Abstract
MethylomeDB (http://epigenomics.columbia.edu/methylomedb/index.html) is a new database containing genome-wide brain DNA methylation profiles. DNA methylation is an important epigenetic mark in the mammalian brain. In human studies, aberrant DNA methylation alterations have been associated with various neurodevelopmental and neuropsychiatric disorders such as schizophrenia, and depression. In this database, we present methylation profiles of carefully selected non-psychiatric control, schizophrenia, and depression samples. We also include data on one mouse forebrain sample specimen to allow for cross-species comparisons. In addition to our DNA methylation data generated in-house, we have and will continue to include published DNA methylation data from other research groups with the focus on brain development and function. Users can view the methylation data at single-CpG resolution with the option of wiggle and microarray formats. They can also download methylation data for individual samples. MethylomeDB offers an important resource for research into brain function and behavior. It provides the first source of comprehensive brain methylome data, encompassing whole-genome DNA methylation profiles of human and mouse brain specimens that facilitate cross-species comparative epigenomic investigations, as well as investigations of schizophrenia and depression methylomes.
INTRODUCTION
DNA methylation is an epigenetic modification that occurs at the 5′-position of cytosine, altering its structure, but not its base pairing properties. In mammalian genomes, 5-methylcytosine occurs predominantly at CpG dinucleotides within differentiated cells, and is faithfully propagated on the daughter strand following DNA replication by the maintenance DNA methyltransferase 1 enzyme (DNMT1). This form of information is flexible enough to be adapted for different somatic cell types, yet stable enough to be retained during mitosis and/or meiosis. DNA methylation is commonly associated with transcriptional silencing because it can directly inhibit the binding of transcription factors or regulators, or recruit methyl-CpG binding proteins (MBPs) with repressive chromatin-remodeling functions (1,2). DNA methylation plays an important role in the protection against intragenomic parasites (3), in genomic imprinting (4) and in X-chromosome inactivation in females. Methylation of CpG dinucleotides is critical in genome defense and chromosomal structural integrity (3,5–7). Errors in DNA methylation establishment or maintenance, or environmentally mediated alterations in DNA methylation patterns may result in phenotypic abnormalities (8). Emerging evidence have revealed that DNA methylation alterations at selected genomic loci may affect social cognition (9), learning and memory (10) and stress-related behaviors (11), and contribute to aberrant gene expression in a range of neurodevelopmental disorders, including autism, schizophrenia, depression and Alzheimer's disease (12–16). Although a multitude of epigenetic marks exist, DNA methylation is the most stable, a crucial factor in studying patterns of epigenetic modifications in human disease.
In recent years, many new approaches have been developed to study genome-wide DNA methylation patterns, providing substantial insight into the role of cytosine methylation in genome organization and function. Some approaches depend on the use of methylation-sensitive or -dependent restriction enzymes (17–21) where the level of DNA methylation is quantified by hybridization to high-density oligonucleotide arrays or sequencing via next-generation sequencing platforms (22). Other approaches capture methylated genomic DNA using immunoprecipitation via an antibody that recognizes 5-methylcytosine, followed by array hybridization or sequencing (23–26). Direct sequencing of bisulfite-treated DNA allows mapping of methylation of individual cytosine nucleotides in a genome-wide fashion (27). We have developed an enzymatic-based method, Methylation Mapping Analysis by Paired-end Sequencing (Methyl-MAPS) (22) to characterize DNA methylation profiles of primary cells from human and mouse post-mortem brain tissues. The Methyl-MAPs method uses a battery of methylation-sensitive and -dependent endonucleases to delineate the methylation status of >80% of CpG sites genome-wide in an unbiased fashion. Fractionation by the methylation-dependent McrBC endonuclease generates the unmethylated compartment (22,28). The methylated compartment is generated by digestion with a panel of all known methylation-sensitive tetranucleotide restriction enzymes termed RE (HpaII, HhaI, AciI, BstUI and HpyCH4V). Genomic DNA from human and mouse is fractionated into methylated and unmethylated compartments, and paired-end libraries are constructed, sequenced and mapped onto the respective human and mouse genomes. To analyze the Methyl-MAPS data, we have developed a data analysis pipeline referred to as Methyl-Analyzer (29) to generate methylation profiles.
The focus of our research is to investigate the role of DNA methylation in central nervous system function, and to identify DNA methylation alterations associated with neurodevelopmental disorders, as depression and schizophrenia. A genome-wide DNA methylation database focusing on brain development and function is an invaluable resource to the community of researchers within the areas of neuroscience, neurobiology, psychiatry and neuro-epigenetics. In this effort, we used the Methyl-MAPS method accompanied by the Methyl-Analyzer pipeline to profile the brain methylome of 29 human samples. Additionally, for comparative epigenetic studies, we profiled the mouse forebrain methylome. The methylation data in its entirety are presented in a novel methylation database referred to as MethylomeDB.
Although a handful of DNA methylation databases exist in the public domain (30–32), they either contain limited methylation data or differ in biological scope. Among these methylation databases, NGSmethDB (30) collects public genome-wide methylation data generated by next-generation sequencing approaches from various species and tissue types. Our database, however, has a focused biological target that aims to characterize the neuroepigenetic landscape of both normal and abnormal human brain. Our study design makes it feasible to identify potential DNA methylation signatures that may be associated with neuropsychiatric disorders, specifically depression and schizophrenia, using rare and well-characterized postmortem human brain specimens, with majority of cases having complete toxicological and psychological autopsy data. In addition to our internally generated data, Methylome DB will include published DNA methylation data from external sources, representing a comprehensive resource for the mammalian brain methylome.
DATA SOURCES
Presently, MethylomeDB provides internal genome-wide DNA methylation profiles of post-mortem brain tissues across both human and mouse species and external age-related DNA methylation profiles. For internal data, a total of 29 human brain specimens are represented from three distinct cortical regions, namely, dorsolateral prefrontal cortex (dlPFC), ventral prefrontal cortex (vPFC) and auditory cortex (AC) (Table 1 and Supplementary Table S1). These regions were selected because they have been implicated in the neuropathology of depression and schizophrenia. Within each human cortical region, both disease and non-psychiatric control samples have been profiled (matching subjects by age and sex in each group). The forebrain region was profiled in a 6-month-old mouse (129S6/SvEv inbred strain). Besides the internal human and mouse methylomes, we included age-related DNA methylation profiles from a study conducted by Hernandez et al. (33), which investigates DNA methylation changes across 90 postmortem brain samples spanning 16–102 years in age. This large number of human samples cover four brain regions: frontal cortex, temporal cortex, pons and cerebellum. These methylation profiles were generated by Infinium HumanMethylation27 Beadchip (Illumina) which covers 27 278 CpG sites in the human genome.
Table 1.
Samples used in MethylomeDB
| Tissue | Category | Sample no. | Age | PMI (h) |
|---|---|---|---|---|
| Brain dlPFC | Control | 4 | 47 ± 8 | 9.5 ± 6.8 |
| Brain dlPFC | Schizophrenia | 5 | 41 ± 15 | 10.4 ± 4.8 |
| Brain vPFC | Control | 6 | 47 ± 6 | 7.8 ± 3.8 |
| Brain vPFC | Depression | 6 | 41 ± 8 | 9.5 ± 3.3 |
| Brain AC | Control | 4 | 47 ± 8 | 7.0 ± 2.8 |
| Brain AC | Schizophrenia | 4 | 39 ± 17 | 9.0 ± 4.2 |
The DNA methylation data in MethylomeDB are represented at single-CpG resolution. We created two MySQL databases to store the human and mouse methylomes, where each table includes data on chromosomal mapping, 5mC chromosomal position, methylation probability and sequence read coverage. The data analysis pipeline, Methyl-Analyzer (29) estimates methylation probabilities based on methylated (RE) and unmethylated (McrBC) digested fragments. The methylation probability provides CpG methylation estimates, ranging from 0 to 1 corresponding to unmethylated to methylated states. The samples in MethylomeDB have methylation profiles with >80% CpG genomic coverage (Supplementary Table S1).
In addition to the human and mouse methylation profile databases, we created annotation databases. We compiled methylation-related CpG annotations characterizing CpG sites with respect to the RE or McrBC enzyme recognition sites, and from public annotations we overlay associated genomic features (e.g. promoter, exon and intron features). Also, we incorporated gene, regulation, variation and conservation annotation tables from UCSC Bioinformatics Genome Browser.
WEB INTERFACE
We have organized the web interface of MethylomeDB into three major functional features that include browse, search and download. The first two functions utilize the basic interface of the UCSC Genome Browser mirror site (with the permission of UCSC Genome Bioinformatics group), which we denote as the MethylomeDB Browser. Methylation data can be viewed in various formats, including (i) microarray (see representative example in Figure 1), (ii) wiggle (Figure 2), (iii) raw reads with methylated/unmethylated fragments produced by the RE and McrBC enzymatic digestions and (iv) read count referring to sequence read coverage of CpGs in wiggle format. Typically, the CpG methylation levels represented in microarray and wiggle formats (or tracks) will likely be the most frequently used tracks by the user community. The wiggle format is quantitative, featuring numerical values, whereas the microarray format is compact, allowing for visualization of DNA methylation data from multiple samples at a glance. The remaining two tracks provide additional technical data specific to the Methyl-MAPS method. The fragment track represents methylated (or unmethylated) sequence fragments that are products of enzymatic digestions. These raw read fragments are used in estimating CpG methylation probabilities. The coverage track represents the combined number of methylated and unmethylated sequence coverage at single CpG resolution. The raw read and CpG coverage tracks together can be used to gain in-depth sequencing information for each sample. These data would also be of utility to evaluate the relative accuracy or confidence associated with the estimated CpG methylation probabilities. Our analyses of biological replicates as well as validation experiments using an independent experimental method for methylation mapping show that sequence coverage of 8× or greater provide robust methylation estimates. Lastly, the download function can be accessed by the user in two ways. The Methylome Browser offers the Table feature for easy download. Users can download whole genome or methylation data by position for selected samples. Advanced users may want to download raw data for more sophisticated bioinformatics analyses. The ‘Download’ page provides links to methylation data for all 30 samples in the current build of the database. These are text files with information on CpG coordinates, chromosome, methylation probabilities and coverage.
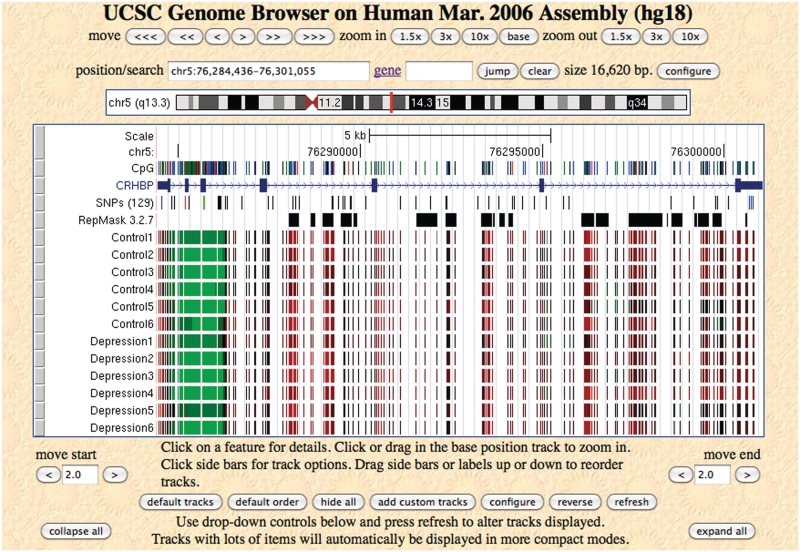
Figure 1.
Methylation profiles in microarray format of 12 brain vPFC samples for gene CRHBP. Methylation probability (0–1) is encoded from green (unmethylated) to black (intermediate methylation) to red (methylated) in continuous color scheme.
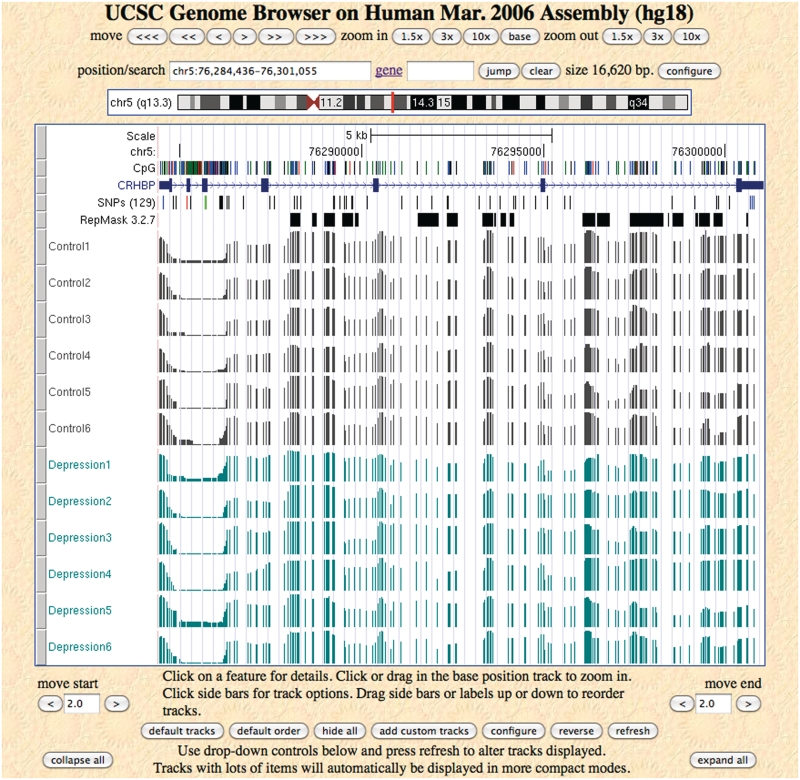
Figure 2.
Methylation profiles in wiggle format of 12 brain vPFC samples for gene CRHBP. The bars for wiggle format range from 0 to 1 representing methylation probability from 0 to 1.
FUTURE WORK
The current version of MethylomeDB is the first release of our database. Although it contains a wealth of brain-specific DNA methylation profiles in both the human and mouse species, the available features and functionality are still limited. However, given the importance of the data as a resource to the scientific community, we have made the MethylomeDB available while we continue to extend the database with new functionality.
We plan to develop more advanced search functions and data analysis tools for the web interface. A unique feature of the Methyl-MAPS method is that it allows for interrogation of DNA methylation patterns genome-wide including repeat sequences that occupy the majority of the human genome. We are able to use the CpG annotations we have compiled to link our DNA methylation data with genomic features, namely, promoter, exon, intron, intergenic and repeat sequences. One useful feature would be to search or browse the methylation distribution of user defined genomic feature(s) for any number of samples. Summary figures representing average methylation by feature and brain region and disease state would be highly appealing for users interested in, for example, the methylation state of a specific gene promoter in different cortical regions or across normal and disease samples. Furthermore, we will create a fast track to search a specific genomic region or a gene. The current version of MethylomeDB Browser nicely displays methylation profiles of a position query. The fast track, however, will show average methylation across multiple samples, and will provide downloadable data files.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
National Institutes of Health (NIH) (HG002915 to F.G.H.) and National Institutes of Mental Health (NIMH) (MH074118 to F.G.H.). Funding for open access charge: NIH (HG002915).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Jay Gingrich for making the mouse methylome data available for inclusion in MethylomeDB and Dr Shruti Rastogi for compiling age-related DNA methylation data.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 4.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 5.Bestor TH. Cytosine methylation mediates sexual conflict. Trends Genet. 2003;19:185–190. doi: 10.1016/S0168-9525(03)00049-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 7.Montagna M, Santacatterina M, Torri A, Menin C, Zullato D, Chieco-Bianchi L, D'Andrea E. Identification of a 3 kb Alu-mediated BRCA1 gene rearrangement in two breast/ovarian cancer families. Oncogene. 1999;18:4160–4165. doi: 10.1038/sj.onc.1202754. [DOI] [PubMed] [Google Scholar]
- 8.Bath KG, Lee FS. Variant BDNF (Val66Met) impact on brain structure and function. Cogn. Affect. Behav. Neurosci. 2006;6:79–85. doi: 10.3758/cabn.6.1.79. [DOI] [PubMed] [Google Scholar]
- 9.Isles AR, Davies W, Wilkinson LS. Genomic imprinting and the social brain. Philos. Trans. R. Soc. Lond., B Biol. Sci. 2006;361:2229–2237. doi: 10.1098/rstb.2006.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 12.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 13.Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc. Natl Acad. Sci. USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–e11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polesskaya OO, Aston C, Sokolov BP. Allele C-specific methylation of the 5-HT2A receptor gene: evidence for correlation with its expression and expression of DNA methylase DNMT1. J. Neurosci. Res. 2006;83:362–373. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- 16.Scarpa S, Fuso A, D'Anselmi F, Cavallaro RA. Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS Lett. 2003;541:145–148. doi: 10.1016/s0014-5793(03)00277-1. [DOI] [PubMed] [Google Scholar]
- 17.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 18.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 21.Yuan E, Haghighi F, White S, Costa R, McMinn J, Chun K, Minden M, Tycko B. A single nucleotide polymorphism chip-based method for combined genetic and epigenetic profiling: validation in decitabine therapy and tumor/normal comparisons. Cancer Res. 2006;66:3443–3451. doi: 10.1158/0008-5472.CAN-05-3739. [DOI] [PubMed] [Google Scholar]
- 22.Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 25.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 26.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 27.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chanrion B, Xin Y, Haghighi F. Enzymatic approaches for genome DNA methylation profiling. In: Craig JM, Wong NC, editors. Epigenetics: A Reference Manual. Norfolk, UK: Caister Academic Press; 2011. [Google Scholar]
- 29.Xin Y, Ge Y, Haghighi FG. Methyl-Analyzer–whole genome DNA methylation profiling. Bioinformatics. 2011;27:2296–2297. doi: 10.1093/bioinformatics/btr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackenberg M, Barturen G, Oliver JL. NGSmethDB: a database for next-generation sequencing single-cytosine-resolution DNA methylation data. Nucleic Acids Res. 2011;39:D75–D79. doi: 10.1093/nar/gkq942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, Chang S, Zhang J, Zhao Q, Xiang H, Kusonmano K, Yang L, Sun ZS, Yang H, Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ongenaert M, Van Neste L, De Meyer T, Menschaert G, Bekaert S, Van Criekinge W. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]