented with curation to represent rRNAs, tRNAs and to provide improved protein names based on curated protein clusters. Eukaryotic genomes are managed based on general taxonomic groups, availability of submitted annotation and existence of an active model organism database. For mammalian genomes included in RefSeq, genome annotation is always provided using the NCBI annotation pipeline. For other organisms, RefSeq genome annotation is propagated from GenBank when available. Otherwise, annotation is provided using NCBI's eukaryotic annotation pipeline if a quality genome assembly is submitted with no intent to annotate, or if annotation is not submitted within a reasonable period of time, or is considered to need updating and the research group is not able to maintain it over time. When possible, the RefSeq group works with research communities and model organism databases to provide a single standard annotation for the reference genome; examples include Drosophila melanogaster, Arabidopsis thaliana, Anopheles gambiae, Saccharomyces cerevisiae and Escherichia coli K-12.
FUTURE DIRECTIONS
One of the short-term goals for the RefSeq group is to be more transparent with regard to curation decisions and support evidence used. The expanded transcript feature annotation mentioned above is a small step in this direction that will be further extended. Curators and programmers supporting the vertebrate RefSeq data set store a wide variety of gene and transcript data attributes that are potentially of use to consumers of the RefSeq data set. Attribute categories, and available stored data, are being reviewed and a subset will be selected for reporting in a structured comment on RefSeq records. Examples of stored attributes include reported RNA editing, potential alternate translation initiation codons, loci reported to be imprinted, use of non-AUG initiation codons and more. In addition, the vertebrate RefSeq group is working on reporting more explicit information about the underlying support for the exon combination that is instantiated in a RefSeq transcript record, to highlight proteins that are highly conserved, and to provide a comparison utility to evaluate putative functional consequence among transcript variants.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pruitt KD, Katz KS, Sicotte H, Maglott DR. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- 2.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karro JE, Yan Y, Zheng D, Zhang Z, Carriero N, Cayting P, Harrrison P, Gerstein M. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic acids research. 2007;35:D55–D60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalgleish R, Flicek P, Cunningham F, Astashyn A, Tully RE, Proctor G, Chen Y, McLaren WM, Larsson P, Vaughan BW, et al. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakash T, Sharma VK, Adati N, Ozawa R, Kumar N, Nishida Y, Fujikake T, Takeda T, Taylor TD. Expression of conjoined genes: another mechanism for gene regulation in eukaryotes. PLoS One. 2010;5:e13284. doi: 10.1371/journal.pone.0013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]