Abstract
The Human Gene and Protein Database (HGPD; http://www.HGPD.jp/) is a unique database that stores information on a set of human Gateway entry clones in addition to protein expression and protein synthesis data. The HGPD was launched in November 2008, and 33 275 human Gateway entry clones have been constructed from the open reading frames (ORFs) of full-length cDNA, thus representing the largest collection in the world. Recently, research objectives have focused on the development of new medicines and the establishment of novel diagnostic methods and medical treatments. And, studies using proteins and protein information, which are closely related to gene function, have been undertaken. For this update, we constructed an additional 9974 human Gateway entry clones, giving a total of 43 249. This set of human Gateway entry clones was named the Human Proteome Expression Resource, known as the ‘HuPEX’. In addition, we also classified the clones into 10 groups according to protein function. Moreover, in vivo cellular localization data of proteins for 32 651 human Gateway entry clones were included for retrieval from the HGPD. In ‘Information Overview’, which presents the search results, the ORF region of each cDNA is now displayed allowing the Gateway entry clones to be searched more easily.
INTRODUCTION
In the post-genomic period, one of the most essential areas of research involves the functional and structural analysis of gene products (proteins). The key element in functional genomics studies is the acquisition of full-length cDNA clones. For this purpose, a number of projects such as the Japanese FLJ project, supported by New Energy and Industrial Technology Development Organization (NEDO) (1–3), and the Kazusa long cDNA project (4,5) have been implemented for the isolation of as many full-length cDNAs as possible and at the highest quality (6,7). To build an infrastructure that facilitates the systematic and comprehensive expression of human proteins, in addition to the availability of full-length cDNA clones, it is vital to have a versatile system for making use of these clones. The Gateway cloning system (Invitrogen, CA, USA) is based on such versatile expression vectors (8); therefore, we adopted this system and subsequently constructed 33 275 human Gateway entry clones from full-length cDNA (9). Accordingly, the Human Gene and Protein Database (HGPD; http://www.HGPD.jp/) was launched in November 2008 (10). Sequence information and protein expression data for the Gateway entry clones can also be retrieved from the HGPD, making it a unique database.
During the post-genomic period, research objectives have focused on the development of new medicines and the establishment of novel diagnostic methods and medical treatments. In addition, a large number of studies using proteins and protein information, which are closely related to gene function, have been undertaken. Thus, in further investigations, the combination of resources and protein information has an important role in accelerating research (11). The HGPD is a unique resource that stores information on a set of human genes, proteins and Gateway entry clones in addition to protein expression and synthesis data.
In this update of the HGPD, we improved sequence information and protein expression data for the human Gateway entry clones. Moreover, we generated new data for the functional classification and subcellular localization of proteins produced by the human Gateway entry clones. In the database interface, the open reading frame (ORF) of each cDNA sequence is visualized in ‘Information Overview’, thereby, allowing users to search the entry clones more easily. In addition, we also redesigned the entire web interface.
DATA IMPROVEMENT
In this update, we improved the sequence information and protein expression data for the Gateway entry clones. In the HGPD, biological data such as the in vitro expression data of human proteins are presented on the frame of cDNA clusters. To build the basic frame, cDNA sequences from FLJ and other public databases (Human ESTs, RefSeq, Ensembl, MGC and so on) are assembled onto the genome sequences. The sequence information of the Gateway entry clones is presented with the source cDNA.
Gateway entry clones
To facilitate the use of full-length cDNA clones, we adopted the versatile Gateway expression system that offers high-throughput gene transfer technology for functional gene analysis and protein expression. For conversion to entry clones, we selected an ORF region in each cDNA that meets one of the following criteria: (i) ORF encoding a product >150 amino acids (although the longest ORF starting with an AUG codon has the highest priority, the selected ORF is finally determined by taking into consideration homology search results of shorter ORFs with BLASTX(nr) and BLASTP against the SwissProt and RefSeq databases); (ii) both 149 amino acids > ORF > 100 amino acids and an ORF with an ATGpr value (12) >0.4 and (iii) both 100 amino acids > ORF and a known gene. The ORF regions were then PCR-amplified with the attB1 and attB2 sequences of the Gateway system at both ends and recombined with the attP1 and attP2 sequences of the Gateway pDONR201 donor vector (Invitrogen) (for details, see http://riodb.ibase.aist.go.jp/hgpd/sys_info/help.html#w120_gw). In this update, we constructed an additional 9974 Gateway entry clones (corresponding to approximately 2000 loci), giving a total of 43 249 clones (Table 1). A sequence summary and the nucleotide acid and amino acid sequence data for these clones (v 2.0) can be downloaded from http://riodb.ibase.aist.go.jp/hgpd/sys_info/download.html. This resource of human Gateway entry clones was named HuPEX. If you inquire our Gateway entry clones, you can order them by using Gateway entry clone IDs which are green character in ‘Protein Info’.
Table 1.
Improvement of entries of HGPD
| Data set | V. 1.0 | V. 2.0 (in this update) |
|---|---|---|
| Gateway entry clones | 33 275 (14 590 locia) | 43 249 (16 484 locia) |
| (N-type: 12 754) | (N-type: 17 802) | |
| (F-type: 20 521) | (F-type: 25 447) | |
| In vitro protein expression (SDS–PAGE) patterns | 13 364 | 17 821 |
| In vivo subcellular localization image | – | 32 651 |
aNumbers of locus in HGPD are represented.
SDS–PAGE patterns of human proteins synthesized in vitro
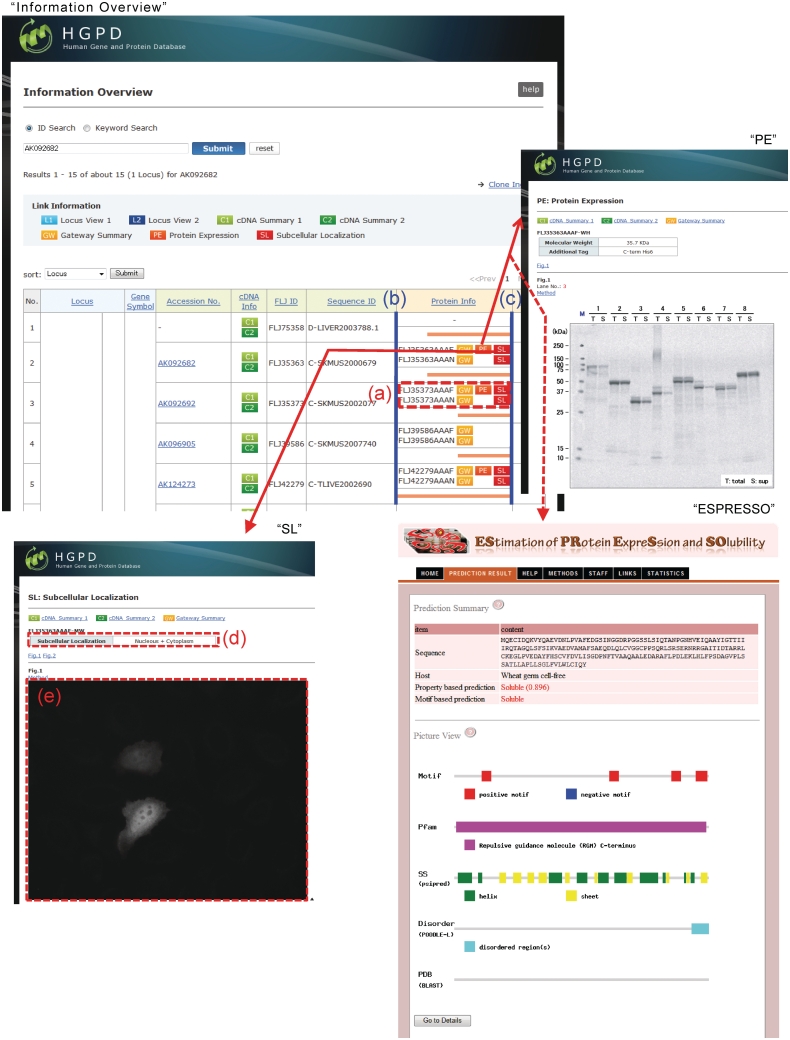
The Gateway system is a versatile expression vector system that can adequately handle large numbers of clones. For the expression of human proteins, we adopted the wheat germ cell-free protein synthesis system (13). In this update, we additionally expressed 4457 human proteins with a C-terminal V5 or His tag and analyzed them using SDS–PAGE (Table 1). The expression data from a total of 17 821 human proteins are stored in the HGPD. These expression patterns are displayed on the ‘PE: Protein Expression’ page (Figure 1; for details, see http://riodb.ibase.aist.go.jp/hgpd/sys_info/help.html#w120_pe).
Figure 1.
Improved display of the HGPD. In ‘Information Overview’, (a) open reading frames (ORFs) of each cDNA clone can be visualized. In ‘Protein Info’, the left endpoint represents the N-terminus of each ‘Locus’ (b) and the right endpoint represents the C-terminus (c). ‘SL’, the subcellular localization of human proteins, is linked by the ‘SL’ button in ‘Protein Info’. (d) Protein subcellular localization by visual observation of the images. (e) The subcellular localization image. ‘PE’ or ‘ESPRESSO’ information is linked by the ‘PE’ button in ‘Protein Info’.
RECENT DEVELOPMENTS
In this update of the database, we provided new classification information for the protein function and subcellular localization of the Gateway entry clones.
Classification under protein function
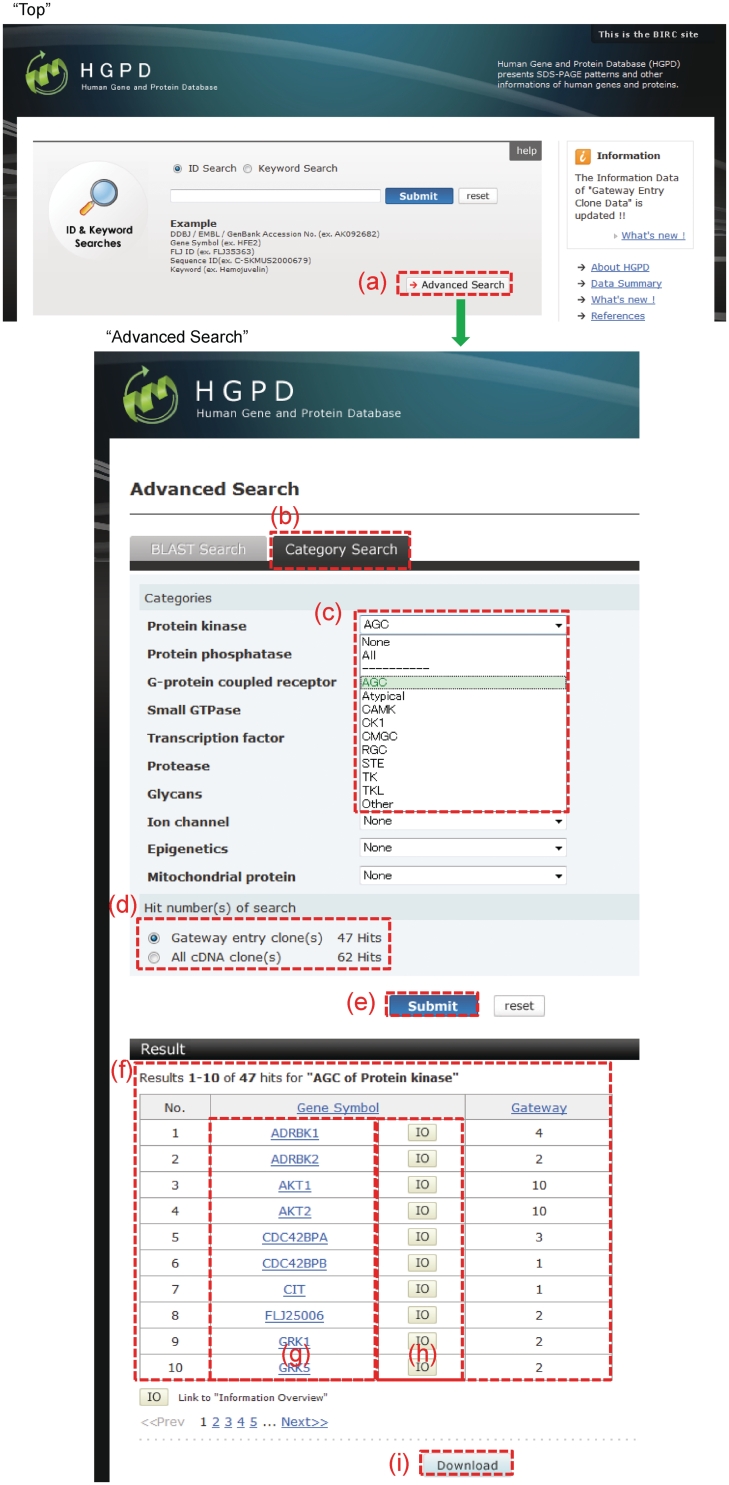
To facilitate searching of the Gateway entry clones, we considered the addition of useful annotation within the HGPD. In the HGPD, certain information on the cDNA clones and Gateway entry clones can be accessed on the ‘Information Overview’ page. We previously stored sequence information and homology search results (i.e. BLAST, Pfam, PROSITE, SignalP, SOSUI and GO) for each cDNA clone. Then, we focused on the protein function of the Gateway entry clones, and matched the entry clones to RefSeq clones using a homology search (BLAST). On the basis of the homology search results, we were able to match the Gateway entry clones to the annotations in public databases (i.e. NCBI Entrez Gene, HPRD, Swiss Prot, OMIM and GO) and published data. In this update, we classified the entry clones into 10 groups according to protein function; these groups were included in the HGPD (Table 2). For example, 407 gene symbols (genes) were classified as ‘Protein kinase’, representing 1322 Gateway entry clones within HuPEX. Thus, a total of 4000 gene symbols (genes) were classified, representing 10 684 Gateway entry clones within HuPEX. These classifications can be retrieved from ‘Category Search’ under ‘Advanced Search’ in the HGPD (Figure 2).
Table 2.
Classification under protein function
| Category | Numbers of gene symbol | Numbers of Gateway entry clone |
|---|---|---|
| Protein kinase | 385 | 1322 |
| Protein phosphatase | 84 | 291 |
| G-protein coupled receptor | 208 | 405 |
| Small GTPase | 134 | 255 |
| Transcription factor | 1704 | 4319 |
| Protease | 399 | 1104 |
| Gricans | 159 | 405 |
| Ion channel | 177 | 395 |
| Epigenetics | 171 | 519 |
| Mitochondrial protein | 637 | 1669 |
Figure 2.
Category Search. This interface is accessed through ‘Advanced Search’ under ‘Top View’ (a). (b) A category search is initiated by clicking the ‘Category Search’ tab. (c) In each category, only one item (term) can be selected. (d) The number of hits from a selected item (term) is then counted, and (f) displayed by clicking the ‘Submit’ button (e). (h) The results are then linked to ‘Information Overview’ by clicking ‘IO’. (g) The ‘Gene Symbol’ interface is linked to NCBI Entrez Gene. (i) The list of search results is obtained by clicking the ‘Download’ button.
Subcellular localization of human proteins fused with fluorescent proteins
To construct image data for the subcellular localization of proteins, 32 651 entry clones were used for the overexpression of proteins with fluorescent protein's tag at the N- or C-terminus in HeLa cells. In this subcellular localization study, there are some differences between the results of native proteins and those of fluorescent fusion proteins (14,15). Therefore, the image data for the subcellular localization of ectopic proteins are constructed in HGPD. These data are displayed in the ‘SL: Subcellular Localization’ page (Figure 1; for details, see http://riodb.ibase.aist.go.jp/hgpd/sys_info/help.html#w120_sl). The protocols of subcellular localization analysis are explained in the method page linked from ‘Method’ button at ‘SL: Subcellular Localization’. The kinds of subcellular localization are shown at ‘SL: Subcellular Localization’ on ‘Help’.
NOVEL DATABASE SERVICES
Category search
In this update, we constructed additional classification information for the Gateway entry clones (Table 2). The interface ‘Category Search’ was developed so that users could obtain useful information on the entry clones (Figure 2). The ‘Category Search’ view is displayed within ‘Advanced Search’ in the ‘Top’ page of the HGPD (Figure 2a), while the ‘Category Search’ tab (Figure 2b) is selected from the ‘Advanced Search’ view. In ‘Categories’, only one category item (term) can be selected (Figure 2c). Each category item (term) has a second level of category items (terms). The number of hit results for gene symbols during a search is counted automatically (Figure 2d), where ‘Gateway entry clone(s)’ counts the number of hit results for gene symbols for which Gateway entry clones have been made. ‘All cDNA clone(s)’ counts the number of all hit results for gene symbols. A results table is then displayed under ‘Results’ (Figure 2f) by clicking the ‘Submit’ button (Figure 2e) after selecting the radio button of ‘Gateway entry clone(s)’ or ‘All cDNA clone(s)’. From this results table, the details of each result are linked to ‘Information Overview’ by clicking ‘IO’ (Figure 2h). The gene symbol provided for each result links to NCBI Entrez Gene (Figure 2g). Moreover, the list of search results is obtained by clicking the ‘Download’ button (Figure 2i).
Visualization of ORFs
In ‘Information Overview’, the ORF of each cDNA clone is visualized in the ‘Protein Info’ column as an orange line (Figure 1a). For the displayed ORF, the left endpoint (Figure 1b) represents the N-terminus of each ‘Locus’ and the right endpoint (Figure 1c) represents the C-terminus. It should be noted that if multiple ‘Locus’ information is indicated by a search in ‘Information Overview’, the left and right endpoints of each ‘Locus’ are different. By visualizing the ORF of each cDNA sequence, a rough homology among the cDNA sequences at each ‘Locus’ can be confirmed.
Images of subcellular localization of human proteins fused with fluorescent proteins
We localized 32 651 proteins of the human Gateway entry clones. The localized images of the proteins are displayed in the ‘SL: Subcellular Localization’ page (Figure 1; ‘SL’). Each subcellular localization is determined by visual observation of the images (Figure 1d), and the actual subcellular localization image is displayed (Figure 1e).
Linkage with other public databases that predict protein expression
In the HGPD, a total of 17 821 human protein expression patterns for the Gateway entry clones have been stored (Figure 1; ‘PE’); however, in this update, not all human protein expression patterns have yet been entered in the database. ESPRESSO (Computational Biology Research Center, National Institute of Advanced Industrial Science and Technology, http://mbs.cbrc.jp/ESPRESSO/), which predicts protein expression and solubility from sequence information, has been launched. To predict the protein expression of the Gateway entry clones that have not been characterized, we linked the HGPD to ESPRESSO using the ORF sequences of the human Gateway entry clones, allowing us to predict the protein expression and solubility of these clones (Figure 1; ‘ESPRESSO’).
IMPROVEMENT OF THE DATABASE SERVICE
In this update, the following database services are improvement.
Redesigned the entire web interface.
Addition of the function of sort for ‘Gene Symbol’ in ‘Information Overview’.
FUTURE DEVELOPMENTS
In the future, we hope to develop the following two areas of the HGPD. One is to further classify the protein function of the Gateway entry clones. The other is to link the accession numbers of the cDNA clones and their gene symbol to their protein accession numbers (e.g. NCBI reference sequence NP and XP numbers, Swiss-Prot entry names and accession numbers and so on), and to improve the ‘ID search’ tool by including protein accession numbers.
FUNDING
Research Information Database of the Tsukuba Advanced Computing Center of AIST (Human Gene and Protein Database); National Bioscience Database Center and Database Center for Life Science (mirror site of Human Gene and Protein Database) and NEDO project, ‘Functional Analysis of Human Proteins and its Application (2001–2005)’ and ‘Development of Basic Technology to Control Biological Systems Using Chemical Compounds (2006–2010)’. Funding for open access charge: AIST.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the Helix Research Institute and the Research Association for Biotechnology for the FLJ cDNA clones.
REFERENCES
- 1.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 2.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi OK, Barrero AR, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:0001–0020. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 5.Ohara O, Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Nomura N. Construction and characterization of human brain cDNA libraries suitable for analysis of clones encoding relatively large proteins. DNA Res. 1997;4:53–59. doi: 10.1093/dnares/4.1.53. [DOI] [PubMed] [Google Scholar]
- 6.Temple G, Lamesch P, Milstein S, Hill DE, Wagner L, Moore T, Vidal M. Proteome: developing expression clone resources for the human genome. Hum. Mol. Genet. 2006;15:R31–R43. doi: 10.1093/hmg/ddl048. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Boehm J, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas S, Alkan O, Bhimdi T, et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartley J, Temple G, Brasch M. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–7895. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goshima N, Kawamura Y, Fukumoto A, Miura A, Honma R, Sato R, Wakamatsu A, Yamamoto J, Kimura K, Nishikawa T, et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- 10.Maruyama Y, Wakamatsu A, Kawamura Y, Kimura K, Yamamoto J, Nishikawa T, Sugano S, Goshima N, Isogai T, Nomura N. Human Gene and Protein Database (HGPD): a novel database presenting a large quantity of experiment-based results in human proteomics. Nucleic Acid Res. 2009;37:D762–D766. doi: 10.1093/nar/gkn872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa T, Ota T, Isogai T. Prediction whether a human cDNA sequence contains initiation codon by combining statistical information and similarity with protein sequences. Bioinformatics. 2000;16:960–967. doi: 10.1093/bioinformatics/16.11.960. [DOI] [PubMed] [Google Scholar]
- 13.Sawasaki T, Ogasawara T, Morishita R, Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl Acad. Sci. USA. 2002;99:14652–14657. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Ehrhardt DW, Rhee SY. Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length arabidopsis proteins. Plant Physiol. 2006;141:527–539. doi: 10.1104/pp.106.078881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian G-W, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al. High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 2004;135:25–38. doi: 10.1104/pp.104.040139. [DOI] [PMC free article] [PubMed] [Google Scholar]