Abstract
Peptidases, their substrates and inhibitors are of great relevance to biology, medicine and biotechnology. The MEROPS database (http://merops.sanger.ac.uk) aims to fulfil the need for an integrated source of information about these. The database has hierarchical classifications in which homologous sets of peptidases and protein inhibitors are grouped into protein species, which are grouped into families, which are in turn grouped into clans. The database has been expanded to include proteolytic enzymes other than peptidases. Special identifiers for peptidases from a variety of model organisms have been established so that orthologues can be detected in other species. A table of predicted active-site residue and metal ligand positions and the residue ranges of the peptidase domains in orthologues has been added to each peptidase summary. New displays of tertiary structures, which can be rotated or have the surfaces displayed, have been added to the structure pages. New indexes for gene names and peptidase substrates have been made available. Among the enhancements to existing features are the inclusion of small-molecule inhibitors in the tables of peptidase–inhibitor interactions, a table of known cleavage sites for each protein substrate, and tables showing the substrate-binding preferences of peptidases derived from combinatorial peptide substrate libraries.
INTRODUCTION
The MEROPS database is a manually curated information resource for proteolytic enzymes, their inhibitors and substrates. The database can be found at http://merops.sanger.ac.uk.
A proteolytic enzyme breaks down a polypeptide or protein by cleaving peptide bonds. Proteolytic enzymes are needed for the survival of all living organisms, and are of importance to mankind in the fields of medicine, nutrition, agriculture and technology (1).
The MEROPS database provides a classification and nomenclature of proteolytic enzymes and their inhibitors that is widely used throughout the academic community. The classification of proteolytic enzymes is derived from the system developed by Rawlings and Barrett (2). When it became apparent that paper publications to update the classification were no longer adequate, the database was developed at the Babraham Institute (3). The database moved to the Wellcome Trust Sanger Institute in 2002 (4). A classification of the protein inhibitors of peptidases (5) was added in 2004 (4) and coverage of the mostly synthetic, small-molecule inhibitors (SMIs) was added in 2008 (6).
Knowledge of the cleavages within protein, peptide and synthetic substrates is important for understanding the specificity and physiological roles of proteolytic enzymes, so the MEROPS database also includes a collection of known cleavage sites in substrates (7). Peptidase specificity is shown as a WebLogo display (8) and as a table of preferences for each substrate-binding pocket (6).
THE MEROPS CLASSIFICATION SYSTEMS
Proteolytic enzymes are frequently multi-domain proteins, with peptidase activity restricted to a single structural domain. Protein inhibitors are also frequently multi-domain proteins, often containing multiple, homologous inhibitor domains. Throughout the MEROPS database, only that portion of the sequence corresponding to a single peptidase domain (the ‘peptidase unit’) or a single inhibitor domain (the ‘inhibitor unit’) is used in sequence and structure comparisons.
The classifications are hierarchical. At the bottom of each hierarchy is the peptidase or inhibitor unit. The protein to which it belongs that has been most fully characterized biochemically is chosen as a representative called a ‘holotype’. Sequences considered to represent the same protein but from different organisms (i.e. orthologues) are grouped as a single protein species according to the criteria set out by Barrett and Rawlings (9). A new holotype (and protein species) is identified when a protein has been biochemically demonstrated to have a different specificity from any other member of the same family. For a peptidase, either it cleaves different substrates, cleaves the same substrates in different places or interacts with a different set of inhibitors; for an inhibitor, it interacts with a different set of peptidases or binds a peptidase much more tightly. A new identifier is also created if the characterized protein has a different architecture, or does not cluster on an evolutionary tree with other characterized proteins. The numbers of identifiers set up for peptidases and inhibitors are shown in Table 1.
Table 1.
Counts of protein species, families and clans for proteolytic enzymes and protein inhibitors in the MEROPS database
| MEROPS 8.5 |
MEROPS 9.5 |
|||
|---|---|---|---|---|
| Peptidases | Inhibitors | Peptidases | Inhibitors | |
| Sequences | 140 313 | 16 337 | 192 053 | 17 451 |
| Protein species | 3243 | 589 | 4202 | 634 |
| Families | 211 | 67 | 225 | 71 |
| Clans | 42 | 32 | 44 | 34 |
The numbers in Release 9.5 of MEROPS (July 2011) are compared to those in Release 8.5 (August 2009).
Homologues [detectable by a sequence similarity search using FastA (10), BlastP (11) or HMMER (12)] are grouped into a family. A family contains any number of homologues. One sequence is chosen as the type example of the family, and all sequences in the family are homologous to this type example, either directly or transitively. A sequence is included in the family if a pairwise alignment with an existing member of the family shows a statistically significant match, i.e. the expect value is <0.001.
The highest level of the hierarchy is that of clan, and all sequences within a clan are believed to be derived from the same ancestor, even if there is no significant sequence similarity. The most rigorous criterion for including proteins in the same clan is a similar tertiary structure. The DALI algorithm and server (13) is used to compare structures, and if the z-score from the DALI comparison to that of an existing member of a clan is >6.0, the sequence is added to that clan. The order of active-site residues is conserved in all members of a clan, and where no tertiary structure is known, a family may be added to a clan if this is the same. A clan can consist of a single family if the tertiary structure of a member is unrelated to that of any other peptidase or protein inhibitor.
Table 1 shows statistics for release 9.5 of the MEROPS database. In the 2 years since the previous article (14), despite the number of sequences in the database having increasing by over a third, only 18 new families and 4 new clans have been added.
RECENT DEVELOPMENTS
The website has been redesigned and improved. Frames have been removed from some HTML pages so that a user can bookmark any page. In addition, a Request Tracker ticketing system has been introduced to allow users to make comments and suggestions and to report errors. This can be accessed via a ‘feedback’ link present in the footer of every page.
The database has been extended to include proteolytic enzymes other than peptidases. Families of self-cleaving proteins that utilize the peculiar chemistry of asparagine to break peptide bonds without hydrolysis, known as ‘asparagine peptide lyases’ (15) have been added to the database.
Indexes are now provided for peptidase substrates (see below) and gene names of peptidases and protein inhibitors. The gene name index also includes synonyms of the names and the locus names from completely sequenced genomes. The names are listed alphabetically, along with the source organism (with a clickable link to the organism page in MEROPS) and the protein name recommended by the MEROPS team (with a clickable link to the summary page in MEROPS).
The MEROPS database includes over 44 000 literature references, and in addition to links to PubMed and to the text of papers from journal websites made available via DOI (digital object identifier), links are now made to the free text articles in PubMed Central (16). We have also implemented a new facility to search our literature collection for a specific PubMed identifier. This is available via the ‘Searches’ option on the left-hand menu. On entering a PubMed identifier the full reference is returned, plus a list of peptidases and inhibitors for which this reference is cited in MEROPS, with a link to the MEROPS summary page for each.
Cross-references to other databases
A number of new cross-references have been established between items in the MEROPS database and other publicly available databases. Over 200 cross-references to Wikipedia articles have been set-up for individual peptidases and inhibitors on the relevant summary pages, with reciprocal links within those Wikipedia pages. New cross-references have been established between SMIs and the ChEBI (17) and DrugBank (18) databases, with 100 and 40 cross-references, respectively.
Sequence features
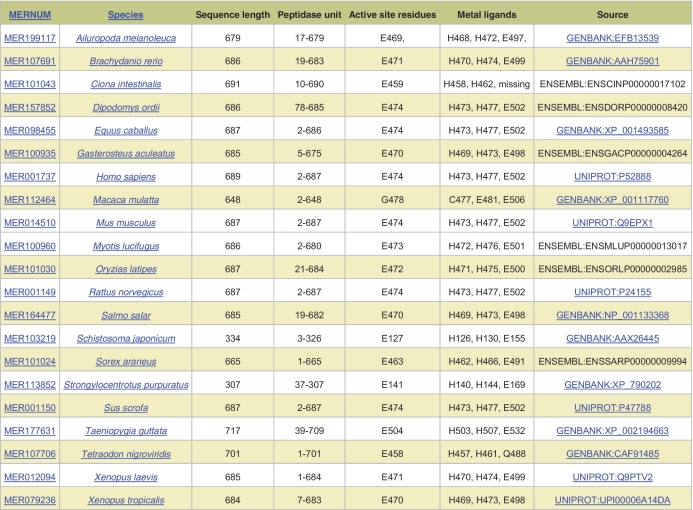
A new page giving details of species variants of peptidases has been created. Whenever a sequence is added to the MEROPS collection, several parameters are calculated from a BlastP pairwise comparison, including the position of active-site residues and the extent of the peptidase unit. The Sequence Features page presents the results as a table (Figure 1). Each row in the table shows the following information: the MEROPS sequence identifier, the scientific name of the source organism, the sequence length, the extent of the peptidase (or inhibitor) unit relative to the complete coding sequence, the predicted active-site residues (and metal ligands for a metallopeptidase) and the source of the sequence included in the MEROPS collection with a link to the relevant database. The organism scientific name is clickable and takes the user to the relevant organism page in MEROPS. For the active-site residues and metal ligands, each amino acid is shown in single letter code next to the residue number derived from the source sequence. If the sequence is a fragment or from a eukaryotic genome sequencing project where the automated gene build has missed an exon, then absent active-site residues are labeled ‘missing’. The items are arranged alphabetically by species scientific name, but can be re-sorted by the MEROPS sequence identifier.
Figure 1.
Sequence features display. The sequence features are shown for orthologues of thimet oligopeptidase.
Tertiary structure displays
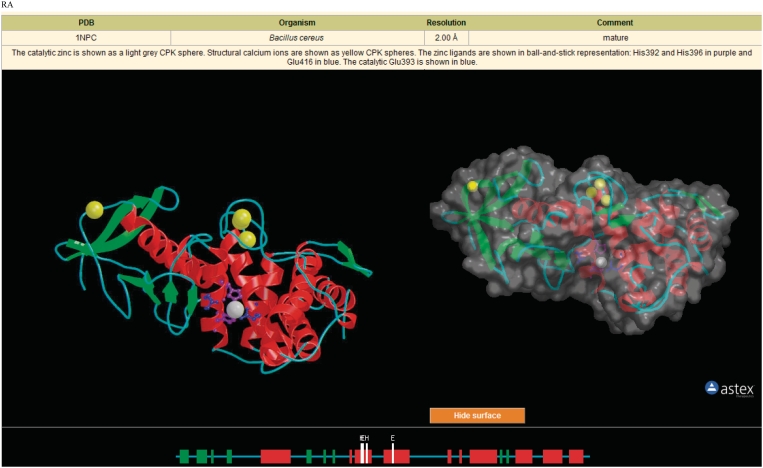
When the tertiary structure of a peptidase or a protein inhibitor has been solved, and the co-ordinates are available from the Protein Data Bank (PDB) (19), a structure page is presented at the MEROPS website. Besides a table of PDB entries, this has also included a fixed Richardson image (20) showing the structure with helices shown as red coils and strands as green arrows, with the active-site residues (and metal ligands for metallopeptidases) in ball-and-stick representation. Metal ions, attached carbohydrates, and inhibitors are also displayed where appropriate. However, a rotating image provides more insight into a protein structure, and so we now present a rotating image using AstexViewer (21) alongside the fixed image. The same structural elements, residues, metals and carbohydrates are shown in both images, because the command line input for the AstexViewer is derived from the input file used for the Richardson image. There is an option to show the surface of the molecule, and the image can be rotated in any direction by clicking on the image and holding down the left mouse button. Various other options are available by clicking on the right mouse button, including changing colors, saving the image and measuring the distances between atoms. To be able to use the AstexViewer, users must have Java installed. An example of the images on a structure page is shown in Figure 2.
Figure 2.
Displays of tertiary structure. The Structure page for thermolysin (M04.001) is shown. The table provides the cross-reference to the PDB entry, source organism, resolution, a comment and a description of the elements displayed in the images below. The image on the left-hand side is a rendered Richardson image generated using the programs RasMol (22), Molscript (23) and Render (24). The image on the right shows the surface of the molecule using the AstexViewer. This image can be rotated in any direction by the user, and the surface hidden by clicking the ‘hide surface’ button. The third image shows secondary structure, active-site residues and metal ligands as they appear in the protein sequence.
Peptidases from model organisms
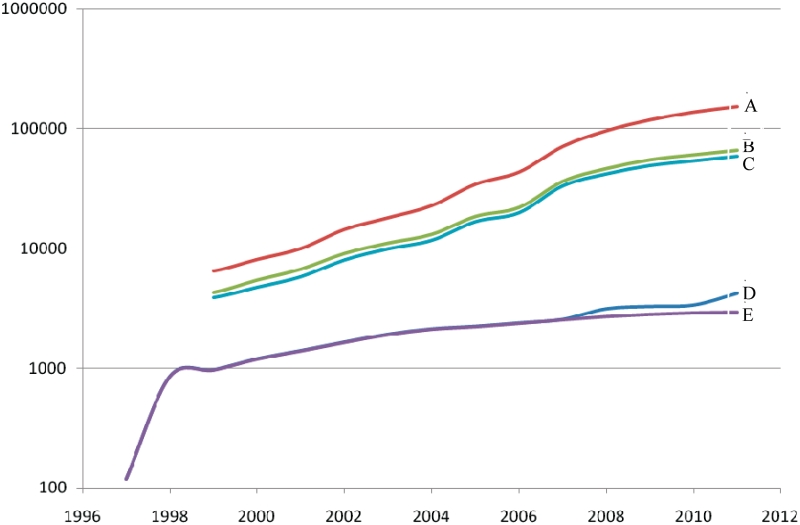
Homologues of peptidases and protein inhibitors are being sequenced much faster than they can be characterized. Consequences of this can be seen in Figure 3, which shows the cumulative totals of homologues of peptidase sequences in the MEROPS database since 1998, and the total number of MEROPS peptidase identifiers per year. It also shows the number of homologues that have been assigned to identifiers. Although MEROPS identifiers can be applied to species variants, the number of sequences that are unassigned is increasing rapidly. Less than half of all putative peptidases can be classified at the peptidase level, because the sequences are too divergent from that of the holotypes or the protein architecture is significantly different. This has led us to search for methods to draw attention to enzymes that may be suited to biochemical study because they come from well-characterized model organisms. An approach that we have adopted is to extend the concept of the holotype to these uncharacterized proteins. Special MEROPS identifiers have been created for all the uncharacterized peptidase homologues from a variety of model organisms: human, mouse, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae and Escherichia coli. These special identifiers resemble the standard MEROPS identifier except that the first character after the dot is the letter ‘A’ or ‘B’ so that it is easy to distinguish an identifier for a characterized peptidase from that of an uncharacterized one. Such special identifiers have not been set up for protein inhibitors or non-peptidase homologues. Table 2 shows the number of peptidases and putative peptidases in each model organism. In total, 1248 special identifiers have been created. Creation of special identifiers is useful if orthologues from other species can be identified, because a widely distributed putative protein is much more likely to become characterized. The special identifiers C26.A17 (GuaA protein, E. coli), C26.A19 (PabA protein, E. coli) and M20.A11 (AbgB protein, E. coli) have each been applied to putative proteins from over 500 species. Figure 3 also shows how these special identifiers have helped us to cluster putative peptidase sequences as species variants. When a peptidase assigned to a special identifier is biochemically characterized, a standard type of MEROPS identifier will be assigned to replace the special one.
Figure 3.
Growth in number of determined putative peptidase sequences. The curves shown are (A) all sequences, (B) sequences assigned to identifiers, (C) sequences assigned to identifiers excluding model organisms, (D) all identifiers and (E) identifiers excluding model organisms.
Table 2.
Counts of peptidase-encoding genes in selected model organisms
| Organism | Sequences assigned to standard MEROPS identifiers | Sequences assigned to special MEROPS identifiers | Total |
|---|---|---|---|
| Homo sapiens | 557 | 1 | 558 |
| Mus musculus | 553 | 14 | 567 |
| Drosophilia melanogaster | 111 | 320 | 431 |
| Caenorhabditis elegans | 74 | 273 | 347 |
| Arabidopsis thaliana | 125 | 452 | 577 |
| Saccharomyces cerevisiae | 76 | 25 | 101 |
| Escherichia coli | 88 | 167 | 255 |
Small-molecule peptidase inhibitors
A new series of identifiers has been created for SMIs. SMIs include naturally occurring compounds such as pepstatin, bestatin and amastatin, as well as synthetic inhibitors generated in a laboratory, and so do not lend themselves to any form of natural classification, unlike the peptidases and protein inhibitors. Instead, each SMI is assigned an identifier consisting of an initial J followed by a five digit number. For example, pepstatin is J00095 and ethylenediaminetetraacetic acid is J00149. This allows users to connect directly to an SMI summary.
The page of inhibitor interactions, available from most peptidase summaries, now includes small molecule as well as protein inhibitors. These interactions have been collected from the literature. For each SMI, a link has been provided to the relevant summary.
Peptidase substrates
Our collection of known cleavage sites in substrates consists of 54 837 cleavages in release 9.5, of which 48 557 (88.5%) were mapped to identifiers in the UniProt database (representing cleavages in 14 446 different proteins). The remaining 6281 (11.5%) represent cleavages in synthetic substrates. This is an increase of 15 191 cleavages (27.7%) since release 8.5 (August 2009). Substrates have been tagged as physiological, non-physiological, pathologic and synthetic, as judged by the original authors, unless there is evidence to indicate otherwise. It is now possible to filter the substrates listed for each peptidase so that only physiologically relevant, pathologic, non-physiological or synthetic substrates are shown.
An index of substrate names is now available. This lists the name, the UniProt accession, the peptidase known to cleave that substrate with a link to the summary for that peptidase, and a count of cleavages performed by each peptidase. On clicking the UniProt identifier, the user is presented with a display showing the cleavages within the sequence and a table of cleavages. The table shows:
the residue number of the amino acid in the P1 position (i.e. on the left of the scissile bond);
the name of the peptidase responsible (with a link to the relevant peptidase summary);
the residue range of the substrate used in the experiment compared to the complete coding sequence that is presented in the UniProt entry (e.g. minus a signal peptide or propeptide for a mature protein);
whether the cleavage is thought to be physiological or not;
how the cleavage was determined (using the following symbols: NT for N-terminal sequencing, MS for mass spectroscopy, MU for site-directed mutagenesis and CS for theoretical cleavages that fit the consensus sequence of a peptidase substrate);
a comment describing the purpose of the cleavage (e.g. ‘release of a signal peptide’); and
a reference.
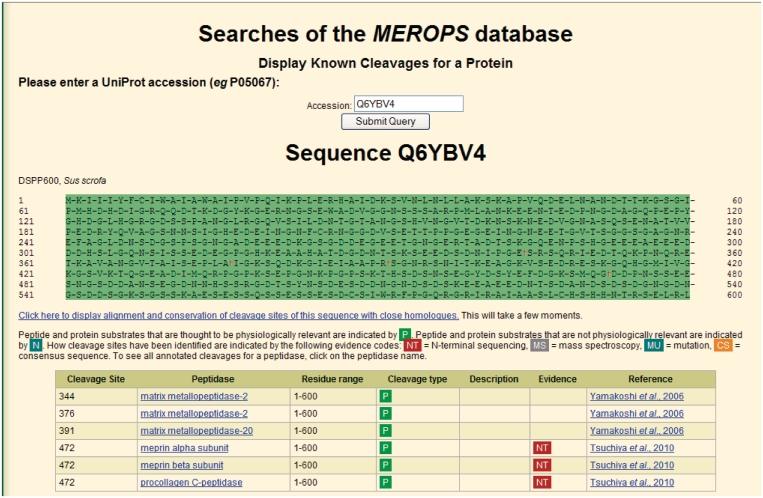
An example of cleavages annotated in a protein substrate is shown in Figure 4.
Figure 4.
Display of cleavages in a protein substrate. Known cleavages in the DSPP600 protein from pig are shown. The full sequence is shown at the top with cleaved bonds indicated by the ‘dagger’ symbol. More details of each cleavage are shown in the table beneath.
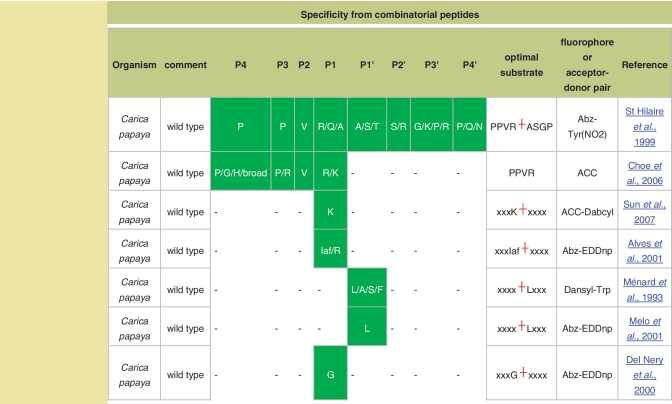
The identification of cleavage sites in substrates is important not only for determining the physiological roles of peptidases, but also for determining the specificity of the peptidase, which can help in the design of better and more selective synthetic substrates and inhibitors. There are now high-throughput techniques for determining peptidase specificity which automatically calculate preferences for each substrate-binding pocket, but do not determine the cleavage position in each synthetic peptide. An array of different peptides is made that is known as a ‘combinatorial library of substrates’. Because the cleavage position and sequence of each substrate is not known, these cannot be entered into the MEROPS collection of substrate cleavages. However, Poreba and Drag (2010) (25) have assembled a collection of peptidase preferences from the available literature, and made them available to MEROPS. These are presented as a table on each relevant peptidase summary. The table lists the source organism of the peptidase, a comment (such as whether the peptidase was wild-type or recombinant), the specificity in terms of substrate-binding pockets P4 to P4’ with the preferred amino acids shown in single letter code, the optimal substrate derived from the study, the fluorophore attached to the substrate or the acceptor–donor pair of a quenched fluorescent substrate, and a reference. Figure 5 shows an example of the new combinatorial peptides display.
Figure 5.
Specificity from combinatorial peptide libraries. The amino acid preferences within each substrate-binding pocket (labeled P4–P4′) are shown for experiments using combinatorial libraries of peptide substrates for the peptidase papain.
FUNDING
Wellcome Trust (grant number WT098051). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank the following: Pfam and Rfam colleagues for helpful discussions, especially John Tate for help with displays; Paul Bevan and Matthew Waller from the Sanger Institute web team for all their help in maintaining this resource and for refactoring the codebase; Molecular Connections (Bangalore, India) who have been employed to collect substrate cleavages from the scientific literature; Matthew Jenner for his help with molecular images and links to Wikipedia, and Jack Feltham for help with collecting substrate cleavages. We would also like to thank those users who have pointed out errors and omissions, or who have suggested changes and improvements.
REFERENCES
- 1.Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. London: Elsevier; 2004. [Google Scholar]
- 2.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem. J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 1999;27:325–331. doi: 10.1093/nar/27.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawlings ND, Tolle DP, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem. J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawlings ND. A large and accurate collection of peptidase cleavages in the MEROPS database. Database. 2009 doi: 10.1093/database/bap015. doi: 10.1093/database/bap015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett AJ, Rawlings ND. ‘Species’ of peptidases. Biol. Chem. 2007;388:1151–1157. doi: 10.1515/BC.2007.151. [DOI] [PubMed] [Google Scholar]
- 10.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 14.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings ND, Barrett AJ, Bateman A. Asparagine peptide lyases: a seventh catalytic type of proteolytic enzymes. J. Biol. Chem. 2011;286:38321–38328. doi: 10.1074/jbc.M111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Matos P, Alcantara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson JS. Schematic drawings of protein structures. Methods Enzymol. 1985;115:359–380. doi: 10.1016/0076-6879(85)15026-3. [DOI] [PubMed] [Google Scholar]
- 21.Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J. Comput. Aided Mol. Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- 22.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 23.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 24.Bacon D, Anderson WF. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph. 1988;6:219–220. [Google Scholar]
- 25.Poreba M, Drag M. Current strategies for probing substrate specificity of proteases. Curr. Med. Chem. 2010;17:3968–3995. doi: 10.2174/092986710793205381. [DOI] [PubMed] [Google Scholar]