Abstract
neXtProt (http://www.nextprot.org/) is a new human protein-centric knowledge platform. Developed at the Swiss Institute of Bioinformatics (SIB), it aims to help researchers answer questions relevant to human proteins. To achieve this goal, neXtProt is built on a corpus containing both curated knowledge originating from the UniProtKB/Swiss-Prot knowledgebase and carefully selected and filtered high-throughput data pertinent to human proteins. This article presents an overview of the database and the data integration process. We also lay out the key future directions of neXtProt that we consider the necessary steps to make neXtProt the one-stop-shop for all research projects focusing on human proteins.
INTRODUCTION
In the last 30 years, massive resources have been deployed to understand the molecular components and processes of human cells, both for clinical and fundamental research applications. While this effort has been first targeted toward the sequencing of the genome and the mapping of its transcriptome, it has now shifted toward the studies of the major actors of life, proteins. The molecular and functional complexity of human proteins is challenging and requires bioinformatics resources specifically aimed at capturing, integrating and maintaining up-to-date the available knowledge about them.
In a step toward this end, the UniProt/Swiss-Prot group has completed the manual annotation of the full set of human proteins, derived from about 20 000 genes, in September 2008 (1). The proteomic space generated from these gene products is enormous, up to an estimated 1 million different protein species derived from DNA recombination, alternative mRNA splicing and the wealth of protein post-translational modifications (PTMs). However, as estimated from the UniProtKB/Swiss-Prot knowledgebase content, ∼25% of those proteins (i.e. around 5000) have not yet been studied experimentally. For the remainder, the information available is often scarce. Many proteins have not been completely analyzed with respect to their abundance, distribution, subcellular localization and interactions with other biomolecules, post-translational modifications or—even more critical—function. The more complete our understanding of human proteins is the better equipped we will be to understand the functioning of the human body at molecular level.
The neXtProt knowledge platform, for and by the researcher community
Data are easier to generate than knowledge. Much undiscovered knowledge is hidden in large sets of heterogeneous and noisy data distributed across a multitude of resources and web sites. The problem is intensified by the fact that databases regularly become obsolete after a few years due to lack of financial support. This trend is especially true for research on human biology, owing to the sheer quantity of resources at the disposal of researchers.
To address these issues, we have created neXtProt (http://www.nextprot.org/), a web-based protein knowledge platform on human proteins (see screenshot of the home page in Figure 1). The ultimate goal for neXtProt is to serve for research on human the same role that Model Organism Databases (MODs) serve for model species. neXtProt is developed within the Swiss Institute of Bioinformatics (SIB) (www.isb-sib.ch), which has extensive expertise in building high-quality protein-centric resources such as UniProtKB/Swiss-Prot (2), PROSITE (3), ENZYME (4), STRING (5) and the Swiss-Model Repository (6).
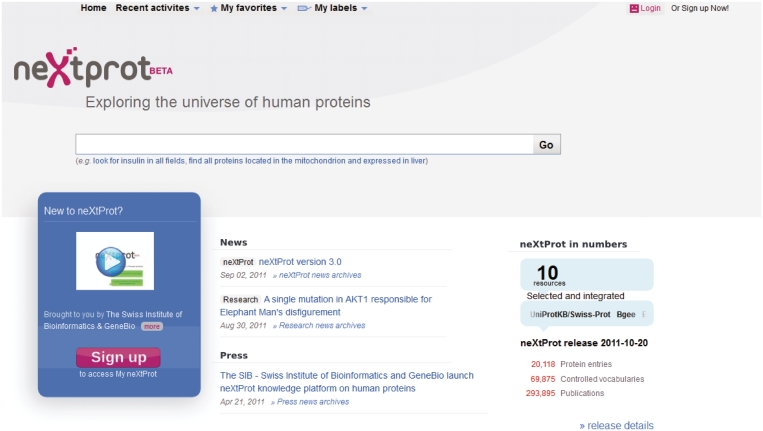
Figure 1.
The neXtProt home page grants access to the database via a search tool. Users can sign-in (top right) to create a personal account that allows them to personalize their usage of the platform by keeping a history of their queries and favoring or tagging search results. The home page also links to pages with more information about the current content of the platform in term of integrated resources (‘release details’ link at the bottom right).
neXtProt is being developed as a service for the community, and is using the knowledge and the expertise of the community to populate it with very high quality data and tools. For each data type we need to incorporate, we identify groups that have expertise in that area and collaborate with them to integrate data. In addition to making neXtProt and its users benefit from expert data in all areas, this philosophy helps us ensure that our data are up to date and helps advertise both neXtProt and our collaborators' resources to our respective user communities via reciprocal cross-links.
neXtProt content: data and ontologies
The primary data set in neXtProt comes from the high-quality solid work that has been the hallmark of UniProtKB/Swiss-Prot since its inception in 1986: we integrate all the information from the Swiss-Prot human entries. The information captured by Swiss-Prot, however, is only a small fraction of what is available. The fact that neXtProt is centered on a single species, human, makes it possible to widen not only the quantity but also the range of data being captured.
While we are still early in the neXtProt development path, we have already integrated a significant amount of additional information relevant to human proteins, notably:
Extensive protein expression information obtained by immunohistochemistry on healthy tissues from the Human Protein Atlas (HPA) (7).
Micro-array and cDNA expression information in healthy tissues originating from ArrayExpress (8) and UniGene (9,10). This RNA-based expression data have been meta-analyzed by the SIB Evolutionary Bioinformatics group and is available in the Bgee resource (11).
Subcellular localization results from two different high-throughput projects: DKFZ GFP-cDNA localization (12,13); and Weizmann Institute of Science's Kahn Dynamic Proteomics Database (14).
We have started to integrate high-quality mass spectrometry-derived proteomics information and, in particular, a number of published sets of N-glycosylation and phosphorylation sites. We also store peptide/protein identification results from experiments carried out in the context of the HUPO plasma (15) and brain (unpublished) initiatives obtained from PeptideAtlas (16), as well as some sets directly submitted to us by a network of collaborators.
The Gene Ontology (GO) (17,18) annotations of all human proteins as captured by GOA (19).
The mapping of proteins to their genomic transcripts on the human genome using Ensembl (20).
Additional single-amino acid polymorphism (SAPs) variants obtained from dbSNP (9) and Ensembl.
Additional identifiers, including cDNA clone names encoding for the proteins, Affymetrix and Illumina DNA probesets; cross-references to CCDS (21) and HPRD (22).
Abstracts of all articles from PubMed that are cited in human Swiss-Prot entries as well as some cited by other resources such as Entrez Gene (GeneRIFs) (9), MINT (23) and PDB (24) and which have been computationally mapped to the relevant protein entry by the UniProt consortium.
Ontologies and controlled vocabularies (CVs) are essential for consistent annotation and powerful data retrieval. A large number of vocabularies exist that cover various areas of biology. It is a challenge to choose the most appropriate vocabularies with respect to completeness, how well it represents the data we are capturing and how much interoperability it provides with other resources. Ontology and CVs are therefore an essential component of neXtProt.
We have imported into neXtProt the Gene Ontology (GO), UniProt disease, keyword, post-translational modification and subcellular location ontologies, UniPathway (25), enzyme classification (ENZYME) and part of the Medical Subject Headings (MeSH) (26). We also created mini-CVs based on UniProtKB annotations to cater for domains, protein families, protein-bound metal ligands and topology.
Available ontologies and controlled vocabularies, including MeSH, eVoc (27), BRENDA tissue ontology (28) and FMA (29), describe human anatomy with different scopes, coverage and precision levels. Since none of them allowed us to integrate and compare data from different resources (e.g. microarrays/ESTs from Bgee and immunohistochemistry from HPA) keeping the original granularity, we developed our own tissue and cell-type ontology.
neXtProt interface and functionalities
Users access the platform through an intuitive, simple interface centered on a Google-like search functionality that enables both simple (free text) and relatively complex queries (through the use of search topics) (Figure 2). Users can choose to search in neXtProt for protein entries, publications or terminologies (ontologies and controlled vocabularies). Once a search has been made, it is possible to filter the results according to a number of criteria. The search results are displayed either as simple lists or as mini-summaries.
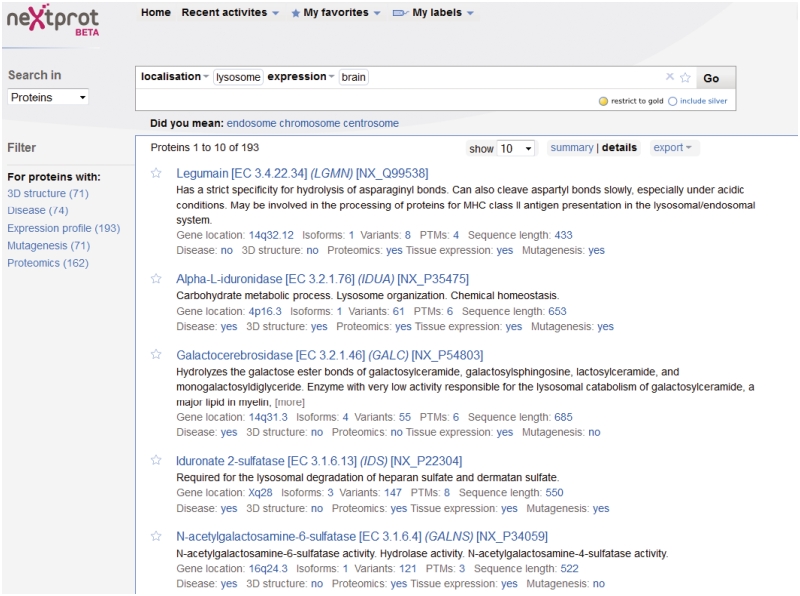
Figure 2.
The Search Results page. neXtProt is indexed across several biological areas, corresponding to the different views found on each entry (some data are present in multiple indexes). This allows users to make complex searches, for example finding all proteins localized to the lysosomes and expressed in the brain. From the search results page, it is possible to do various exports: obtain the list of proteins as an Excel file; the protein sequences as FASTA or PEFF, or the entire entry in XML format. When a user is logged in, checkboxes appear next to each entry, so that s/he is able to select specific entries for which the data are exported.
Users of neXtProt can sign-in to create a personal account that allows them to personalize their usage of the platform by keeping a history of their queries and favoring or tagging the search results.
neXtProt provides an original way of visualizing proteins entries: they can be seen from three different perspectives: the ‘Protein’, the underlying ‘Gene’ and the ‘References’ used to annotate it. The protein and gene perspectives are further subdivided in views that put the available information in context: function, medical, expression (Figure 3), interactions, localization, sequence (Figure 4), proteomics, structures, exons (Figure 5) and protein and gene identifiers. Special efforts have been made to document specific information on splice isoforms. For example, in the ‘sequence view’, the different splice isoforms can be graphically compared, highlighting the shared and specific sequence features (domains, sites, etc.) of each form.
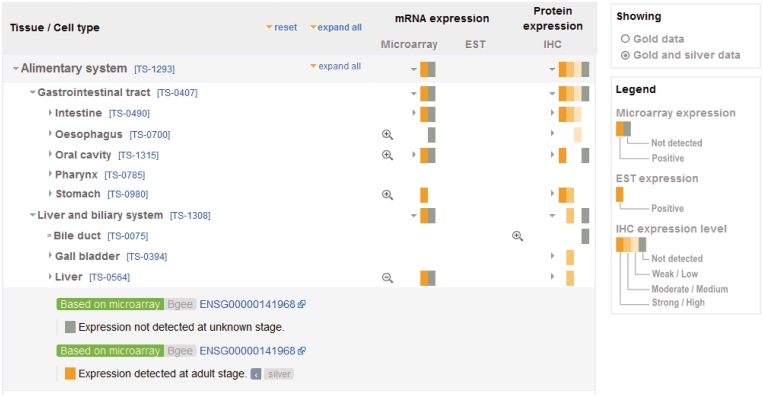
Figure 3.
The Expression Data view displays data via a browser of the neXtProt human anatomy ontology. Currently, the data presented come from two different sources: Bgee and HPA (see text). Data are captured and displayed at the original granularity level (Loupe symbols), and is propagated to higher levels using the ontology to be comparable across the data sets. The tool provides a menu to toggle between showing only Gold data and showing both Gold and Silver data. An icon next to the annotation indicates the silver data. Unmarked data is gold.
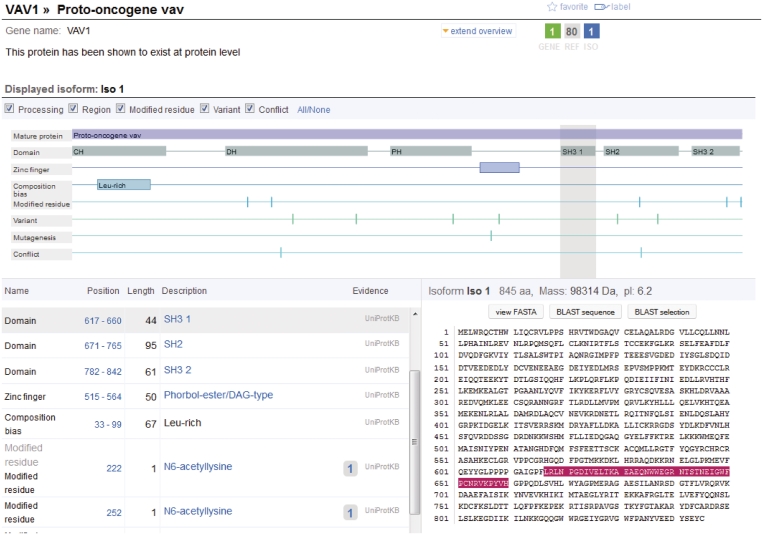
Figure 4.
The Sequence Viewer, accessible from the Sequence view, displays - in addition to the sequence itself, the different features of the sequence (processing, regions, modified residues, topological information, variants, sequence conflicts, etc.) as a graphical overview and a table view. When a feature is selected, either from the graphical viewer or from the feature table, the corresponding sequence is immediately highlighted on the right. The sequence viewer also provides direct access to the BLAST tool, which has the option of using the full sequence, a selection corresponding to a sequence feature or any other sub-sequence selected by the user.
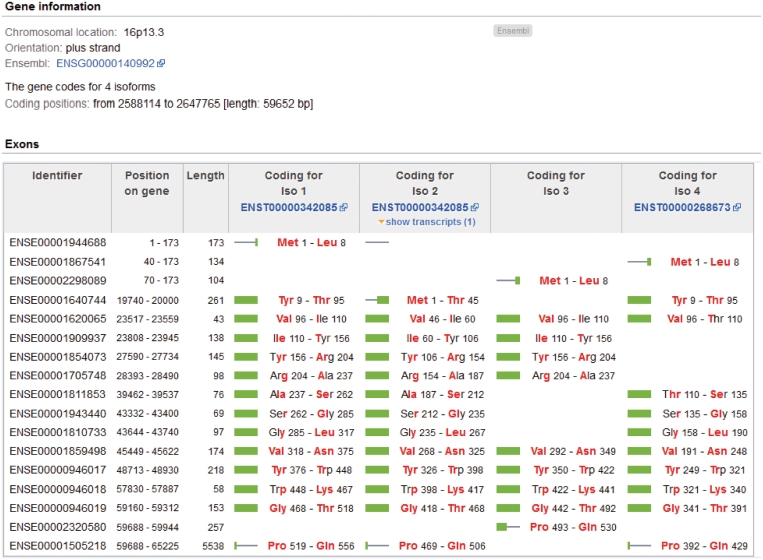
Figure 5.
The Exon view, available from the Gene perspective, gives the precise coordinates of all protein isoforms that can be mapped on Ensembl transcripts, based on exons. For each exon, its position on the gene is shown, as well as the length if the exon in nucleotides. The coding regions are represented by a small glyph, in which coding fragments are shown with a large green line, and non-coding sequences by a thin gray line. (Strictly non-coding exons are not shown.) The first and the last amino acid of each exon are shown, and the reading frame of each exon is indicated by red labeling of the amino acids. For example, Val 96–Ile 110 means that the first amino acid of that exon (Val) is completely encoded within that exon, while only the first nucleotide of the last one (Ile) is encoded in that exon.
neXtProt also provides a dedicated page for each term from our controlled vocabularies and ontologies. These pages display graphical and tree representations of the ontologies, as well as links to proteins annotated with these terms or their children (Figure 6). Similarly, there are pages for publications: these pages display the full publication record, including the abstract as well as the list of proteins that were annotated with that publication.
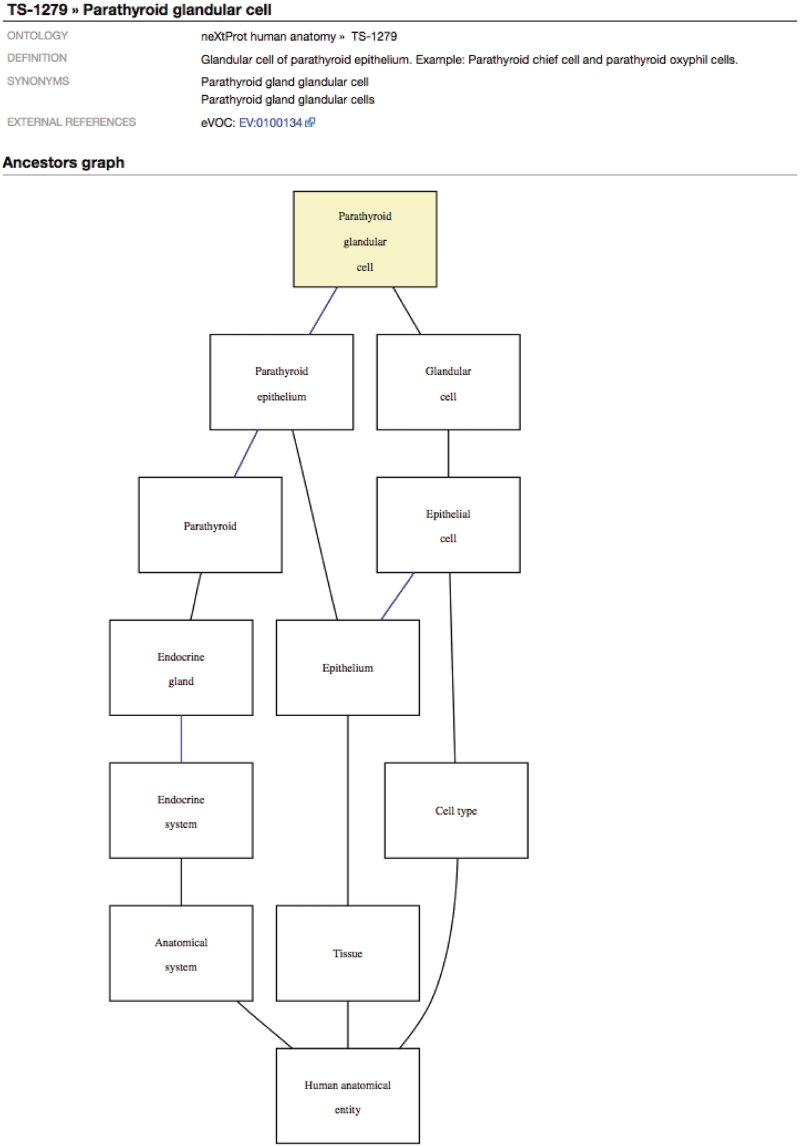
Figure 6.
For each term within the ontologies and hierarchical controlled vocabularies, a dedicated page shows its definition, synonyms and cross-references to other ontologies. In addition, the Graphical Ontology Viewer shows the relationship of that term to all its parents.
In term of tools, neXtProt provides access to a simple BLAST (30) implementation and we are currently beta-testing a tool to analyze enrichment of lists of proteins in term of various categories of annotations such as GO terms, domains, subcellular locations, etc.
neXtProt provides export functionality, namely, the download of lists of protein entries as text or Excel files, the corresponding sequences in FASTA format and the complete set of annotations in XML. To cater the needs of the proteomics community, we are the first resource to have implemented export of sequences and annotations of PTMs and variants in the PEFF format (31) which has been developed in the context of the HUPO Proteomics Standards Initiative. Bulk download of the full complement of sequence and annotations is also available through our anonymous ftp site (ftp.nextprot.org). Through the ftp site, users can also download our CVs and our ontology for human anatomy.
neXtProt's unique approach to data quality
Not all data published or available in public repositories are of the same quality. However, this fact has rarely been captured in databases, whose attitude is often that the user should be able to view all data to make a judgment on the reliability of the information s/he is presented with. This attitude tends to overwhelm the user with too much information, often making it simply impracticable to evaluate it; and requires that all users have expertise in all fields. In an attempt to overcome this problem, we are providing neXtProt users with a data integration philosophy based on a three-tier quality system:
Gold: highest quality data, corresponding to error rates of <1%.
Silver: good quality data, corresponding to error rates of <5%. Silver data are marked as such in the annotations.
Bronze: data deemed of a lower quality that we do not integrate in neXtProt.
Within neXtProt, users can choose to view and search only ‘Gold’ data (the default option), or view both ‘Gold’ and ‘Silver’. The grading of experimental data is not a trivial process and there is no simple rule that can be applied across the large landscape of high-throughput technologies that produce the data that need to be integrated into neXtProt. To make our quality-grading criteria transparent to users, we are documenting these criteria in a metadata information record linked to the relevant experiments. Whenever possible, we establish the quality thresholds—bronze, silver and gold—with the group who has produced the data. We expect that quality grading will be a dynamic process where users’ feedback will play an important role.
FUTURE DEVELOPMENTS
neXtProt aims to act as a central hub for all knowledge on human proteins. To achieve this, we are constantly integrating new data from widely used resources. Some key developments planned for the near future are described here.
neXtProt has been selected to be the knowledge platform for the newly launched HUPO Human Proteome Project (HPP) (32). To this end, neXtProt will need to integrate data and tools aimed to support the HPP. Among other developments, this means increasing the amount of proteomics data (post-translational modifications and peptide identification) and extending its scope toward quantification results obtained from selected reaction monitoring (SRM) experiments.
We are collaborating with the STRING group (http://string-db.org/) to integrate human protein network information (5). This, together with an increase of protein–protein interaction data provided by Intact (33) and other members of the IMEx consortium of interaction databases (34), will allow neXtProt users to explore graphically the functional protein complexes and their dynamic and spatial regulation through a Cytoscape plugin (35). Information on protein networks will be complemented by data on interactions between proteins and small molecules (such as drugs) and between proteins and nucleic acids.
While neXtProt only caters for human proteins, we want to provide the phylogenetic range of species in which a given human protein exists. We will also extract from Swiss-Prot experimental information carried out in organisms other than human but providing information directly relevant to the cognate human protein(s). For example, selected phenotypes from knock out or knock down experiments in mouse or zebrafish or enzyme characterization of bovine or pig counterparts.
In terms of tools and interface, we want to build an intuitive and powerful system, having capacities that are not yet available in other life sciences platforms. This is why we want to add a number of tools to neXtProt. Among them, we are planning to provide an advanced search option that will allow to specifically retrieve any stored data item and to carry out complex (including Boolean and analytical) queries; a multiple sequence aligner with a user-friendly interface and a 3D structure viewer that enables protein sequence annotations (PTMs, domains, variants, etc.) to be displayed overlaid on the structural view.
We are also exploring how we can allow users who have created personal accounts to customize our platform and to allow them to participate in group discussions and data sharing activities. Currently, URLs for searches and displayed pages are REST-compatible but this is not sufficient to allow third party developers to make full use of our platform and of the data available in neXtProt. This is why we are currently developing an Application Programming Interface (API) for neXtProt. This API will be used to integrate the future 3D structure viewer developed by BIONEXT (http://www.bio-next.com) in the context of a collaborative research project.
CONCLUSIONS
We have created neXtProt, a new protein knowledge platform on human proteins. It extends the high-quality UniProtKB/Swiss-Prot annotations for human proteins to include several new data types. The development of neXtProt is just beginning and will continue to expand with respect to the quantity and scope of data presented. We are convinced that the comprehensive biocuration of human proteins is a community endeavor. With this in mind, neXtProt is being built as a participative platform and we look forward to receiving users' input for its future development.
FUNDING
The SIB; Genebio SA; the Swiss Confederation's Commission for Technology and Innovation (CTI, grant 10214.1 PFLS-LS); the neXtProt server is hosted by VitalIT; the bioinformatics competence center that supports and collaborates with life scientists in Switzerland. Funding for open access charge: SIB.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the UniProt groups at SIB, EBI and PIR for their dedication in providing up-to-date high-quality annotations for the human proteins in Swiss-Prot thus providing neXtProt with a solid foundation. The authors thank Laurent-Philippe Albou, Frédéric Bastian, Pierre-Alain Binz, Christine Carapito, Eric Deutsch, Nasri Nahas, Marc Robinson-Rechiavi, Mathias Uhlen, Christian von Mering for stimulating discussions, advices and/or providing us data. From 2009 to 2011, neXtProt has been jointly developed by the Swiss Institute of Bioinformatics (SIB) and GeneBio SA.
REFERENCES
- 1.The UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 8.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pontius JU, Wagner L, Schuler GC. Ch. 21. In: McEntyre J, Ostell J, editors. The NCBI Handbook. Bethesda, MD: National Center for Biotechnology Information; 2003. [Google Scholar]
- 11.Bastian FPG, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Data Integration in the Life Sciences. Vol. 5109. Berlin/Heidelberg: Springer; 2008. pp. 124–131. [Google Scholar]
- 12.Liebel U, Starkuviene V, Erfle H, Simpson JC, Poustka A, Wiemann S, Pepperkok R. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 2003;554:394–398. doi: 10.1016/s0014-5793(03)01197-9. [DOI] [PubMed] [Google Scholar]
- 13.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigal A, Danon T, Cohen A, Milo R, Geva-Zatorsky N, Lustig G, Liron Y, Alon U, Perzov N. Generation of a fluorescently labeled endogenous protein library in living human cells. Nat. Protocols. 2007;2:1515–1527. doi: 10.1038/nprot.2007.197. [DOI] [PubMed] [Google Scholar]
- 15.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell. Proteomics. 2011;10:M110 006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutsch EW. The PeptideAtlas Project. Methods Mol. Biol. 2010;604:285–296. doi: 10.1007/978-1-60761-444-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goel R, Muthusamy B, Pandey A, Prasad TS. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol. 2011;48:87–95. doi: 10.1007/s12033-010-9336-8. [DOI] [PubMed] [Google Scholar]
- 23.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgat ACE, Coudert E, Axelsen KB, Keller G, Bairoch A, Bridge A, Bougueleret L, Xenarios I, Viari A. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Res. 2012;40:D761–D769. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sewell W. Medical subject headings in Medlars. Bull. Med. Libr. Assoc. 1964;52:164–170. [PMC free article] [PubMed] [Google Scholar]
- 27.Kelso J, Visagie J, Theiler G, Christoffels A, Bardien S, Smedley D, Otgaar D, Greyling G, Jongeneel CV, McCarthy MI, et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gremse M, Chang A, Schomburg I, Grote A, Scheer M, Ebeling C, Schomburg D. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mejino JV, Jr, Agoncillo AV, Rickard KL, Rosse C. Representing complexity in part-whole relationships within the foundational model of anatomy. AMIA Annu. Symp. Proc. 2003:450–454. [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Orchard S, Hoogland C, Bairoch A, Eisenacher M, Kraus HJ, Binz PA. Managing the data explosion. A report on the HUPO-PSI Workshop. August 2008, Amsterdam, The Netherlands. Proteomics. 2009;9:499–501. doi: 10.1002/pmic.200800838. [DOI] [PubMed] [Google Scholar]
- 32.Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, et al. The human proteome project: current state and future direction. Mol. Cell Proteomics. 2011;10:M111 009993. doi: 10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orchard S, Aranda B, Hermjakob H. The publication and database deposition of molecular interaction data. Curr. Protoc. Protein Sci. 2010 doi: 10.1002/0471140864.ps2503s60. Chapter 25, Unit 25 23. [DOI] [PubMed] [Google Scholar]
- 35.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]