Abstract
The University of California Santa Cruz Genome Browser (http://genome.ucsc.edu) offers online public access to a growing database of genomic sequence and annotations for a wide variety of organisms. The Browser is an integrated tool set for visualizing, comparing, analyzing and sharing both publicly available and user-generated genomic data sets. In the past year, the local database has been updated with four new species assemblies, and we anticipate another four will be released by the end of 2011. Further, a large number of annotation tracks have been either added, updated by contributors, or remapped to the latest human reference genome. Among these are new phenotype and disease annotations, UCSC genes, and a major dbSNP update, which required new visualization methods. Growing beyond the local database, this year we have introduced ‘track data hubs’, which allow the Genome Browser to provide access to remotely located sets of annotations. This feature is designed to significantly extend the number and variety of annotation tracks that are publicly available for visualization and analysis from within our site. We have also introduced several usability features including track search and a context-sensitive menu of options available with a right-click anywhere on the Browser's image.
INTRODUCTION
The University of California Santa Cruz (UCSC) Genome Browser (1,2) at http://genome.ucsc.edu is a web-based set of tools providing access to a database of genome sequence and annotations for visualization, comparison and analysis by the scientific, medical and academic communities. Our primary mission is to provide timely and convenient open access to high-quality human genome sequence and annotations in a framework that enables easy exploration from genome-wide down to the base level. Annotation datasets, or ‘tracks’, on the human genome cover conservation and evolutionary comparisons, gene models, regulation, expression, epigenetics and tissue differentiation, variation, phenotype and disease associations. Our mission extends to a number of additional organisms including 4 other primates, 14 additional mammals including a marsupial and a monotreme, 10 non-mammalian vertebrates and 24 non-vertebrates, each with varying degrees of genome-specific annotation. Many of the genomes in our database have multiple assembly versions dating back to the early years of the UCSC Genome Browser when it was one of the few tools providing public access to the human genome (3).
The convenience and versatility of our tool set is continually challenged by the accelerating abundance of new data sets and analyses, fed by new technologies and research collaborations, which in turn are built upon the critical mass of publicly accessible biological data. Given the Genome Browser’s established role within the public square of genome science, there is increasing pressure to accept ever more data and ensure that the critical mass is not fragmented. To accommodate this growth, there is an ongoing effort to support decentralized data models and enable data contributors to more fully integrate their own data into the UCSC Genome Browser. With the addition of ‘track data hubs’ this year, we allow remote hosting of data that can be integrated into the browser in very sophisticated ways. To further accommodate this growing complexity we have also introduced several new features to improve the usability and intuitiveness of the browser.
LOCAL DATA SETS
The Genome Browser locally hosts mapping and sequence annotation tracks that describe assembly, gap and GC content for all organisms in the browser database. Additionally, for most organisms we show alignments from RefSeq genes (4), mRNAs and ESTs from GenBank (5), and other gene or gene prediction tracks such as Ensembl Genes (6). For human and mouse assemblies, we also offer a locally generated UCSC Genes track based upon RefSeq, GenBank, CCDS and UniProt data (7,8). About half of the genomes hosted at UCSC include a multiple sequence alignment (multiz) track (9) and pairwise genomic alignments between assemblies to further comparative and evolutionary investigations. Expression, regulation, variation and phenotype tracks are available for many of the assemblies. Most locally hosted tracks include descriptions with references and links to the original contributors or research upon which the annotations are based.
In addition to our main site, we have recently introduced a preview site (http://genome-preview.ucsc.edu) to allow early access to data sets that have not been fully vetted or may not meet the criteria for release on our main browser. As of September 2011 our preview site contained 10 additional mammalian genomes and, within the human GRCh37/hg19 assembly, almost 1800 additional annotation tracks. Data on our preview site should be used with the understanding that it may not be of release quality. (Note that we refer to genome assemblies first by the combination of official/UCSC names, then by UCSC name alone in this text.)
New genome assemblies
In the past year, we have added four new genome assemblies to the Genome Browser. The latest chimpanzee (Pan troglodytes) assembly (CGSC 2.1.3/panTro3) from the Chimpanzee Sequencing and Analysis Consortium (10) includes standard mapping and sequence tracks, RefSeq Genes, N-SCAN gene predictions (11), and a 12-species multiple alignment and conservation scoring by phastCons and phyloP (12,13). Also, we added the first sheep (Ovis aries) assembly (ISGC Ovis_aries_1.0/oviAri1), which was produced by the International Sheep Genomics Consortium (ISGC) (14). This assembly has RefSeq gene annotations and pairwise alignments to five other species. The second lizard (Anolis carolinensis) assembly (Broad AnoCar2.0/anoCar2) from the Broad Institute (15) includes annotation tracks for Ensembl Genes, human protein mappings by tBLASTn (16), and conservation and pairwise alignments to six other species. The latest assembly for zebrafish (Danio rerio) (Zv9/danRer7) from the Wellcome Trust Sanger Institute and Genome Reference Consortium was also released with RefSeq Genes, Ensembl Genes, human proteins, GenBank mRNAs and an eight-species conservation track, among others.
We anticipate the public release of four more genome assemblies by the end of the year: a cow (Bos taurus) assembly (Bos_taurus_UMD_3.1/bosTau6) from the Center for Bioinformatics and Computational Biology, University of Maryland (17); the microbat (Myotis lucifugus) draft assembly (Broad Myoluc2.0/myoLuc2) from the Broad Institute; the turkey (Meleagris gallopavo) draft assembly (TGC Turkey_2.01/melGal1) from the Turkey Genome Consortium (18); and the baker's yeast (Saccharomyces cerevisiae) assembly (SacCer_Apr2011/sacCer3) from the Saccharomyces Genome Database project (19).
New and updated annotations
During the past year, many of our users have migrated from the NCBI36/hg18 human genome assembly to the hg19 assembly, yet a great deal of research that was performed on hg18 will not be repeated on the new assembly. Therefore, we have remapped approximately 30 of the hg18 annotations to the corresponding sequence locations on hg19. Tracks generated by this ‘lift-over’ method may not be of as high of quality as those that are originally mapped to the reference genome; therefore, all such tracks in the hg19 assembly have been prominently flagged. Several original contributors of popular hg18 annotations were able to completely regenerate them for the hg19 reference genome.
In addition to the remapped annotations, a number of existing data sets underwent major revisions and many new data sets were added this year. Several highlights deserve additional attention. We released more than 7000 tracks and downloadable files as the Data Coordination Center for the Encyclopedia of DNA Elements (ENCODE) Project (20,21), described in a companion paper in this issue. The Genome Reference Consortium's incident database, which covers genome assembly problem notes or resolutions, is now included on hg19, NCBI37/mm9 and danRer7 and updated daily. The DECIPHER (22) database of chromosomal microdeletions/duplications/insertions, translocations and inversions has been added to hg19. The re-engineered Online Mendelian Inheritance in Man (OMIM) data sets from Johns Hopkins University (23) have been released on human assemblies hg18 and hg19 and are updated weekly. These tracks, colored by phenotype class, cover gene associations and phenotypes with no known gene associations; a separate track focuses on phenotypes associated with dbSNP identifiers. We released a major update of the UCSC Genes track (7) on the mm9 mouse assembly, and an updated release of this data set on the hg19 human assembly is anticipated by the end of 2011. Gencode Genes version 7 (24) was released, as were three releases of the Ensembl Genes (6,25) data set. A track of Human RNA Editing from the DARNED (26) database has been added. UCSC's Integrated Regulation track derived from ENCODE data was released on hg19 and expanded to include transcription factor binding evidence.
With the exponential growth of clinical sequencing, there is an urgent need for filters to remove common and spurious variants from the large sets of discovered variants. To aid in the development of such filters, as of dbSNP build 132 (27) we have added new data fields from dbSNP to our database representation: allele frequencies (when available), several properties assigned by dbSNP such as ‘clinically associated’ and ‘genotype conflict’, and submitter handles. The new data fields, as well as UCSC's annotations of unusual conditions, can be used to color and filter track items. In addition, we have extracted subsets of dbSNP variants to create several new tracks. We still provide a comprehensive dbSNP track (now named All SNPs), but variants whose flanking sequences map to more than one genomic location are hidden by default. The Mult. SNPs track contains only those variants mapped to more than one genomic location. Common SNPs contains uniquely mapped SNPs for which allele frequencies are available and for which the major allele frequency is at most 99%. The Flagged SNPs track contains uniquely mapped SNPs not already included in Common SNPs that are flagged by dbSNP as ‘clinically associated’.
DECENTRALIZED DATA
The foundation of the Genome Browser's data decentralization is our support for remotely hosted data files: bigBed/bigWig format (28) and BAM format (29). Unlike distributed annotation system (DAS) servers (30), remote data files need only be Internet-accessible by URL. The indexed files are queried by byte range and cached locally on UCSC servers. This allows the Browser to treat remote data sets nearly identically to locally stored data and ensures excellent query times to popular annotations. While one strength of DAS served annotations is the flexible XML format, remote data files are compressed data in native formats allowing efficient access to substantially greater amounts of data. We extended our Table Browser this year to fully support the BAM format in addition to the two ‘big’ formats. Continuing this trend, we have added support for Variant Call Format (VCF) (31) compressed and indexed by tabix (32). VCF was developed by the 1000 Genomes Project (33) for the interchange of single nucleotide variants, indels, copy number variations and structural rearrangements discovered by the project.
The Genome Browser custom track feature has long been available and, combined with remotely hosted data sets, is a relatively quick and easy avenue for researchers to view and share their genomic data within the browser. However, custom tracks have several configuration and organization limitations, and are not designed to be full extensions to the browser database hosted at UCSC. With this in mind, it is not surprising that there are over 200 Genome Browser mirrors, which have allowed many research groups to set up their own tracks within a fully local instance of the browser. However, even when made publicly available, mirrors are easily overlooked by the larger research community, and the overhead of maintaining a mirror site can lead to degradation of those sites, even when the unique data sets at the sites remain invaluable.
Extending the browser with track data hubs
Building upon the foundation of remotely hosted data formats, this year we have introduced ‘track data hubs’ to the Genome Browser. Data hubs allow researchers at institutions around the globe to combine and expertly configure large numbers of data sets into meaningful arrangements for advanced analyses, and fully integrate them into the Genome Browser. Data hubs can be used for temporary efforts or limited collaboration, but with greater flexibility than custom tracks and much less overhead than mirrors require. Their real value however, lies in the opportunity to publicly release full extensions of the browser database. Hub tracks are visible and configurable in the browser in a nearly identical way as UCSC-hosted tracks. The hub tracks are also fully integrated into the Table Browser, allowing the same sort of filtering, intersecting, downloading and forwarding to third parties as is available for locally hosted tracks.
The premier example of a data hub made publicly available in the browser is the Human Epigenome Atlas hosted at Washington University in St Louis, which has operated a mirror site for years. The hub provides two releases from the NIH Roadmap Epigenomics Mapping Consortium (34) on the hg19 human assembly (with an additional one aligned to hg18) consisting of close to 3000 distinct data sets covering over 1000 experiments. On hg19 this vast array of data has been organized into 19 major groups and 116 different collections allowing the selection of subsets of tracks based upon experiment type, cell/sample type and assay, among others. A large number of summary tracks use the browser's multi-colored overlay of wiggles method, introduced last year (2). This facilitates the display of multiple data sets into a single visualization track for focused comparisons. Such organization and visual comparison is unachievable with custom tracks or DAS served annotations. The new data hub feature extends the utility of the Genome Browser to a vast amount of research beyond its previous scope.
NEW USABILITY FEATURES
Another consequence of the increasing amount and complexity of genomic data is the difficulties users can experience in finding specific data and viewing it in meaningful ways. This year we have introduced new search and navigation capabilities to make data sets easier to find and configure, and to make the browsing experience more intuitive.
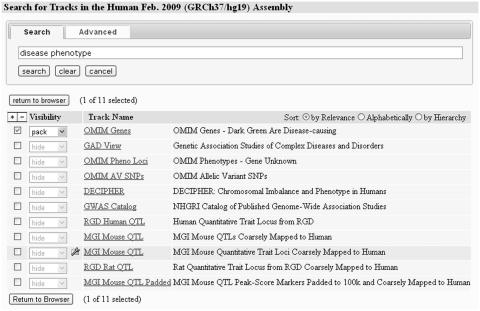
Until recently the number and groupings of Genome Browser data sets were small enough that simple exploration from the main browser page was sufficient to discover what was available. But in the past few years, especially on the human and mouse genomes, simple exploration has become overwhelmed by the thousands of annotation and analysis tracks now spread through multiple layers of organization. The browser's new track search utility allows users to find data sets by entering a few key words into a simple dialog, then selecting the resulting tracks for immediate viewing in the browser using any of the standard visualizations (Figure 1). An advanced option allows searching by key terms for those tracks described by ‘controlled vocabulary’ terms such as cell lines and antibody targets (currently limited to tracks associated with the ENCODE project). Advanced search provides selectable lists of these terms enabling a guided search to available data. Multiple terms can be applied in both ‘and’ and ‘or’ combinations. For example, in a single advanced search, a user can find all tracks covering cell lines ‘H1 and HeLa’ that show evidence of transcription factors CTCF and p300 bindings.
Figure 1.
A simple search has been performed to find annotation tracks related to disease phenotype. One of the search results is a container of related tracks.
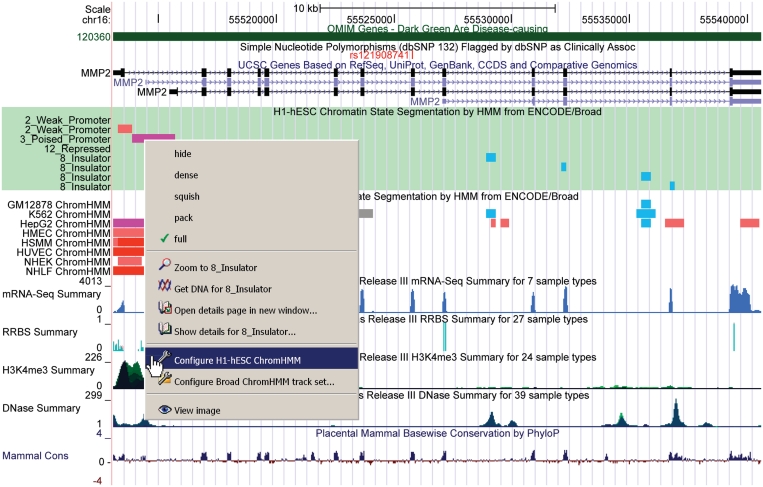
This year we enabled a user to directly target and configure track features in the browser image through a context-sensitive menu displayed with a simple right click on the image (control click on the Mac). This right-click menu provides a quick way to change visibilities and other configuration options, and even allows the configuration of individual subtracks which otherwise are only configurable as a group. The right-click menu also provides options for viewing feature detail, getting DNA sequence, zooming in on a selected feature, as well as recovering the current multi-part image as a simple .png file (Figure 2). We have also added the ability to drag the browser image left or right to reposition the viewable portion of the image on the chromosome. We have extended the drag-reorder configuration feature to allow reordering of whole groups within the image by vertically dragging the associated section of the sidebar. Finally, the existing drag-zoom feature has been expanded to shift-drag-zoom, which allows the user to drag-zoom from anywhere on the browser image to more easily focus on features of interest. Together, the track search, right-click menu and the three dragging features make it much easier for users to find, visualize and configure their discoveries in the Genome Browser.
Figure 2.
Annotation tracks from OMIM, dbSNP, UCSC Genes, ENCODE Histone HMM and the Epigenomics Roadmap data hub combined in one image. The new context-sensitive menu is shown with the highlighted track about to be configured.
Along with the data hub and usability features added to the browser in the past year, we have focused on maintaining efficiency and response time. For data hubs, this effort was especially critical. Only the portions of remote data sets that are viewed are acquired and cached locally at UCSC. Once cached, the response time is not affected, but the initial retrieval can depend upon network latencies from multiple remote web servers. By parallelizing the retrieval, response times have been dramatically improved. At the same time, we examined the Genome Browser CGI processes, database configuration and hardware, and identified and removed several bottlenecks and inefficiencies. This effort has allowed us to keep the Genome Browser server response time within one second for the vast majority of requests.
FUTURE DIRECTIONS
Over the course of the next year we plan to continue making user interface improvements and developing track data hubs. We would like to add more flexibility to the configuration of large track sets, and in how multiple tracks from different sources can be combined. We plan to extend the track search mechanism to work with hubs much like it works with locally hosted tracks. We are also in the midst of extending track hub support to additional data types.
The genomics science field, now and going forward, does not suffer from a lack of data or the tools to investigate that data, but no data set can be fully understood without context. The critical mass of publicly accessible genomic data is providing the context by which the contours of biology may be illuminated. Through the Genome Browser, UCSC plans to continue to provide a reliable platform for bringing many multiple strands of research together to facilitate scientific examination, interpretation and collaboration.
FUNDING
National Human Genome Research Institute (grant number P41HG002371 supporting G.P.B., H.C., M.D., P.A.F., A.S.H., F.H., D.K., V.K., W.J.K., R.M.K., C.H.L., L.R.M, A.P. B.J.R., B.R., G.R. and A.S.Z.; U41HG004568 supporting M.S.C., T.R.D., M.G., F.H., W.J.K., K.L., V.S.M., B.J.R., K.R.R., C.A.S., and M.W.; and subcontracts from P01HG5062 supporting G.P.B., W.J.K. and B.R; U54HG004555 supporting M.D. and R.A.H.; U41HG004269 supporting A.S.H. and W.J.K.; U01HG004695 supporting W.J.K.); subcontracts from the National Institute of Dental and Craniofacial Research (U01DE20057 supporting G.P.B. and R.M.K.]; National Institute of Child Health and Human Development [RC2HD064525 supporting H.C., A.S.H. and R.M.K.); and the National Institute of Environmental Health Sciences (U01ES017154 to W.J.K). Support from Howard Hughes Medical Institute to D.H. Funding for open access charge: Browser/ENCODE DCC.
Conflict of interest statement. P.A.F., B.R., A.S.Z., A.S.H., D.K., G.P.B., H.C., M.D., T.R.D., B.M.G., R.A.H., F.H., V.K., R.M.K., K.L., C.H.L., L.R.M., A.P., B.J.R., K.R.R., D.H. and W.J.K. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
ACKNOWLEDGEMENTS
The authors would like to thank the many data contributors whose work makes the Genome Browser possible, our Scientific Advisory Board for steering our efforts, our users for their consistent support and valuable feedback, and our outstanding team of system administrators: Jorge Garcia, Erich Weiler, Victoria Lin and Gary Moro.
REFERENCES
- 1.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, Wallis J, Sekhon M, Wylie K, Mardis ER, Wilson RK, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 4.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;18:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 8.Karolchik D, Kuhn R, Baertsch R, Barber G, Clawson H, Diekhans M, Giardine B, Harte R, Hinrichs A, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chimpanzee Sequencing & Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 11.Gross SS, Brent MR. Using multiple alignments to improve gene prediction. J. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- 12.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou MM, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Archibald AL, Cockett NE, Dalrymple BP, Faraut T, Kijas JW, Maddox JF, McEwan JC, Hutton Oddy V, Raadsma HW, Wade C, et al. The sheep genome reference sequence: a work in progress. Anim. Genet. 2010;41:449–453. doi: 10.1111/j.1365-2052.2010.02100.x. [DOI] [PubMed] [Google Scholar]
- 15.Alfoldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg Le A, Bouffard P, Burt DW, Crasta O, Crooijmans RP, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 20.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM®) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 27.Sherry S, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski E, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000_Genome_Project_Data_Processing_Subgroup The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkinson AM, Albrecht M, Birney E, Blankenburg H, Down T, Finn RD, Hermjakob H, Hubbard TJP, Jimenez RC, Jones P, et al. Integrating biological data – the Distributed Annotation System. BMC Bioinformatics. 2008;9:S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.1000 Genomes_Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]