Abstract
In animals, RNA binding proteins (RBPs) and microRNAs (miRNAs) post-transcriptionally regulate the expression of virtually all genes by binding to RNA. Recent advances in experimental and computational methods facilitate transcriptome-wide mapping of these interactions. It is thought that the combinatorial action of RBPs and miRNAs on target mRNAs form a post-transcriptional regulatory code. We provide a database that supports the quest for deciphering this regulatory code. Within doRiNA, we are systematically curating, storing and integrating binding site data for RBPs and miRNAs. Users are free to take a target (mRNA) or regulator (RBP and/or miRNA) centric view on the data. We have implemented a database framework with short query response times for complex searches (e.g. asking for all targets of a particular combination of regulators). All search results can be browsed, inspected and analyzed in conjunction with a huge selection of other genome-wide data, because our database is directly linked to a local copy of the UCSC genome browser. At the time of writing, doRiNA encompasses RBP data for the human, mouse and worm genomes. For computational miRNA target site predictions, we provide an update of PicTar predictions.
INTRODUCTION
The regulation of gene activity on the RNA level has been at the heart of intensive research efforts since the description of the operon (1). Post-transcriptional regulation is highly versatile and adaptable by controlling RNA availability in cellular time and space. Messenger RNA stability, transport, storage and translation are largely determined by the interaction of mRNA with microRNAs (miRNAs) and RNA-binding proteins (RBPs). We have just begun to understand the extent and dynamics of transcriptome-wide binding events that lead to the temporal formation of functional ribonucleoprotein complexes.
Within doRiNA, we focus on two key players of post-transcriptional regulation: miRNAs and RBPs.
microRNAs
miRNAs originate from long stem–loop containing primary transcripts (pri-miRNAs) that are generally transcribed by RNA Polymerase II. pri-miRNAs are substrates of the RNAse III enzyme Drosha and its binding partner, the dsRNA-binding protein DGCR8/Pasha. In the nucleus, a complex of Drosha and DGCR8 cleaves pri-miRNAs into ∼70 nt precursor hairpins (pre-miRNA), which are exported to the cytoplasm. In the cytoplasm, the pre-miRNA is further cleaved by another RNAse III enzyme Dicer into a mature miRNA and its partner strand, the miRNA* (microRNA star). The mature miRNA is defined as the strand, which is loaded into the RNA-Induced Silencing Complex (RISC) complex. Krol et al. (2) give an excellent overview on miRNA biogenesis.
The mature miRNA identifies its mRNA target by binding to partially complementary sites within 3′UTRs (3), resulting in mRNA degradation and translational repression of the RNA target (4). This drastically differs from the short-interfering RNA mechanism, which requires perfect complementarity, and leads to RNA-directed cleavage of the target transcript.
The doRiNA database offers computational miRNA target site predictions for man, mouse and worm. These predictions constitute the long awaited update of PicTar predictions (5–7).
RNA-binding proteins
Nascent RNAs are co-transcriptionally bound by RBPs leading to the formation of ribonucleoprotein complexes. RBPs are characterized by containing one or multiple RNA recognition domains (RBDs), which cooperate to recognize RNA sequences (8). RBPs do not only recognize simple RNA sequence motifs, but can also integrate the structural context into the recognition process. This becomes evident for the simple case of double-strand binding as opposed to single-strand binding RBPs.
The in silico prediction of RBP target sites is still in its infancy (9). That is why we decided to exclude computational predictions and constrain our data set to RBP target sites from high-resolution, transcriptome-wide cross-linking and immunoprecipitation (CLIP) experiments (10). One variant of CLIP, called PAR-CLIP (photoactivatable-ribonucleoside enhanced cross-linking and immunoprecipitation), relies on the incorporation of photo-reactive nucleotide analoga into newly synthesized RNA (11). Successful incorporation and cross-linking induces characteristic base substitutions in the sequenced cDNA reads. These base substitutions support target site identification at nucleotide-level resolution. For example, an incorporation of 4-thiouridine into RNA and subsequent cross-linking yields characteristic T → C base transitions in sequencing reads.
RBPs binding sites from our own experiments are all based on the PAR-CLIP method and were processed in the same way (see ‘Materials and Methods’ section for details). Additionally, we collect and integrate target sites from published HITS-CLIP experiments (12) and other variants into doRiNA as long as they provide precise positional target site information.
Entering doRiNA
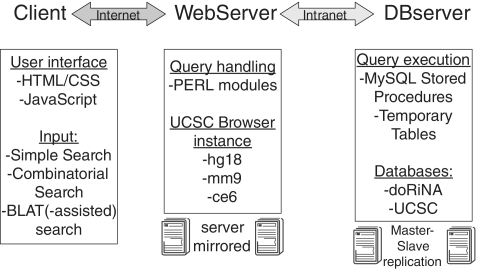
The doRiNA database integrates miRNA and RBP target site sets from different species into one framework. We have mainly turned our attention to service availability, query speed and query capability. Service availability is achieved by mirroring the web and database servers (Figure 1). We enable high query speed and complexity by pre-computing several important data characteristics. In doRiNA, users are able to enter the available post-transcriptional regulatory network from a target centric (‘Which regulators target gene X?’) or regulator centric (‘Which genes are regulated by Y?’) view. Complex queries using set operations over subregions of genes (e.g. 3′ UTR or 5′ UTR) have been realized without compromising speed. We deem doRiNA a one-stop solution to transcriptome-wide mining of regulatory interactions in post-transcriptional gene regulation.
Figure 1.
Schematic doRiNA overview. The doRiNA concept is implemented as a linear chain of three: client, web server and database server. We mainly turned our attention to service availability, query speed and query capability.
MATERIALS AND METHODS
The doRiNA infrastructure
To ensure short query response times, we have setup a powerful 12-core web server, which is coupled to a dedicated 8-core MySQL 5.1 database server. A local installation of the UCSC genome browser (13) was directly placed onto the web server. The database server handles requests from the doRiNA user interface as well as from the local UCSC browser installation. Result sets are returned in tabular form (for browsing or download) via the web server and are depicted within the genome browser on a locus-by-locus basis. An overview on the infrastructure is given in Figure 1.
Web server
On the client side, the doRiNA web interface is implemented as a blend of HTML, CSS and Javascript components. The Javascript component utilizes the JQuery library (http://jquery.com/) and the JSON data interchange format (http://www.json.org/). The doRiNA web service is divided into individual species sections (man, mouse and worm). Each species section has a tripartite structure: Simple Search, Combinatorial Search and BLAT(-assisted) search. The user is assisted in formulating a query by several features: autocompletion of gene names, gene name validity checks, conditional activation of web interface control elements and extensive help documents. Search requests can only be submitted if the user input has passed these initial checks. Search requests are processed on the server side by custom PERL modules and scripts. The PERL layer mainly interfaces with the database engine where the actual queries are executed by MySQL Stored Procedures.
Database server
The doRiNA database is built on top of the UCSC genome browser databases. To this end, we have added custom tables to species-specific databases (e.g. hg18). These tables contain precomputed information (e.g. host gene) for each target site to speed up queries. We have put the main work horse of doRiNA, MySQL Stored Procedures in combination with temporary tables, into a separate database (Figure 1). Search requests trigger the execution of cascading stored procedures, which assemble result tables in memory, send them back via the PERL layer to the client and subsequently discard the temporary tables. Concurrent user access is guaranteed by a database inherent session management.
Target sites of RNA-binding proteins
One central question has not been addressed yet: how do we collect and integrate target site information? For RBPs, we follow a 2-fold strategy: first, PAR-CLIP data sets from Hafner et al. (11) and data sets that were produced in-house (i.e. by one of the co-authors) are subject to a processing pipeline (PCP, details see below). This pipeline infers target sites based on a nucleotide conversion score and an entropy measure over read stacks in continuously covered transcript regions (read clusters).
Second, other CLIP data sets (HITS-CLIP, PAR-CLIP, iCLIP and variants thereof) are retrieved from external publications and integrated as is. Interested users find details on data acquisition and processing in the corresponding UCSC genome browser track descriptions.
Analysis pipeline for in-house PAR-CLIP data
All in-house PAR-CLIP tracks were produced with our computational pipeline to determine RBP binding sites at an estimated 5% false positive rate (14). The pipeline performs all steps of the PAR-CLIP analysis taking raw reads and producing cluster sets and lists of target genes, in a largely automated and unbiased way. The emphasis is on stringent filtering and controlling the false positive rate in the identification of binding sites.
Briefly, PAR-CLIP reads are aligned to the human transcriptome (mRNAs or pre-mRNAs) or genome (user choice), allowing for up to one mismatch, insertion or deletion. Only uniquely mapping reads are retained.
Next, we identify clusters of aligned PAR-CLIP reads continuously covering regions of reference sequence and assign two quality scores based on the characteristics of the PAR-CLIP protocol. Efficient cross-linking leads to specific nucleotide conversion events during reverse transcription and next-generation sequencing of RNA from each experiment: cross-linked 4-thiouridine (4SU) and 6-thioguanosine (6SG) residues are converted into C and A, respectively. These conversions mark the RBP binding site on the target RNA (11). The number of these mismatches therefore serves as a cross-link score. The other score addresses problems that may be encountered in a sequencing-based assay: we assign an entropy score based on the number and positions of distinct reads contributing to the cluster to guard against PCR or mapping artifacts.
Finally, the pipeline automatically selects cutoffs on both quality scores by using the reverse complement of the annotated transcripts as a decoy. As PAR-CLIP reads should originate from RBP-bound transcripts, we may regard clusters aligning antisense to the annotated direction of transcription as false positives. We are thus able to select cutoffs on the estimated false positive rate. After filtering by these cutoffs, remaining antisense clusters are dropped. We expect each retained cluster to harbor at least one RBP binding site with a false positive probability ≤5%.
Additional details can be found in Supplementary data.
External target site data
We have collected several published CLIP data sets from the literature (see web site for details). Target sites were either extracted from Supplementary data or obtained from the corresponding author. Some authors did not assign a score to each target site, which does not allow a score-base ranking of these sites. In that case, target sites are assigned a default score and the rank is set to N/A.
PicTar miRNA target site predictions
We have updated the PicTar miRNA target site predictions to the respective UCSC genome releases of man (hg18), mouse (mm9) and worm (ce6). PicTar 2.0 (7) predicts miRNA target sites in 3′ UTRs and utilizes multiple genome sequence alignments to boost its precision. Briefly, all 3′ UTR alignments for a given species set are scanned for perfect and imperfect seed sequences. Perfect seeds consist of a 7 nt perfect match starting at position 1 or 2 from the 5′-end of a mature miRNA. Imperfect seeds contain one insertion/deletion or mismatches to the 3′ UTR sequence. All candidate sites are subject to probabilistic scoring by an Hidden Markov Model (HMM).
For example, human miRNA targets for mature and star sequences from Mirbase v16 were predicted based on UCSC's 44-way Vertebrate Genome alignment. We have incorporated three conservation levels for human target sites into doRiNA: (i) Mammals, chicken and fish—seed conservation across Pan troglodytes, Mus musculus, Rattus norvegicus, Canis lupus, Gallus gallus, Fugu rubripes and Danio rerio. (ii) Mammals, chicken—seed conservation is not required in Fugu rubripes and Danio rerio. (iii) Mammals—seed conservation is not required in Gallus gallus, Fugu rubripes and Danio rerio.
These conservation levels provide a convenient way to choose the optimal sensitivity level while controlling for false positives.
Integration with the UCSC Genome Browser
All target site information for miRNAs or RBPs are integrated into our local installation of the UCSC genome browser as additional local tracks. This guarantees full access to all genome browser features and simultaneous availability of other genome browser tracks (Variation, Regulation and other tracks). In addition, the genome browser interface is commonly used by biologists world-wide and does not require any additional training.
EXAMPLE APPLICATIONS
In the following section, we will present a few example applications of doRiNA. These examples serve as an entry point to doRiNA and outline three main use cases.
Target centric queries—retrieve all regulators of a predefined gene set.
Regulator centric queries—retrieve all genes that are targeted by a predefined set of regulators.
Complex / Combinatorial queries—set operations on regulator target gene sets.
Target centric queries
Several questions in biology focus on a particular gene or gene set of interest. Frequently, questions like ‘which regulators target my gene or gene set of interest?’ arise in scientific discussions. We denote these kind of queries as ‘target centric’.
Setting up the query
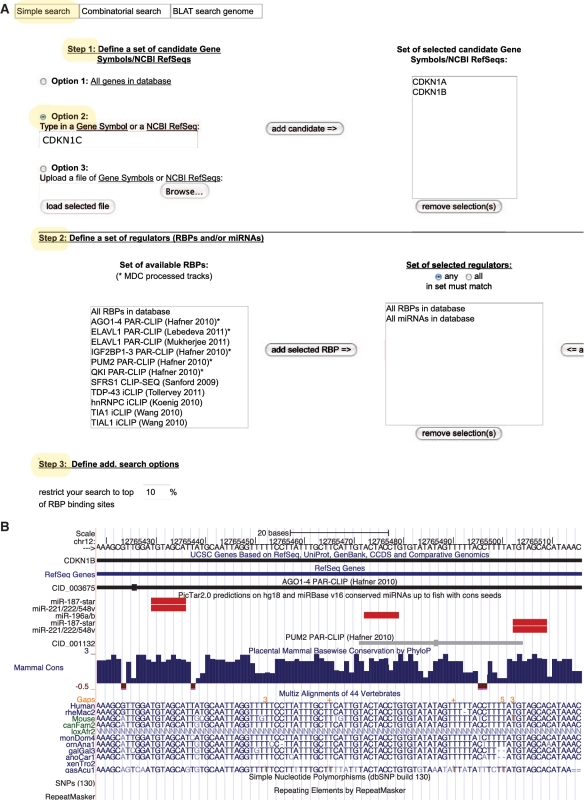
Generally, doRiNA accepts gene symbols and NCBI RefSeq identifiers to define target gene sets. The Simple Search Function (Figure 2A), which is used in this context, offers two different approaches to compile candidate gene lists. The user could either manually define a subset of genes/transcripts (Option 2 in Step 1) or upload a list of gene identifiers (Option 3 in Step 1). For completeness, Option 1 selects the complete available gene set in the corresponding species databases. By using one of these options, the user defines a gene set of interest and subsequently selects post-transcriptional regulators (RBPs and/or miRNAs) to match against (Step 2). All available regulators are conveniently selected by the ‘All RBPs in database’ and ‘All miRNAs in database’ records. A score-based ranking cutoff for RBP target sites is finally set in the last step (Step 3) of the user interface. The search submission button becomes activated if all input passes the online syntax checks.
Figure 2.
Example of a target centric query — the ‘Cip/Kip’ family. (A) Web interface — Simple Search Tab. (B) Visualization of the 3′ UTR of CDKN1B.
Interpretation of results
Search results are reported back in tabular format. A summary on the number of found target sites and genes is shown at the top of the results page. Each table row corresponds to one target site and contains information in a self-explanatory format. Please note that each column can be used to sort the entire tables. If target site scores are provided for a CLIP experiment, we use them to order the table output via the column (top-percent value). Otherwise, the score is set to a default value. Each row offers links to the UCSC genome browser for the entire gene locus (gene symbol location) or the corresponding target site (target site location).
Example: the CDKN1 gene family
Let us assume that we are interested in RBPs as post-transcriptional regulators of the ‘Cip/Kip’ family, which encompasses cyclin-dependent kinase inhibitor 1 coding genes (CDKN1). Intriguingly, Kedde et al. (15) have shown that p27 (CDKN1B) is post-transcriptionally regulated by mir-221 and mir-222 conditional on an Pumillio-induced RNA structure switch. We already know that there are only three gene family members (CDKN1 A to C). That is why we enter these three gene names manually via option 2 in Step 1 of the Simple Search Tab. We are assisted by the autocompletion function of doRiNA. Since we are interested in any regulator of at least one of the three CDKN1s, we select all RBPs and all miRNAs in database in Step 2. We increase search sensitivity to the maximal level by setting the RBP score rank percentile cutoff to 100%. All other settings are left untouched (default values).
The result page summarizes all query results: there are 209 target sites in total of which 194 are RBP target sites and 15 are conserved miRNA target sites (mammals–chicken–fish). Indeed, two mir-221/222 sites and three PUM2 sites have been reported for CDKN1B in the results table. We navigate to the corresponding UCSC view by clicking on one CDKN1B location link, which opens up an UCSC genome browser view that nicely recapitulates the published target site configuration (Figure 2B).
Users may also inspect individual PAR-CLIP target sites at nucleotide-level resolution by clicking on them. This opens a summary page, which links to an in-depth read cluster display where characteristic mutations (e.g. T → C) are indicated in the corresponding sequencing reads.
Regulator centric queries
In a different application context, scientists frequently have to define the target gene set of a particular regulator or set of regulators. More specifically, one could be interested in either genes that are co-targeted by all selected regulators (intersection of target sets) or just at least one of the selected regulators (union of target sets). We refer to this view as regulator-centric. The difference in search strategy is mainly to leave the target gene set unconstrained (Option 1) and select a confined set of regulators. The set of regulators is conveniently defined from two available lists: one for RBPs and one for miRNAs. The simple search function provides two set operations (all ≡ intersection and any ≡ union) on the selected list of regulators. A radio button toggles between the intersection and union modes.
We will continue with our previous example of the PUM2 & mir-221/222 module and search for all its co-targets.
Example: co-targets of PUM2 and miR-221 / miR-222
We retrieve the requested co-targets by selecting the entire gene set in the database (Option 1). The regulator set is constrained to PUM2 and mir-221/222 in step 2. Since we are looking for co-targets, we switch to the intersection mode by choosing the ‘all’ radio button. Subsequently, we set the RBP score rank percentile cutoff to 100% and leave all other settings untouched. Our query returns 13 target genes (data not shown, one is CDKN1B). We repeat this search with relaxed miRNA conservation criteria and either obtain 60 co-target genes for mammal–chicken conserved miRNA sites or 141 co-targets for mammal-only conserved sites, respectively.
Combinatorial search options
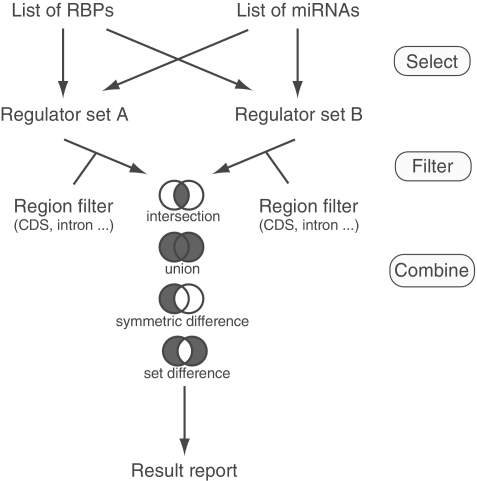
The power of doRiNA becomes eminent in the case of combinatorial search option. This option differs from the aforementioned simple search. It combines the results from two independent simple searches (A and B, Figure 3). Initially, target site positions can be individually confined for sets A and B to a particular gene feature region (CDS, 5′ UTR, 3′ UTR, intron or intergenic). The two filtered sets are subsequently combined by four possible set operations: union, A ∪ B′ intersection, A ∩ B′ symmetric difference, AΔB; and set difference, A∖B. Please bear in mind that both, the A set and the B set, are themselves the outcome of either a union or intersection step. Figure 3 summarizes the query capabilities of the combinatorial search option.
Figure 3.
Combinatorial search options.
DISCUSSION
A better understanding and dissection of post-transcriptional regulation is of paramount importance to molecular biology. With the advent of robust high-throughput methods for target site delineation, either computationally or experimentally, we face the challenge of efficient data organization, representation and analysis. doRiNA is our contribution to meet this challenge by providing a biologist-friendly access to the available target site data for miRNA and RBP regulators. Within doRiNA, we consolidate three different needs in data mining: data exploration, querying and retrieval.
We discern three features of doRiNA as especially important: first, the doRiNA database unifies two protagonists of post-transcriptional regulation, RBPs and miRNAs, in one service. Second, the doRiNA web service provides unparalleled query capabilities with minimal response times. Finally, users benefit from doRiNA's integration with other genome-wide data via the UCSC browser (e.g. SNP data could be intersected with miRNA or RBP target sites).
Comparison to related work
The doRiNA database differs from previously published database solutions like starBase (16) and CLIPZ (17) in several aspects. The starBase database is a very data-rich resource but offers only limited query capabilities (e.g. complex set operations are not supported). This is the same for the CLIPZ database, which has been mainly designed as a service for collaborative CLIP data analysis. Moreover, doRiNA contained more genome-wide data sets at the time of writing and is linked to the UCSC genome browser.
CONCLUSIONS
doRiNA does not merely provide rich data sets for browsing and download but empowers users to flexibly specify hypothesis-driven queries. Users may freely define their target site search space by providing gene lists. Complex combinations of regulators may be submitted as search queries. Without loss of speed, doRiNA is able to operate on different data zoom levels ranging from target gene sets down to individual target site nucleotides.
doRiNA benefits from its seamless integration with a local copy of the UCSC browser, which is very popular among computer-affine biologists.
AVAILABILITY
The doRiNA database is freely available at http://dorina.mdc-berlin.de. There are no access restrictions for academic and commercial use. We kindly ask all users to cite the doRiNA manuscript if they employ search results in their publications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods.
FUNDING
MDC Systems Biology Network (MSBN) as a participant of the Helmholtz-Alliance on Systems Biology (to S.D.M.); Deutsche Forschungsgemeinschaft for a fellowship in the International Research Training Group Genomics and Systems Biology of Molecular Networks (GRK 1360 to M.J.). Federal Ministry for Education and Research (BMBF) and the Senate of Berlin, Berlin, Germany (to M.L. and C.D.). Funding for open access charge: MDC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
All authors wish to acknowledge fruitful discussions with members of the Berlin Institute for Medical Systems Biology.
REFERENCES
- 1.Rajewsky N. MicroRNAs and the Operon paper. J. Mol. Biol. 2011;409:70–75. doi: 10.1016/j.jmb.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Net. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 6.Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 8.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol. Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 16.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khorshid M, Rodak C, Zavolan M. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res. 2011;39:D245–D252. doi: 10.1093/nar/gkq940. [DOI] [PMC free article] [PubMed] [Google Scholar]